Abstract
During phosphate starvation, Snf1-related kinase 1 (SnRK1) activity significantly decreases compared with plants growing under normal nutritional conditions. An analysis of the expression of the genes encoding for the catalytic subunits of SnRK1 showed that these subunits were not affected by phosphate starvation. Transgenic Arabidopsis (Arabidopsis thaliana) plants overexpressing the AKIN10 and AKIN11 catalytic subunits fused with green fluorescent protein (GFP) were produced, and their localizations were mainly chloroplastic with low but detectable signals in the cytoplasm. These data were corroborated with an immunocytochemistry analysis using leaf and root sections with an anti-AKIN10/AKIN11 antibody. The SnRK1 activity in transgenic plants overexpressing AKIN11-GFP was reduced by 35% to 40% in phosphate starvation, in contrast with the results observed in plants overexpressing AKIN10-GFP, which increased the activity by 100%. No differences in activity were observed in plants growing in phosphate-sufficient conditions. Biochemical analysis of the proteins indicated that AKIN11 is specifically degraded under these limited conditions and that the increase in AKIN10-GFP activity was not due to the phosphorylation of threonine-175. These results are consistent with an important role of AKIN10 in signaling during phosphate starvation. Moreover, akin10 mutant plants were deficient in starch mobilization at night during inorganic phosphate starvation, and under this condition several genes were up-regulated and down-regulated, indicating their important roles in the control of general transcription. This finding reveals novel roles for the different catalytic subunits during phosphate starvation.
Phosphorus (P) is an essential element required for plant growth and development (Bieliski, 1973). The P concentration in soil solutions is high but the assimilable form, inorganic phosphate (Pi), is always in a limiting concentration ranging from 1 to 10 μm (Poirier and Bucher, 2002); thus, plants are frequently growing in Pi-limited conditions. Plants have evolved two broad strategies to cope with limiting Pi: (1) responses designed to increase Pi availability or acquisition, and (2) metabolic adaptations to decrease Pi requirements (Poirier and Bucher, 2002). Processes that lead to enhanced uptake include increased production and secretion of phosphatases, exudation of organic acids, and greater root growth along with modified root architecture (Bates and Lynch, 1996; Ma et al., 2001; López-Bucio et al., 2002). There are also processes that conserve the use of Pi, such as the remobilization of internal Pi, modifications in carbon metabolism that bypass Pi-requiring steps, and alternative respiratory pathways (Plaxton, 2004). During Pi-limited conditions, a reduction in cytoplasmic Pi, ATP, and ADP directly affects glycolysis, since they are used as cosubstrates of enzymes, such as ATP-dependent phosphofructokinase, NAD-dependent glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and pyruvate kinase. However, despite low concentrations of cytosolic Pi and nucleoside phosphates, pyrophosphate (PPi) concentrations remain high and PPi can serve as an energy donor under certain conditions (Duff et al., 1989; Theodorou and Plaxton, 1993). In this situation, in order to continue with the production of carbon skeletons to conserve the carbon flux, several PPi-dependent enzymes are activated, such as UDP-Glc phosphorylase and PPi-dependent phosphofructokinase (Duff et al., 1989; Ciereszko et al., 2001). Another alternative glycolytic pathway known in plants is catalyzed by the action of a nonphosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, which is induced during Pi starvation and bypasses Pi-dependent NAD-dependent glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase (Duff et al., 1989; Theodorou and Plaxton, 1993; Plaxton and Carswell, 1999).
Signal transduction mechanisms that produce adaptive changes during Pi deficiency are not clearly known. Modifications in gene expression in response to Pi deficiency can be grouped into the early genes that respond rapidly and often nonspecifically to Pi deficiency and the late genes that alter the morphology, physiology, or metabolism of plants upon prolonged stress. These late genes generally increase the acquisition of Pi or promote the efficient use of Pi within the plant (Vance et al., 2003). In bean (Phaseolus vulgaris) plants, we identified early and late genes that encode proteins that might be involved in signal transduction (Camacho et al., 2008; P. Coello, A. Ramírez, and E. Martínez-Barajas, unpublished data). One of the late genes has high homology with the α-subunit of the AMP-dependent protein kinase (AMPK) complex in animal cells. The protein kinases SNF1/AMPK/Snf1-related kinase 1 (SnRK1) are a conserved family of kinases that play an important role in energy balance control, as demonstrated by the mammalian complex AMPK (Hardie and Sakamoto, 2006). In yeast, Snf1 (for Suc nonfermenting 1) is known to be a key player in the diauxic shift from fermentative to oxidative metabolism in response to Glc deprivation (Rolland et al., 2006). In plants, SnRK1 seems to have important roles in controlling metabolic homeostasis and stress signaling (Baena-González et al., 2007; Baena-González and Sheen, 2008) and during plant development (Zhang et al., 2001; Lu et al., 2007). The SnRK1 kinases are heterotrimeric complexes formed by an α catalytic subunit and β and γ regulatory subunits. In Arabidopsis (Arabidopsis thaliana), each subunit is encoded by multiple genes, giving rise to a large variety of possible heterotrimeric combinations (Polge and Thomas, 2006). To explore the possibility that SnRK1 complexes are involved in Pi-starvation responses, we characterized the expression and the activity of two of the catalytic subunits (AKIN10 and AKIN11) in Arabidopsis plants growing in Pi-sufficient and Pi-starvation conditions. Our results indicate that both catalytic subunits are mainly located in the chloroplast. They are differentially regulated during Pi starvation. Whereas complexes formed with the AKIN10 catalytic subunit increase in activity, those formed with AKIN11 are specifically degraded. To explore the role of AKIN10 during Pi starvation, we isolated a mutant with a T-DNA insertion in akin10. No differences were observed between wild-type and mutant plants during their development and growth. However, at the biochemical level, starch metabolism, especially starch degradation, was impaired in the mutant during Pi starvation. The transcriptional activation of genes coding for proteins directly involved in carbon metabolism and chloroplast biogenesis was up-regulated in the akin10 plants, and genes involved in light signaling, protein degradation, and GTP signaling were down-regulated. All of these results suggest that the AKIN10 catalytic subunit of the SnRK1 complex might be involved in the regulation of some of the metabolic changes that are observed during Pi starvation.
RESULTS
Expression and Activity of SnRK1 during Phosphate Starvation
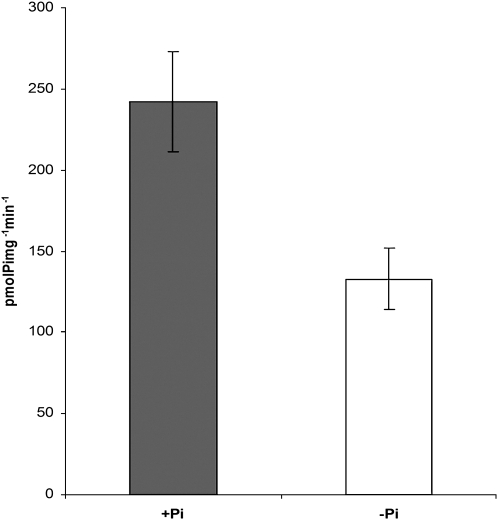
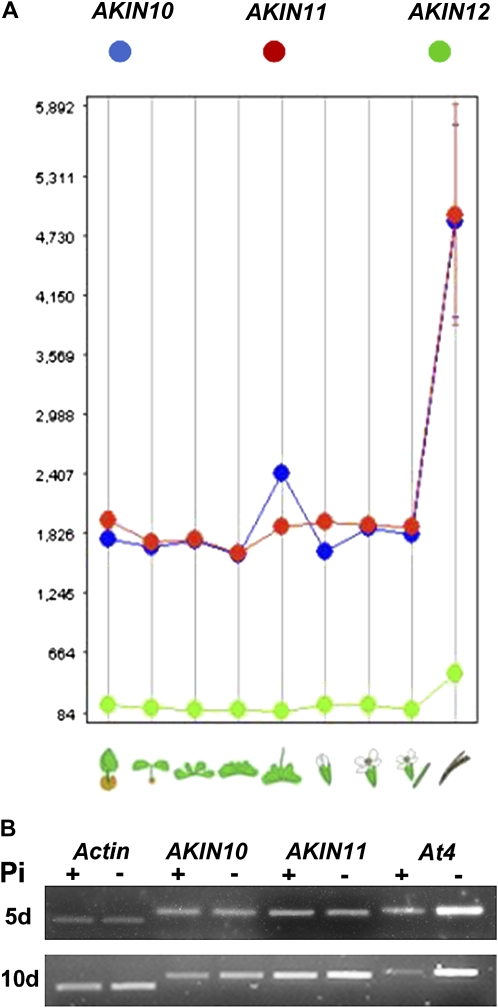
It has been very well documented that kinases of the AMPK/SnRK1/SNF1 family are activated in low-energy conditions and when Glc is absent from the medium (Baena-González and Sheen, 2008). In plants, Pi starvation is a condition that produces an important reduction in the amount of ATP. To get information about the regulation of SnRK1 during Pi deficiency, its kinase activity was measured by the phosphorylation of the SAMS peptide. The SnRK1 activity was lower in leaf extracts from Pi-starved plants (Fig. 1). To evaluate if the difference in activity was due to differential expression of the catalytic subunits, we searched the Genevestigator database and found that only two of the three genes that encode the catalytic subunits (AKIN10 and AKIN11) were expressed in rosette leaves. No changes in gene expression were found in plants grown under complete nutritional conditions (Fig. 2A). In addition, after 5 and 10 d of Pi starvation, semiquantitative reverse transcription (RT)-PCR indicated that no visible changes occurred in the amount of the AKIN10 and AKIN11 transcripts. Amplification of actin was used as an internal control to indicate equal loading. At4 is a Pi starvation-inducible gene and was used as an indicator of the Pi status (Burleigh and Harrison, 1999). In contrast with AKIN10 and AKIN11, At4 induction was higher in Pi-starved plants and was observed after 5 d of treatment (Fig. 2B).
Figure 1.
SnRK1 activity in plants grown for 10 d in Pi-sufficient (+Pi) and Pi-starvation (−Pi) conditions. Activity was estimated using the SAMS peptide kinase assay. Bars represent means ± sd (n = 3).
Figure 2.
A, Average signal of gene expression for AKIN10, AKIN11, and AKIN12 derived from the microarray data available at the Genevestigator database (Zimmermann et al., 2005) and arranged according to tissue. B, Effect of Pi starvation on the accumulation of AKIN10 and AKIN11 transcripts. RT-PCR of total RNA from rosette leaves from plants grown for 5 and 10 d under Pi-sufficient (+) and Pi-starvation (−) conditions. At4 is a Pi-starvation control gene, and actin was used as an internal control. [See online article for color version of this figure.]
Phosphate Starvation Induces Differential Changes in AKIN10 and AKIN11 Catalytic Subunits
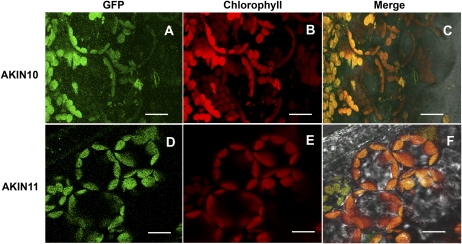
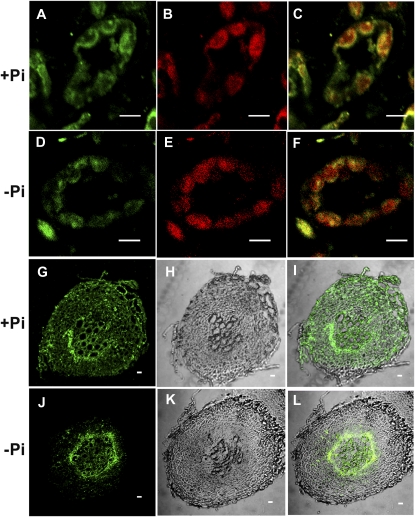
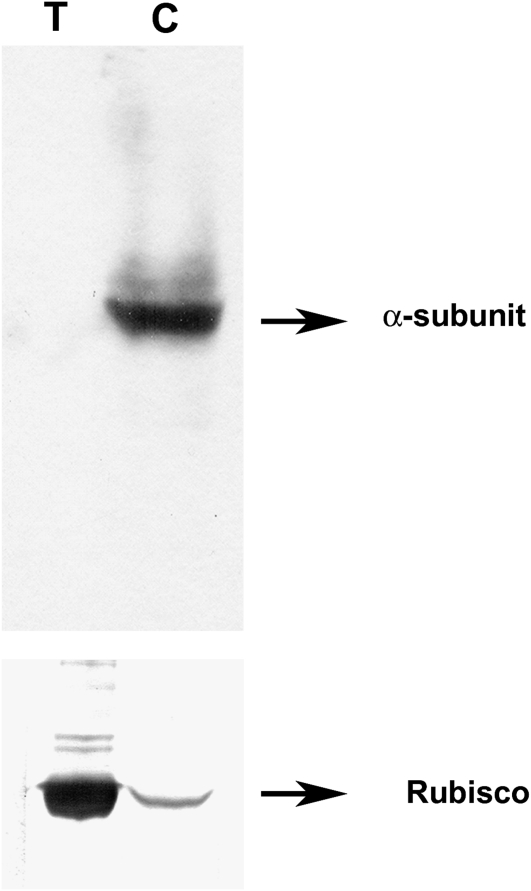
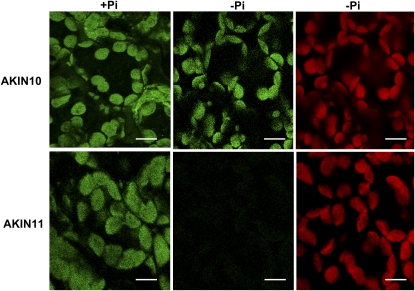
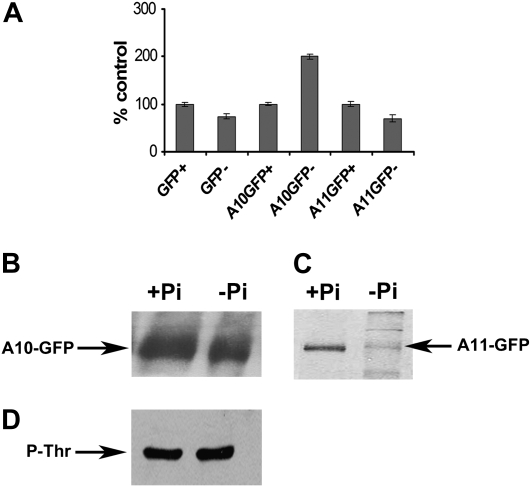
Having established that no changes in gene expression were observed in the catalytic subunits during Pi starvation to explain the reduction in activity, we analyzed the protein levels. Transgenic Arabidopsis plants carrying AKIN10-GFP and AKIN11-GFP fusion proteins were evaluated. Localization of GFP fusion proteins by confocal microscopy showed that both catalytic subunits colocalized with the chlorophyll, suggesting a chloroplast localization (Fig. 3, A and D). In the case of AKIN10, a low but detectable signal was also observed in the cytoplasm (Fig. 3C). To confirm the localization of the catalytic subunits, we raised polyclonal antibodies using a polypeptide that recognized AKIN10 and AKIN11 to detect the subunits in root and leaf sections. Immunolocalization analysis indicated that the catalytic subunits are present in both organs (Fig. 4, A and G). In leaf cells, the catalytic subunits are localized in the chloroplast but the signal was also detected in the cytoplasm (Fig. 4, A–C). In roots, the catalytic subunits were present in all cells, from the epidermis to the vascular tissue (Fig. 4, G–I). To probe the chloroplast localization of the catalytic subunits at the biochemical level, proteins obtained from purified chloroplasts were separated with SDS-PAGE and transferred to a nitrocellulose membrane. Western-blot analysis identified a polypeptide band around 58 kD, which is the expected molecular mass for AKIN11 and AKIN10. The same blot was probed with Rubisco antibodies (Fig. 5). Under Pi starvation conditions, the detection of GFP fusion proteins showed no important changes in AKIN10. Interestingly, the fluorescence corresponding to AKIN11-GFP almost completely disappeared (Fig. 6). This reduction in the fluorescence corresponding to one of the catalytic subunits was also observed in the leaf and root sections of Pi-starved plants, visualized using the anti-α-antibody (Fig. 4, D and J). In leaf cells, the amount of the catalytic subunits decreased notably, since the chlorophyll signal was more intense than the Alexa 568 signal (Fig. 4, D–F). In roots, the catalytic subunits were confined to the surrounding vascular tissue and the signal in the cortex and epidermis disappeared (Fig. 4, J–L). In addition, the activity of SnRK1 in transgenic plants overexpressing AKIN10-GFP growing in Pi-starvation conditions was higher than in plants growing in Pi-sufficient conditions, whereas in AKIN11-GFP plants, Pi starvation reduced SnRK1 activity (Fig. 7A). The determination of AKIN10-GFP and AKIN11-GFP levels in transgenic plants using anti-GFP antibodies showed no changes in AKIN10 and a significant reduction in AKIN11 during Pi starvation (Fig. 7, B and C). These results strongly suggest a differential regulation of both catalytic subunits under this nutritional condition. Since no changes in protein levels for AKIN10 were found, the increase in its activity might be explained by a posttranslational modification. Activation by phosphorylation of the Thr-175 present at the T-loop was evaluated using an antibody that recognized the mammalian phosphorylated Thr-172. The results showed that plants growing in Pi starvation had the same level of phosphorylated AKIN10 than plants growing in Pi-sufficient conditions, indicating that additional mechanisms are activating the AKIN10 complexes (Fig. 7B).
Figure 3.
Confocal fluorescence images of AKIN10 and AKIN11 fusion proteins showing localization at the chloroplast. The GFP signal and the chlorophyll autofluorescence are indicated in green and red, respectively. The merged images (Merge) of chlorophyll autofluorescence, GFP, and light field are shown. Bars = 10 μm. [See online article for color version of this figure.]
Figure 4.
Immunolocalization of the catalytic subunits. Leaf and root sections of plants grown with or without Pi were immunolocalized with anti-α-antibodies. A to C, Leaf sections of plants grown under +Pi conditions (60×). D to F, Leaf sections of plants grown under −Pi conditions (60×). G to I, Roots grown under +Pi conditions (40×). J to L, Roots grown under −Pi conditions (40×). A, D, G, and J show goat anti-rabbit Alexa 568 fluorochrome staining (green signal). B and E show chlorophyll (red signal). H and K show bright-field images of the transverse sections of roots. C and F are overlaid images of Alexa 568 fluorochrome and chlorophyll for colocalization. I and L are overlaid images of Alexa 568 and bright field. Bars = 10 μm. [See online article for color version of this figure.]
Figure 5.
Western-blot detection of the α-catalytic subunit. A, Total protein extracts (T) and chloroplast-extracted proteins (C) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunodetection of AKIN10 and AKIN11 was performed using the enhanced chemiluminescence kit. B, The membrane in A was reblotted using antibodies against Rubisco.
Figure 6.
Confocal fluorescence images of AKIN10-GFP and AKIN11-GFP. Plants were grown in Pi-sufficient (+Pi) or Pi-starved (−Pi) conditions. GFP signal is indicated in green. Chlorophyll autofluorescence is indicated in red. Bars = 10 μm. [See online article for color version of this figure.]
Figure 7.
Characterization of transgenic Arabidopsis plants overexpressing AKIN10-GFP and AKIN11-GFP fusion proteins. A, SnRK1 activity of transgenic plants measured by the phosphorylation of the SAMS peptide. Plants expressing GFP were used as controls. Bars represent means ± sd (n = 3). B, Protein extracts from plants overexpressing AKIN10-GFP were separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunodetection was carried out using anti-GFP antibodies. C, Protein extracts from plants overexpressing AKIN11-GFP were separated and immunodetected as described above. D, Protein extracts from transgenic plants overexpressing AKIN10-GFP were separated and immunodetected using a specific P-Thr antibody.
Silencing of akin10 Affects Starch Metabolism during Pi Starvation
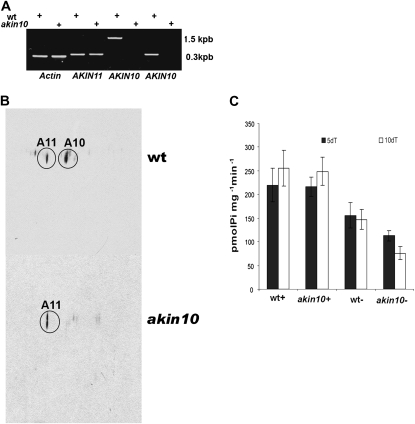
The results presented so far suggest that SnRK1 complexes are formed with the AKIN10 catalytic subunit under Pi starvation. To better understand the role of AKIN10 during Pi starvation, a T-DNA insertion line in akin10 was identified and characterized. In addition, RT-PCR, using gene-specific primers, indicated that no AKIN10 expression was found in the mutant. actin and AKIN11 were expressed in both the wild-type and mutant lines (Fig. 8A). When the presence of AKIN10 and AKIN11 was evaluated by western blot using two-dimensional SDS-PAGE, AKIN10 (pI = 8.36) was observed only in the wild type, whereas AKIN11 (pI = 7.4) was identified in both wild-type and mutant plants (Fig. 8B). No differences in SnRK1 activity were observed in the wild-type and mutant plants growing in Pi-sufficient conditions. However, SnRK1 activity decreased more in the mutant than in the wild-type plants when they were grown in Pi-starvation conditions (Fig. 8C).
Figure 8.
Characterization of akin10 mutants. A, RT-PCR of wild-type (wt) and akin10 plants detecting AKIN10 and AKIN11 transcripts. actin was used as a control. B, Protein extracts obtained from wild-type and mutant plants were separated by two-dimensional SDS-PAGE. Proteins were transferred to nitrocellulose membranes, and the catalytic subunits were identified using the anti-α-antibody. C, SnRK1 activity was measured in protein extracts obtained from wild-type and mutant plants grown for 5 and 10 d in Pi starvation. Bars represent means ± sd (n = 3).
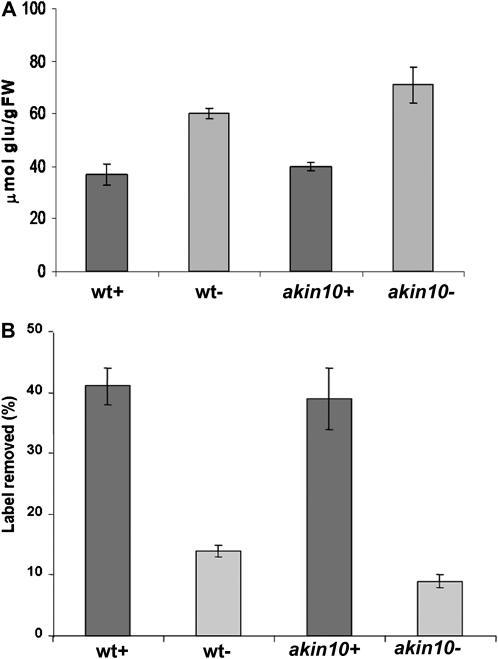
In plants, SnRK1 is directly involved in the regulation of carbohydrate metabolism (Halford et al., 2003; Polge and Thomas, 2006). Antisense expression of SnRK1 in barley (Hordeum vulgare) produces pollen with no starch and arrests at the binucleate state of development (Zhang et al., 2001). In Arabidopsis, starch degradation was impaired in a transient akin10, akin11 double mutant (Baena-González et al., 2007). During Pi starvation, starch accumulation in leaves is one of the most consistently observed biochemical responses (Ciereszko and Barbachowska, 2000; Bernal et al., 2005). To determine if AKIN10 has a regulatory role in starch metabolism, the amount of starch was measured in wild-type and mutant plants. There was no difference in the starch levels between plants growing in Pi-sufficient conditions. However, starch accumulation was higher in the mutant than in the wild-type plants during phosphate starvation (Fig. 9A). To determine if starch mobilization was affected, plants were incubated with 14CO2 and the distribution of the label was analyzed. The differences between the labels associated with the starch granules at the end of the day and at the end of the night are indications of starch mobilization. The results showed that less mobilization occurred in akin10 during Pi starvation than in the wild type (Fig. 9B).
Figure 9.
Starch metabolism in wild-type and akin10 plants. A, Starch accumulation in wild-type (wt) and akin10 plants grown in Pi-sufficient and Pi-starvation conditions. FW, Fresh weight. B, Starch mobilization measured as the difference in the 14C label associated with the starch granules at the end of the day and at the end of the night. Bars represent means ± sd (n = 5).
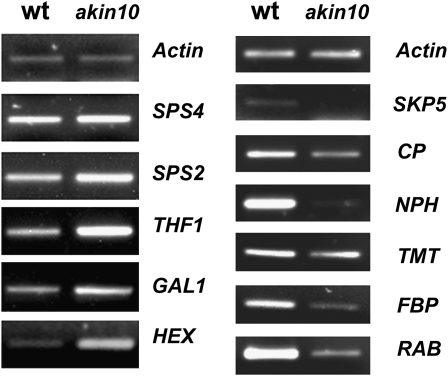
It has been demonstrated that SnRK1 also regulates gene expression (Baena-González et al., 2007). To determine if akin10 mutants also had a transcriptional effect during Pi starvation, we did a microarray analysis to identify genes that could be regulated by SnRK1. We evaluated the transcript accumulation of some genes that had changed levels of expression. The results showed that some of these genes, which are directly involved in carbohydrate metabolism, increased transcript levels in mutant plants grown under Pi starvation. In contrast, other genes related to general stress were down-regulated (Fig. 10).
Figure 10.
AKIN10 modifies the transcription of different genes. RT-PCR of total RNA from rosette leaves from wild-type (wt) and akin10 plants growing in Pi starvation. Genes involved in carbon metabolism, such as SUCROSE PHOSPHATE SYNTHASE4 (SPS4), SUCROSE PHOSPHATE SYNTHASE2 (SPS2), GALACTOKINASE1 (GAL1), and HEXOKINASE (HEX); genes involved in Glc sensing, such as THYLAKOID FORMATION1 (THF1); and genes involved in general stress signaling, such as SKP1-INTERACTING PARTNER5 (SKP5), CYSTEINE PROTEINASE (CP), PHOTOTROPIC RESPONSIVE (NPH), HEXOSE TRANSPORTER2 (TMT2), F-BOX PROTEIN (FBP), and RAB-RELATED GTP-BINDING PROTEIN (RAB) were evaluated using specific primers. actin was used as an internal control. The accession number of each gene is indicated in “Materials and Methods.”
DISCUSSION
We have identified genes that are up-regulated in Pi-starvation conditions, and some of them encode proteins that are involved in signal transduction (Camacho et al., 2008; Martinez et al., unpublished data). One of these proteins has high homology with the catalytic subunit of the AMPK from mammalian cells and with the catalytic subunit of the Snf1 from yeast. In plants, these kinases have been classified as SnRK1. AMPK has been directly implicated in the regulation of the cellular energetic balance, acting as a sensor (Hardie, 2007). Therefore, plant SnRK1 might be involved in signaling changes during Pi starvation.
SnRK1 Activity during Pi Starvation
By analyzing the Genevestigator database, we observed that the expression of AKIN12 was very low during Arabidopsis development. Therefore, we focused our study on AKIN10 and AKIN11. The activity of SnRK1 decreases during Pi starvation, and this difference is not due to changes in the expression of the catalytic subunits during normal or Pi-starvation conditions. Overexpressing both catalytic subunits as GFP fusion proteins indicated that during Pi starvation, AKIN11-GFP complexes decreased in activity whereas AKIN10-GFP complexes increased. In the AKIN11 transgenic plants, this catalytic subunit is degraded during Pi starvation, and its degradation produces a reduction in activity. Interestingly, in yeast, the catalytic subunit of Snf1 interacts with the Srb10/Srb11 protein kinase. After phosphorylation of the C-terminal region of RNA polymerase II, its activity is inhibited and Srb11 is degraded in a proteasome-dependent manner. Moreover, Snf1 has been directly implicated in its degradation (Cooper et al., 1999). In the same way, Arabidopsis AKIN10 and AKIN11 interact with the Skp1/ASK1 subunit of an E3 ubiquitin ligase and with the α4/PAD1 subunit of the 20S proteasome (Farrás et al., 2001). It has been proposed that SnRK1 is part of an SCF complex that phosphorylates its substrates. Recently, it was demonstrated that AKIN10 interacts with PRL1, which is a receptor substrate of the complex DDB1-CUL4 E3 ligase, leading to its degradation (Lee et al., 2008). We are currently investigating if AKIN11 is similarly degraded and if Pi starvation is the only signal that triggers its destruction. Furthermore, we evaluated if the activation of AKIN10 was due to the specific phosphorylation of the Thr-175, which is equivalent to the mammalian Thr-172 that is phosphorylated at the T-loop during the activation of the complex (Sugden et al., 1999). In AKIN10, the increase in activity could not be explained by the phosphorylation of the Thr-175 present at the T-loop since it is phosphorylated all the time, indicating that Pi starvation is not a stimulus that activates the complex. It is possible that AKIN10 is associated with other regulatory subunits during Pi starvation and that those complexes have differences in activity. In mammalian cells, γ1, associated either with α1 or α2, forms complexes with higher activity than with the other two γ-subunits (Cheung et al., 2000), indicating that the subunit composition regulates their activity. Altogether, these results suggest that Arabidopsis plants growing in Pi-sufficient conditions mainly have complexes formed by the AKIN11 catalytic subunit. When plants are transferred to Pi starvation, AKIN11 is degraded, with a concomitant reduction in activity. On the other hand, there are few complexes formed by AKIN10, and their activation during Pi starvation is not enough to produce an increase in the total activity of SnRK1. However, when AKIN11 is degraded, the complexes formed by AKIN10 might be important in regulating some metabolic changes.
Localization of AKIN10 and AKIN11 Is Mainly Chloroplastic
The subcellular localization of AKIN10 and AKIN11 indicated that both enzymes are chloroplastic and cytoplasmic. Cytoplasmic localization was assumed, since SnRK1 has been directly involved in the regulation and inactivation of enzymes such as Suc phosphate synthase, nitrate reductase, and HMG-CoA reductase (Hey et al., 2005; Polge and Thomas, 2006; Polge et al., 2008). However, the chloroplastic localization was unexpected. An analysis of the N-terminal sequence in AKIN10 and AKIN11 using the Wolf PSort subcellular localization predictor (Horton et al., 2007) indicated cytoplasmic, nuclear, and chloroplastic localizations, although the last two had low scores. The diverse roles of the SNF1/AMPK/SnRK1 family of kinases raise the question of how such versatility is achieved. Genetic studies in yeast suggest that isoforms of the β-subunits are important for functional specificity. In fact, subcellular localization of Snf1 kinase is regulated by specific β-subunits (Vincent et al., 2001). In the case of Arabidopsis, the regulatory subunit βγ has a recognizable chloroplast transit peptide, so interaction with that subunit could direct the catalytic subunits to the chloroplast. On the other hand, the subcellular localization prediction for β1 and β2 regulatory subunits (Horton et al., 2007) indicated that they are mainly nuclear and nuclear and chloroplast, respectively. We can hypothesize that the interaction of the catalytic subunits with those regulatory subunits would localize the complex in different organelles. We are currently evaluating the subcellular localization of the catalytic subunits by making deletions of the N-terminal sequence of AKIN10/AKIN11-GFP fusion proteins and observing the final GFP localization. In addition, we are evaluating if a direct interaction with the βγ-subunit directs the complex to the chloroplast.
Although the chloroplastic localization has not been completely explained, starch metabolism, especially at night, is impaired in Arabidopsis plants with a transient reduction in AKIN10 and AKIN11 proteins (Baena-González et al., 2007), opening the possibility that SnRK1 directly or indirectly regulates enzymes that are present in the chloroplast. This is, to our knowledge, the first report indicating the chloroplastic localization of SnRK1.
AKIN10 Is Involved in Starch Degradation and in Gene Expression during Pi Starvation
In order to obtain more information about the role of AKIN10 during Pi starvation, we identified and characterized a mutant plant lacking this subunit. The evaluation of its role in starch metabolism indicated that mutant plants mobilized less starch at night than wild-type plants during Pi starvation. Starch accumulation during Pi deficiency has been directly associated with less degradation at night (Bernal et al., 2005). Enzymes involved in starch degradation are not fully understood, and the importance of phosphorylation in modulating their activities has not been investigated. This is in contrast with the modulation of starch synthesis, where isoforms of starch-branching enzyme are activated by phosphorylation in the chloroplast (Tetlow et al., 2004). Even more, the overexpression of a SnRK1 potato (Solanum tuberosum) catalytic subunit increased starch levels up to 30%, and there is no evidence that changes in the activation state of the ADP-Glc pyrophosphorylase exist in these transgenic plants, indicating that some other mechanism, such as direct phosphorylation of other enzymes, could be involved (McKibbin et al., 2006). However, starch degradation was reduced in a sex4 mutant, which has an impaired protein phosphatase. This phosphatase has a glycogen-binding domain, which in plants is recognized to bind starch. The model proposes that SEX4 interacts with starch in vivo and is directly necessary for its metabolism. Thus, SEX4 may directly regulate the activity of starch-degrading enzymes or may modulate the activity of a protein kinase, which in turn may change the activity of enzyme(s) involved in starch degradation (Niittylä et al., 2006). Additionally, SnRK1 forms complexes with β-subunits that contain a putative glycogen-binding domain (P. Coello, unpublished data), so we can hypothesize that SnRK1 may be bound directly to starch through the β-subunits and either be modulated directly by SEX4 or in an independent manner. We are currently investigating some of these possibilities.
Research on plant SnRK1 has revealed its additional role as a central regulator of the transcriptome under multiple types of stress signals (Baena-González et al., 2007; Baena-González and Sheen, 2008). Our results with the akin10 mutant showed that transcription was affected during Pi starvation. The induction of genes potentially involved in Suc synthesis (SPS4 and SPS2), sugar metabolism (GAL1 and HEX), and vesicle-mediated formation of thylakoid membranes (THF1) occurred under these conditions. In contrast, other genes involved in the monosaccharide transportation in tonoplast (TMT2), protein degradation (FBP and SKP), GTP signaling (RAB), and light signal transduction (NPH) were down-regulated. Some of these changes in gene expression might reflect multiple types of stress convergence produced by energy deficiency. This is supported by the fact that some of the Pi-starvation responses are mediated by sugar signaling (Liu et al., 2005; Karthikeyan et al., 2007). The involvement of SnRK1 in Pi starvation signal transduction is in accordance with the idea that different stresses are sensed as energy deficiencies, so future work is needed to delimit these convergent transcriptional stress responses in order to be able to manipulate crop responses to Pi limitation.
MATERIALS AND METHODS
Growth of Plants and Isolation of Mutants
Arabidopsis (Arabidopsis thaliana) plants were grown in a growth chamber at 21°C to 23°C under short-day conditions (8 h of light/16 h of dark) at a light intensity of 120 μmol photons m−2 s−1 (fluorescent bulbs). akin10 mutant was isolated from the SIGnAL T-DNA collection (line SALK_ 127939). Homozygous mutants were identified by PCR genotyping using the following gene-specific primers: LP (5′-ACCACACGTTGGAAACTTTTG-3′) and RP (5′-CAGATGGGTTCCTAACAGCAG-3′) in combination with the T-DNA-specific primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′). For the phosphate-starvation treatment, seedlings were transferred to Agrolite and watered every third day with a Hoagland II solution (Jones, 1982), either containing 500 μm ammonium phosphate (+Pi) as the only source of P or no phosphate (−Pi), depending on the experiment.
RNA Isolation and RT-PCR
Total RNA was isolated using the Trizol reagent according to the manufacturer's instructions (Invitrogen). cDNA was synthesized using ImProm-II Reverse Transcriptase according to the manufacturer's instructions (Promega). The gene-specific primer pairs (forward and reverse) for detecting the following genes were as follows: SPS4 (At4g10120), 5′-GCTAGCCTCAGGTTCAAGCT-3′ and 5′-TGTGGAGGCCACCCAGTAAG-3′; SPS2 (At5g11110), 5′-TTCCGCTGGGGTGGAGAGAG-3′ and 5′-AGAGCGACCTTAATGGCGTC-3′; HEX (At2g19860), 5′-TGGCGATATCGTCCCACCTA-3′ and 5′-TGAGAGTGAGAAGCAGCAAG-3′; GAL1 (At3g06580), 5′-CCATACACGGCTGAGGAGAT-3′ and 5′-AGAGCGACCTTAATGGCGTC-3′; THF1 (At2g20890), 5′-CAAGCGACCCATCCCGAGTA-3′ and 5′-TGCTTCTTCTTCTCCCTCTC-3′; TMT2 (At4g35300), 5′-TTGAGCAGGCGGGTGTCGGG-3′ and 5′-GCAGCACGGGGAGACTGTAA-3′; FBP (At5g10770), 5′-CAGGGATCTTCGACGGCTCT-3′ and 5′-CGTCCACACAAATTCAGGCG-3′; RAB (At3g09900), 5′-GATATGGGACACTGCTGGAC-3′ and 5′-TGATCCCTTGGGGCTCGGCT-3′; SKP (At3g54480), 5′-TTGCCTGGTCGGTGGAGGCG-3′ and 5′-CGAACACGCTGCAGAACCAA-3′; CP (At3g19400), 5′-AACGCTGGATGTGATGGAGG-3′ and 5′-GGGTAAGAAGGCATCATTGC-3′; and NPH (At1g03010), 5′-AGCTTGTAGCCACGCAGCTC-3′ and 5′-GCCTTGCCACTCAAGGAAGG-3′. As an internal control, the following forward and reverse primers of the ACTIN gene were designed: 5′-TGGGGCAGAAGGATGCGTATG-3′ and 5′-CAATACCAGTTGTACGGCCAC-3′. For a phosphate-starvation control, the At4 cDNA was amplified using the following primers: forward (5′-CCCCTAAAGAAACTGAAGCTCAAGAA-3′) and reverse (5′-GGAACATAAACATATAAGAGCTAT-3′). To analyze gene expression, equivalent amounts of total RNA were input in cDNA synthesis and the same amount of cDNA was included in PCR. The resultant PCR products were resolved on a 1% agarose gel for comparison.
Transgenic Plant Construction
The coding regions of AKIN10 (At3g01090) and AKIN11 (At3g29160) were amplified by PCR using the primers for AKIN10 (5′-CCCATGGATGGATCAGGCACA-3′ and 5′-GAGGACTCGGAGCTGAGCAAG-3′) and AKIN11 (5′-CACCATGGATCATTCATCAAAT-3′ and 5′-GATCACACGAAGCTCTGTAAG-3′). The fragment with the coding sequence was introduced into the pENTR/D-TOPO vector (Invitrogen) and then recombined with an N-terminal GFP fusion binary vector (pEarlyGate103) using recombinant Gateway technology (Earley et al., 2006). Arabidopsis transformation using the Agrobacterium tumefaciens strain GV3101 was performed by the floral dip method (Clough and Bent, 1998).
Kinase Activity Assay
Frozen leaves were homogenized in a chilled mortar with ice-cold extraction buffer containing 50 mm HEPES, pH 8.2, 50 mm NaF, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 50 mm NaCl, 10 mm β-glycerophosphate, 8% (v/v) glycerol, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, and 50 μm leupeptin. The homogenate was centrifuged at 20,000g for 10 min at 4°C. Proteins obtained in the crude extracts were precipitated by adding 11% (w/v) polyethylene glycol 10,000. The solution was gently stirred for 20 min and centrifuged at 20,000g for 30 min. The pellet was dissolved in 30 μL of extraction buffer, and the total protein was evaluated according to the Bradford method (Bradford, 1976). One or 2 μg of total protein was used in the assay. The SAMS peptide kinase activity was determined as described by Davies et al. (1989), but with some modifications. Peptide phosphorylation was assayed in a final volume of 25 μL containing kinase buffer (40 mm HEPES, pH 7.0, 50 mm sodium fluoride, 25 mm β-glycerophosphate, 50 mm NaCl, 8% [v/v] glycerol, 0.8 mm EDTA, 1 mm dithiothreitol, 5 mm MgCl2,1 mm EGTA, and 1 mm Na3VO4), 200 μm SAMS peptide (HMRSAMSGLHLVKRR), 200 μm AMP, and [γ-32P]ATP (200 μm, 5 μCi μL−1, 3,000 Ci mmol−1). After incubation for 10 min at 30°C, 15-μL aliquots were removed and spotted on 1-cm × 1-cm squares of phosphocellulose paper (P81; Whatman), which were dropped into 500 mL of 1% (v/v) H3PO4. The squares were washed three times for 20 min each in 1% H3PO4 and then once with acetone, air dried, and counted by immersion in 5 mL of a toluene-based scintillation fluid.
Chloroplast Extraction
Chloroplasts were isolated from Arabidopsis leaves and purified by centrifugation through Percoll (Pharmacia) gradients as described by Weigel and Glazebrook (2002).
Antibodies and Western Blots
A peptide corresponding to 10 conserved amino acids in the AKIN10 and AKIN11 proteins was synthesized by Sigma. Peptide antigens were made by linking synthetic peptide to keyhole limpet hemocyanin by a standard protocol (Harlow and Lane, 1988). The hemocyanin-conjugated peptides were used to raise antibodies in a female New Zealand White rabbit. For the initial immunization, 200 μg of conjugate in Freund's complete adjuvant was delivered via several subcutaneous injections. The rabbit received two additional boosts, using 200 μg of hemocyanin-conjugated peptides in Freund's incomplete adjuvant at 2 and 6 weeks after initial injections. The final bleed was performed by cardiac puncture 9 d after the final injection.
Antibodies that recognized the mammalian phosphorylated Thr at the T-loop of the catalytic subunit were purchased from Santa Cruz Biotechnology. Antibodies that recognized the N terminus of the GFP were purchased from Sigma.
Soluble or chloroplastic proteins were subjected to electrophoresis after mixing with 2× Laemmli SDS-PAGE sample buffer and boiling for 5 min. After electrophoresis, proteins were transferred to nitrocellulose membranes using a mini gel apparatus (Bio-Rad). The blot was developed using an enhanced chemiluminescence kit (GE Healthcare). Two-dimensional SDS-PAGE was performed using the Multiphor II Electrophoresis Unit (Amersham) according to the manufacturer's instructions.
Microscopy and Immunolabeling
Roots and leaves of Arabidopsis grown with or without phosphate were harvested and fixed in 4% (v/v) formaldehyde in phosphate-buffered saline (PBS), dehydrated in an ethanol series, and embedded in Paraplast Plus (Polysciences). Sections of 6 to 7 μm were blocked with PBS plus 3% bovine serum albumin, 0.01% sodium azide, and 0.1% Triton X-100 for 4 h at 4°C. Sections were simultaneously incubated with the primary rabbit anti-α-subunit antibody (1:250 dilution) at 4°C overnight. Sections were then rinsed with PBS and then incubated with secondary goat anti-rabbit Alexa 568-fluorochrome-conjugated antibody for 4 h at 4°C. Sections were examined by confocal fluorescence
For the study of GFP fusion proteins, leaf sections were mounted on slides and the GFP fluorescence was observed using a FluoView 1000 confocal microscope (Olympus). Images were captured with a digital camera, and photographs were processed for optimal presentation using Photoshop 8.0
14CO2 Labeling, Starch Isolation, and Determination
Mature leaves were labeled with 14CO2 as described by Viola et al. (2001). Five hours after the light period started, plants were incubated with the 14CO2 liberated from 50 μCi (2.18 GBq mmol−1) of sodium [14C]bicarbonate by the addition of 200 μL of 3 m lactic acid. Plants were exposed to 14CO2 for 1 h before injecting an excess of 3 m KOH to neutralize the acid and to absorb any remaining 14CO2. One group of plants was analyzed 4 h later, and another group was incubated in the dark for 18 h. For starch isolation and quantification, leaf samples were homogenized in 2 to 3 mL of 80% ethanol and extracted twice at 80°C for 30 min. The soluble fraction was discarded, and the ethanol-insoluble fraction was resuspended in 1.5 mL of water and incubated at 90°C for 4 h. After cooling, 220 units of amyloglucosidase (from Rhizopus; EC 3.2.1.3) in 1.5 mL of 200 mm acetate buffer (pH 4.5) was added to each sample and incubated at 37°C overnight. Glc and the label associated with the starch were determined in the supernatant (Bernal et al., 2005).
Acknowledgments
We thank Lorena Chávez González, Simón Guzmán León, José Luis Santillán Torres, and Jorge Ramírez for assistance in the microarray determination; Gerardo Coello, Gustavo Corral, and Ana Patricia Gómez for computer assistance; and Laurel Fabila and Carmen Parra for technical assistance.
This work was supported by the DGAPA-UNAM (grant no. IN202206), PAIP (grant no. 6290–13), and CONACyT (grant no. 52072). S.F. and L.E. received a scholarship from CONACyT, Mexico.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Patricia Coello (pcoello@servidor.unam.mx).
Some figures in this article are displayed in color online but in black and white in the print edition.
Open Access articles can be viewed online without a subscription.
References
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcriptional network in plant stress and energy signalling. Nature 448 938–943 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J (2008) Convergent energy and stress signaling. Trends Plant Sci 13 474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19 529–538 [Google Scholar]
- Bernal L, Coello P, Martínez-Barajas E (2005) Possible role of R1 protein on starch accumulation in bean seedling (Phaseolus vulgaris L.) under phosphate deficiency. J Plant Physiol 162 970–976 [DOI] [PubMed] [Google Scholar]
- Bieliski R (1973) Phosphate pools, phosphate transport and phosphate availability. Annu Rev Plant Physiol 24 225–252 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho Y, Martınez-Castilla L, Fragoso S, Vázquez S, Martínez-Barajas E, Coello P (2008) Characterization of a type A response regulator in the common bean (Phaseolus vulgaris) in response to phosphate starvation. Physiol Plant 132 272–282 [DOI] [PubMed] [Google Scholar]
- Cheung PCF, Salt IP, Davies SP, Hardie DG, Carling D (2000) Characterization of AMP-activated protein kinase c-subunit isoforms and their role in AMP binding. Biochem J 346 659–669 [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Barbachowska A (2000) Sucrose metabolism in leaves and roots of bean (Phaseolus vulgaris L.) during phosphate deficiency. J Plant Physiol 156 640–644 [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski LA (2001) Phosphate status affects the gene expression, protein content and enzymatic activity of UDPglucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212 598–605 [DOI] [PubMed] [Google Scholar]
- Clough FJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Strich R (1999) Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol 19 3338–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SP, Carling D, Hardie DG (1989) Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studies using a specific and sensitive peptide assay. Eur J Biochem 186 123–128 [DOI] [PubMed] [Google Scholar]
- Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC (1989) Phosphate starvation inducible “bypasses” of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol 90 1275–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Farrás R, Ferrando A, Jaásik J, Kleinow T, Ocres L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf-1 related (SnRK1) protein kinase. J Exp Bot 54 467–475 [DOI] [PubMed] [Google Scholar]
- Hardie DG (2007) AMP activated/Snf1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8 774–785 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Sakamoto K (2006) AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology (Bethesda) 21 48–60 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 53–77
- Hey SJ, Powers SJ, Beale MH, Hawkins ND, Ward JL, Halford NG (2005) Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol J 4 219–229 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Naka K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35 W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JB (1982) Hydroponics: its history and use in plant nutrition studies. J Plant Nutr 5 1003–1030 [Google Scholar]
- Karthikeyan A, Varadarajan DK, Jain A, Held MA, Carpita NC, Raghothama KG (2007) Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225 907–918 [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Denga X (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Samac DA, Bucciarelli B, Allan DL, Vance CP (2005) Signaling of phosphorus deficiency-induced gene expression in white lupin requires sugar and phloem transport. Plant J 41 257–268 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MR, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP (2001) Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant Cell Environ 24 459–467 [Google Scholar]
- McKibbin RS, Muttucumaru N, Paul MJ, Powers SJ, Burrell MM, Coates S, Purcell PC, Tiessen A, Geigenberger P, Halford NG (2006) Production of high-starch, low-glucose potatoes through over-expression of the metabolic regulator SnRK1. Plant Biotechnol J 4 409–418 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281 11815–11818 [DOI] [PubMed] [Google Scholar]
- Plaxton W (2004) Plant responses to stress: biochemical adaptations to phosphate deficiency. In Encyclopedia of Plant and Crop Sciences. Taylor and Francis, New York, pp 976–980
- Plaxton WC, Carswell MC (1999) Metabolic aspects of the phosphate starvation response in plants. In HR Lerner, ed, Plant Responses to Environmental Stresses, from Phytohormones to Genome Reorganization. Marcel Dekker, New York, pp 350–372
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0024, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Polge C, Jossier M, Crozet P, Gissot L, Thomas M (2008) β-Subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities: physical interaction with nitrate reductase specifically occurs via AKINβ1-subunit. Plant Physiol 148 1570–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M (2006) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12 20–28 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halfors NG, Hardie DG (1999) Two SNF1 related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase and sucrose phosphate synthase in vitro. Plant Physiol 120 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ (2004) Protein phosphorylation in amyloplast regulates starch branching enzyme activity and protein-protein interaction. Plant Cell 16 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou ME, Plaxton WC (1993) Metabolic adaptation of plant respiration to nutritional phosphate deprivation. Plant Physiol 101 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157 423–447 [DOI] [PubMed] [Google Scholar]
- Vincent O, Townley R, Kuchin S, Carlson M (2001) Subcellular localization of the Snf1 kinase is regulated by specific b subunits and a novel glucose signaling mechanism. Genes Dev 15 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola R, Roberts AG, Haupt S, Gazzani S, Handcock RD, Marmiroli N, Machray GC, Oparka KJ (2001) Tuberization in potato involves a switch from apoplastic to symplastic phloem unloading. Plant Cell 13 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 217–219
- Zhang Y, Shewry PR, Jones H, Barcelo P, Lazzeri PA, Halford NG (2001) Expression of antisense SnRK1 protein kinase sequence causes abnormal pollen development and male sterility in transgenic barley. Plant J 28 431–441 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10 407–409 [DOI] [PubMed] [Google Scholar]