Abstract
Sterols become functional only after removal of the two methyl groups at C4 by a membrane-bound multienzyme complex including a 3β-hydroxysteroid-dehydrogenase/C4-decarboxylase (3βHSD/D). We recently identified Arabidopsis (Arabidopsis thaliana) 3βHSD/D as a bifunctional short-chain dehydrogenase/reductase protein. We made use of three-dimensional homology modeling to identify key amino acids involved in 4α-carboxy-sterol and NAD binding and catalysis. Key amino acids were subjected to site-directed mutagenesis, and the mutated enzymes were expressed and assayed both in vivo and in vitro in an erg26 yeast strain defective in 3βHSD/D. We show that tyrosine-159 and lysine-163, which are oriented near the 3β-hydroxyl group of the substrate in the model, are essential for the 3βHSD/D activity, consistent with their involvement in the initial dehydrogenation step of the reaction. The essential arginine-326 residue is predicted to form a salt bridge with the 4α-carboxyl group of the substrate, suggesting its involvement both in substrate binding and in the decarboxylation step. The essential aspartic acid-39 residue is in close contact with the hydroxyl groups of the adenosine-ribose ring of NAD+, in good agreement with the strong preference of 3βHSD/D for NAD+. Data obtained with serine-133 mutants suggest close proximity between the serine-133 residue and the C4β domain of the bound sterol. Based on these data, we propose a tentative mechanism for 3βHSD/D activity. This study provides, to our knowledge, the first data on the three-dimensional molecular interactions of an enzyme of the postoxidosqualene cyclase sterol biosynthesis pathway with its substrate. The implications of our findings for studying the roles of C4-alkylated sterol precursors in plant development are discussed.
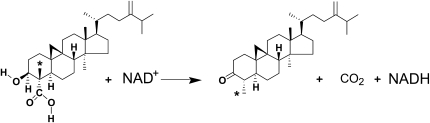
Sterols are essential components of all eukaryotic cell membranes, and their biosynthetic pathways differ significantly between plants, animals, and fungi. The sterol molecule becomes functional only after removal of the two methyl groups at C4 (Lees et al., 1995; Benveniste, 2004; Bouvier et al., 2005). We have characterized the activities of a sterol C4-methyl oxidase (SMO), a 4α-carboxysterol-3β-hydroxysteroid-dehydrogenase/C4-decarboxylase (3βHSD/D; Fig. 1), and a NADPH-dependent 3-oxosteroid reductase from partially purified enzymes in order to define the steps involved in plant sterol C4-demethylation (Pascal et al., 1993, 1994; Rondet et al., 1999). The first step in this process is initiated by SMO, which converts the C4α methyl group of C4-methylated sterol substrate to produce a 4α-carboxysterol derivative (Fig. 2) that is subsequently decarboxylated by 3βHSD/D to produce a C4-monodemethylated 3-oxosteroid. Finally, the C4-monodemethylated 3-oxosteroid intermediate is stereospecifically reduced by the 3-ketoreductase. Two distinct families of SMO genes have been identified in Arabidopsis (Arabidopsis thaliana; Darnet and Rahier, 2004), and we recently characterized molecularly and enzymatically two bifunctional 3βHSD/Ds (Fig. 1) from Arabidopsis (Rahier et al., 2006). These hydroxysteroid-dehydrogenases (HSDs) constitute novel members of the short-chain dehydrogenase/reductase (SDR) family in plants.
Figure 1.
Reaction catalyzed by 3βHSD/D. The C4β-methyl group of the 3β-hydroxy,4α-carboxy-4β,14α-dimethyl-9β,19-cyclo-5α-sterol (identified with an asterisk) is isomerized to the C4α position during the reaction.
Figure 2.
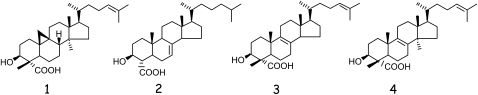
Structures of the compounds considered in this work: [1], 4α-carboxy-4β,14α-dimethyl-9β,19-cyclo-5α-cholest-24-en-3β-ol; [2], 4α-carboxy-5α-cholest-7-en-3β-ol; [3], 4α-carboxy-4β-methyl-5α-cholest-8,24-dien-3β-ol; [4], 4α-carboxy-4β,14-dimethyl-5α-cholest-8,24-dien-3β-ol.
The SDRs have been divided into two major families, referred to as classical and extended, that differ in size: about 250 amino acids for the classical SDRs and 350 for the extended SDRs (Jörnvall et al., 1995). For both groups, the cofactor binding site is localized in the N-terminal part and the substrate binding site in the C-terminal part. Probably because of its unique bifunctionality and membrane association, 3βHSD/D protein shows low sequence identity with other members of the SDR family and forms a differentiated cluster in phylogenetic analysis (Rahier et al., 2006; Wu et al., 2007). According to a recent classification, 3βHSD/D would be a member of the extended SDR family (Kallberg et al., 2002) for which the number of experimentally solved three-dimensional (3D) structures is lower than for the classical family. In fact, there are only two structures for the NAD(H)-preferring enzymes of this type, including dTDP-Glc-4,6-dehydratase (DesIV; Allard et al., 2004).
3βHSD/D is essential for the biosynthesis of sterols in all organisms. Yeast mutants lacking the C4-demethylation process are not viable (Bard et al., 1996), and nonfunctional 3βHSD/D is lethal in the yeast erg26 mutant (Gachotte et al., 1998). In animals, deficiency in 3βHSD/D and the C4-demethylation process is lethal for the embryo (König et al., 2000). Mutations in the corresponding ortholog Nsdhl gene are associated with the X-linked, male-lethal dominant mutations bare patches (Bpa) and striated (Str) in mouse (Liu et al., 1999) and cause the embryo-lethal CHILD syndrome in humans (Bornholdt et al., 2005). Remarkably, mutants of plants deficient in 3βHSD/D and C4-demethylation have not been reported so far, possibly reflecting the vital importance of this pathway-specific step.
Structural data for SDRs in plants is particularly scarce. In plants, the most thoroughly investigated SDRs are tropinone reductases I and II from Datura stramonium and Hyoscyamus niger (Nakajima et al., 1998, 1999) and progesterone 5β-reductase from Digitalis lanata (Thorn et al., 2008), for which determination of the x-ray structure and site-directed mutagenesis of putative substrate-binding residues revealed the decisive amino acid residues controlling the reaction of these enzymes. Recently, a homology model of a SDR involved in plant secondary metabolism, salutaridine reductase from Papaver bracteatum, was built. The data were taken as a basis to elucidate the reaction mechanism, the cofactor preference, and the substrate binding by site-directed mutagenesis (Geissler et al., 2007).
During the past decade, direct studies of plant sterol biosynthesis enzymes have been facilitated by the cloning of most of the corresponding genes. However, details of the 3D structure for membrane enzymes involved in the postoxidosqualene segment of sterol synthesis in plants, animals, and fungi are not available. In particular, the structure, the substrate-binding characteristics, and the detailed catalytic mechanism of membrane-bound 3βHSD/D are unknown. Due to the difficulty of crystallizing membrane proteins, homology modeling provides a convenient means for understanding their functions.
In this study, we constructed a model of Arabidopsis 3βHSD/D based on its similarity to the structure of the previously crystallized SDR DesIV from Streptomyces venezuelae (Allard et al., 2004). Furthermore, 4α-carboxysterol substrate as well as the cofactor NAD+ were docked into the protein model. The model provides novel information on substrate and cofactor 3D interactions with the 3βHSD/D active site. The predictions of catalytically important amino acids were verified experimentally using mutant proteins generated by site-directed mutagenesis. Based on these data, we propose a catalytic mechanism for the Arabidopsis 3βHSD/D.
RESULTS
Homology Modeling of the 3D Structure of 3βHSD/D from Arabidopsis
Overall Description of the Protein Model
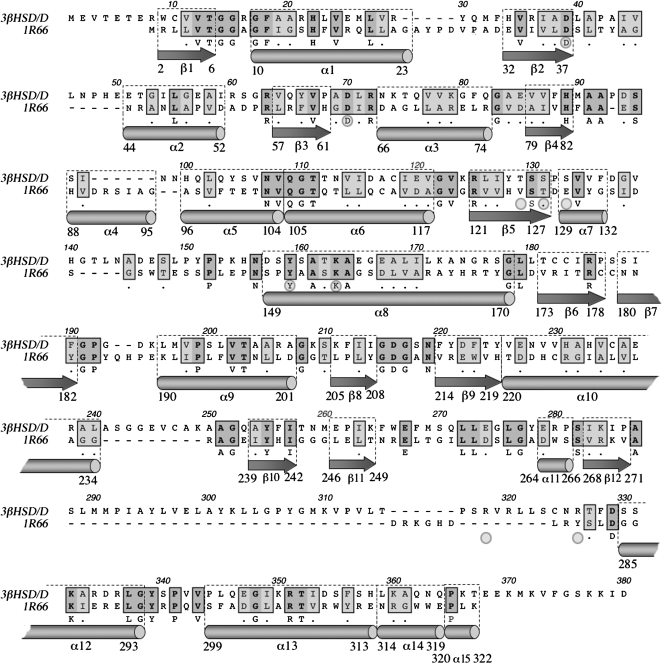
The 3D structure of any ortholog of Arabidopsis 3βHSD/D protein is unknown at present. Thus, the search for the structure of the closest paralog led to the structure of DesIV from S. venezuelae (Allard et al., 2004). Based on this evidence, DesIV was taken as a template for 3βHSD/D (as described in “Materials and Methods”; Figs. 3 and 4). The peptide sequence of DesIV displayed 24% identity and 40% similarity out of 376 positions when aligned with that of 3βHSD/D (Fig. 3). In addition, the molecular masses of 3βHSD/D (42 kD) and DesIV (37 kD) are particularly higher than those of other HSDs and both have strict NAD+ cofactor specificity (Rondet et al., 1999).
Figure 3.
Alignment and secondary structure comparison between 3βHSD/Ds from Arabidopsis and the template protein DesIV (1R66) from S. venezuelae. The secondary structure elements are indicated as arrows for β-strands and as cylinders for α-helices. Amino acids further analyzed by mutagenesis are shown with gray circles. The nomenclature used for helices and strands corresponds to that of 1R66 pdb file.
Figure 4.
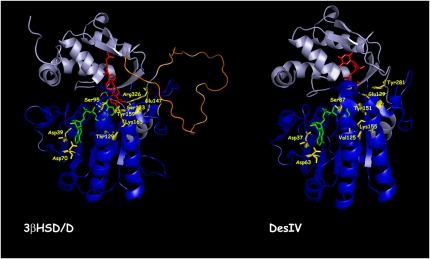
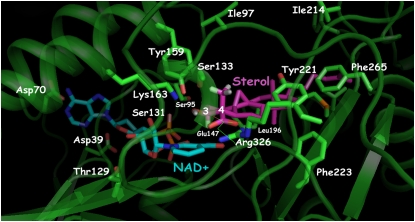
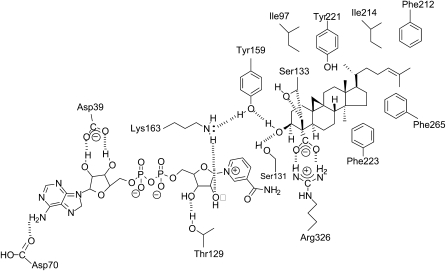
Ribbon diagrams of Arabidopsis 3βHSD/D based on homology modeling and of DesIV used as a template, with key amino acids identified. The N-terminal domain, responsible for NAD docking (dark blue), and the C-terminal domain, responsible for carboxysterol or DAU positioning (light blue), are shown. The loop in orange including the putative hydrophobic membrane-interacting domain could not be modeled. The NAD+ (green), carboxysterol, and DAU (red) structures are included. For 3βHSD/D, the key catalytic Tyr-159 and Lys-163 residues for 3β-dehydrogenation, the carboxysterol-stabilizing residue Arg-326, and the carboxysterol 4β-methyl proximal residue Ser-133 are also shown (yellow). Asp-39, Asp-70, Ser-95, and Thr-129 residues are shown hydrogen bonding to NAD+. Glu-147 possibly interacting with Arg-326 in the absence of substrate is also shown. The homolog residues are shown for DesIV. [See online article for color version of this figure.]
The structure of DesIV consists of two distinct domains (Fig. 4): a large N-terminal domain, from Met-1 to Tyr-182, composed of five α-helices and eight β-strands that is involved in NAD+ binding; and a small C-terminal domain with only four α-helices and two β-strands, which is involved in substrate recognition and positioning (Allard et al., 2004). The 3βHSD/D model is very similar, and all of the secondary elements could be preserved except one α-helix labeled α4.
As predictable from the functions of their domains, the sequences of 3βHSD/D and DesIV are more similar in the N-terminal regions, which have identical functions, than in the C-terminal regions, which bind different substrates. All of the residues whose side chains are involved in NAD+ binding are conserved: Asp-37, Asp-63, Ser-87, Tyr-151, and Lys-155 in DesIV, which correspond to Asp-39, Asp-70, Ser-95, Tyr-159, and Lys-163, respectively, in 3βHSD/D.
DesIV is known to be a dimer, and three regions from its N-terminal domain contribute to the dimeric interface: Ile-93 to Gln-108, Pro-144 to Ser-149, and Leu-160 to Tyr-169. Within these segments, spanning 32 amino acids, seven are strictly conserved in 3βHSD/D (the five-amino acid stretch 103NVQGT107, Pro-144, and Asn-148), four are replaced by similar ones (Val-98/Leu-102,Leu-160Val-161Ala-162/Ala-168Leu-169Ile-170), and the 21 remaining amino acids are different. This observation of a relatively weak sequence conservation in the contact region could be related to the fact that 3βHSD/D has been shown to be active as a monomer (Rondet et al., 1999), thus lowering the conservation pressure upon these chain segments.
Finally, one should note a very long insertion of about 45 amino acids into the C-terminal part of 3βHSD/D (Fig. 3). This region with its hydrophobic patch of about 24 amino acids is thought to be the membrane anchor. It is situated at the surface of our model, away from the NAD+ binding site but close to the carboxysterol proposed binding site (Fig. 4). Because of the lack of a corresponding anchor in DesIV, which is not membrane bound, this region was not constrained during modeling and resulted in an unstructured loop.
Based on the above analysis, our 3βHSD/D model displayed enough accuracy to be able to explore the geometry of the substrate-binding cavity for identification of residues in contact with the NAD+ and 4α-carboxysterol substrates as well as putative catalytic residues.
Hypothetical Mode of Substrate Binding of 3βHSD/D
4α-Carboxy-4β,14α-dimethyl-9β,19-cyclo-5α-cholest-24-en-3β-ol [1] (Fig. 2), the substrate of Arabidopsis 3βHSD/D, was docked into the substrate pocket of the 3βHSD/D model (Figs. 4 and 5). The docking arrangement of [1] in the active site of 3βHSD/D shows that the carboxysterol substrate is completely buried in the enzyme, with the sterol side chain pointing to the entrance of the active site cavity. As can be seen (Figs. 4 and 5), the tetracyclic steroid nucleus and side chain can be modeled into a large pocket featuring a variety of hydrophobic amino acid residues. In particular, side chains of Tyr-221, Phe-265, and Leu-196 make direct contacts with the B, C, and D rings of the steroid nucleus. They replace the hydrophilic residues Arg-215, Asn-250, and Lys-190, respectively, in the Des IV active site, where Arg-215 forms a salt bridge with one of the β-phosphoryl oxygens, Asn-250 interacts with the 3-hydroxyl group of the Rib, and Lys-190 interacts with the sugar O-2 of the substrate dTDP (Allard et al., 2004). Further hydrophobic interactions can be observed between the side chain Phe-223 and the 14α-methyl group and the C and D rings of the sterol substrate. Moreover, additional hydrophobic interactions are possible between Ile-213, Ile-214, Leu-201, and Met-197 and the side chain of [1]. Weaker hydrophobic interactions are imaginable between Ile-97 and Phe-212 and the β-face and side chain of the sterol nucleus.
Figure 5.
Closeup view of active site residues in 3βHSD/D determined by homology modeling in relation to modeled NAD+ and steroid substrate (4α-carboxy-4β,14α-dimethyl-cholesta-9β,19-cyclo-24-en-3β-ol) molecules. Residues analyzed in this study are mentioned. Tyr-159 and Lys-163 are at catalytic distance to the substrate and at a distance to interact with each other. Substrate is further stabilized by a salt bridge with Arg-326. Ser-133 is in close proximity to the C4β-methyl group of the steroid substrate. Asp-39, Asp-70, Ser-95, and Thr-129 residues are interacting with NAD+. Hydrophobic residues that are in the immediate surroundings of the carboxysterol in its binding pocket are also shown, as well as Glu-147 possibly interacting with Arg-326 in the absence of substrate. This figure was created with PyMOL (DeLano, 2002). [See online article for color version of this figure.]
Importantly, the model shows that Arg-326 is predicted to interact with the carboxysterol substrate and that its NH2 groups are correctly positioned to form a salt bridge with its carboxylic group. In the absence of bound carboxysterol, the model suggests that Arg-326 could be stabilized by the formation of a salt bridge with Glu-147. This Arg residue is replaced by the neutral Tyr-281 in Des IV. Furthermore, the model localizes Ser-133 in close proximity (<3.2 Å) to the C4β-methyl group of [1]. Most importantly, the 3β-hydroxyl group of [1] is within the range of hydrogen-bond interactions to the hydroxyl group of Tyr-159. It could also form a weak hydrogen bond with the hydroxyl group of Ser-131. Moreover, the amino group of Lys-163 is located in close proximity (<2.5 Å) to Tyr-159. In addition, the 3α-hydrogen of [1] is in the correct geometry and proximity to the accepting carbon of the NAD+ cofactor, that is an angle [C3(1)-C4(NAD+)-N1(NAD+)] of about 95°, a value compatible with a hydride transfer from the C3-carbon atom of the substrate and the nicotinamide ring, and a C4(NAD+)-C3αH [1] distance of approximately 2 Å.
NAD+ was docked into the substrate pocket of the 3βHSD/D model in a similar conformation as that found in the DesIV template, with both Ribs in the C2-endo conformation and the nicotinamide ring in the syn conformation. The model predicts that several conserved side chains are in close contact with the NAD+ to the 3βHSD/D, including Asp-39, Asp-70, Ser-95, Tyr-159, and Lys-163, corresponding to Asp-37, Asp-63, Ser-87, Tyr-151, and Lys-155, respectively, in DesIV. The model indicates that the carboxylate of Asp-70 is in proximity to hydrogen bonds to the C-6 amino group of the adenine ring and that the carboxylate of Asp-39 bridges the 2′- and 3′-hydroxyl groups of the adenine Rib. Although the model shows that the distance of the ε-amino group of Lys-163 to Tyr-159 is shorter than the distance to the 2′-hydroxyl group of the nicotinamide Rib, it may participate in proper dinucleotide positioning. In addition, the model predicts that the hydroxyl side chain of Thr-129 (Val-129 in DesIV) could form a hydrogen bond with the 3′-hydroxyl of the nicotinamide Rib. Finally, the central diphosphate group of NAD+ interacts mainly with the Gly-rich motif 13TGGRGFAA20.
Site-Directed Mutagenesis Analysis
Site-Directed Mutagenesis, Expression, and Characterization of the Wild-Type and Mutant 3βHSD/Ds
Based on the interactions observed in the model, we generated site-directed mutant proteins in which selected single amino acid residues supposed to be involved in substrate binding or catalytic activity were modified. In order to have minimal effects on secondary structure and on conformation, we chose to mutate the different amino acids by sterically conservative hydrophobic and electrically neutral residues. The wild-type and mutated 3βHSD/Ds were cloned into the pVT102U shuttle vector under the control of the constitutive alcohol dehydrogenase promoter and expressed in the Saccharomyces cerevisiae erg26 null mutant. The pVT transformants were plated on complementation selection plates in a medium supplemented with δ-aminolevulinic acid (δ-ala) required for heme synthesis but devoid of sterol. Heme auxotrophy was required to facilitate sterol uptake for erg26 mutants. As shown previously (Gachotte et al., 1998; Rahier et al., 2006), the erg26 strain transformed with pVT-At3βHSD/D was capable of growing aerobically without ergosterol supplementation, while erg26 null and erg26/pVT-VOID transformants as well as transformants obtained with nonfunctional 3βHSD/D mutants could grow only on an ergosterol (cholesterol)-supplemented medium. To confirm the authenticity of the complementation assay, the strains were picked from the selection plate and the prototrophic strains were grown in a δ-ala-containing liquid medium devoid of sterol and the auxotrophic strains were grown in a cholesterol-containing medium. After sterol extraction, the sterol profiles were analyzed by gas chromatography (GC) and GC-mass spectrometry (MS). The erg26 strain transformed with the wild-type 3βHSD/D accumulated more than 75% of C4-demethylated sterols including ergosterol (53%) as the major sterol and minor amounts (10%) of residual 4,4-dimethylsterols (Table I). In comparison, the auxotrophic erg26 null strain grown in a cholesterol-containing medium accumulated lanosterol (86%) and small amounts of 4α-carboxy-4β,14-dimethyl-cholest-8,24-dien-3β-ol [4] (Fig. 3; 14%), as shown previously (Gachotte et al., 1998; Rahier et al., 2006). In the presence of δ-ala and cholesterol, the erg26 null strain accumulated 4α-carboxy-4β-methyl-cholest-8,24-dien-3β-ol [3] but no ergosterol, as shown previously (Gachotte et al., 1998).
Table I.
Sterol composition of the erg26 null mutant and transformants carrying Arabidopsis wild-type and mutagenized 3βHSD/D
Sterols were analyzed as described in “Materials and Methods.” Data shown are means ± sd of two to four experiments.
| Enzyme Used to Transform the erg26 Null Mutant | Cholesterol Added in the Medium | Total C4-Demethylated Sterolsa or Complementation Rate of the erg26 Mutant | Total 4,4-Dimethylsterolsb | Total 4α-Carboxysterolsc | Other Sterolsd = 4α-Methylsterols |
|---|---|---|---|---|---|
| erg26 null mutant | Yes | 0e | 86 ± 6 | 14 ± 6 | 0 |
| Wild type | No | 76 ± 3 | 10 ± 1 | 0.5 ± 0.04 | 13.5 ± 2 |
| D39V | Yes | 0 | 89 ± 6 | 11 ± 6 | 0 |
| D70V | Yes | 0 | 96 ± 1 | 4.2 ± 0.4 | 0 |
| D70A | Yes | 0 | 98 ± 1 | 2.3 ± 0.3 | 0 |
| T129V | Yes | 0 | 96 ± 1 | 3.9 ± 0.2 | 0 |
| Y159F | Yes | 0 | 89 ± 3 | 10.8 ± 3.5 | 0 |
| K163I | Yes | 0 | 86 ± 2 | 14.3 ± 2.5 | 0 |
| R326I | Yes | 0 | 97 ± 1 | 3.0 ± 0.9 | 0 |
| R318I | No | 82 ± 4 | 12.5 ± 7 | 1.2 ± 0.3 | 5 ± 0.5 |
| Δ[288–303] | Yes | 0 | 97 ± 1 | 2.4 ± 0.3 | 0 |
| S131A | No | 64 ± 1 | 19 ± 1 | 1.1 ± 0.4 | 16 ± 2 |
| S133A | No | 63 ± 2 | 19 ± 1 | 0.5 ± 0.04 | 17.5 ± 2 |
| S133T | No | 45 ± 2 | 43 ± 3 | 6.6 ± 0.5 | 5.4 ± 0.9 |
| S133Y | Yes | 0 | 91 ± 1 | 9.2 ± 0.8 | 0 |
C4-Demethylated sterols were a mixture of zymosterol (tR = 1.055), ergosterol (tR = 1.084), ergosta-7,22-dien-3-ol (tR = 1.094), 14α-methyl-ergosta-8,24(28)-dien-3-ol (tR = 1.112 ), ergost-7-en-3-ol (tR = 1.117), ergosta-8,24(28)-dien–3-ol (tR = 1.141), and ergosta-7,24(28)-dien-3-ol (tR = 1.158).
4,4-Dimethylsterols were a mixture of lanosterol (tR = 1.207) and 4,4-dimethyl-cholesta-8,24-dien-3-ol (tR = 1.216).
4α-Carboxysterols were a mixture of 4α-carboxy-4,14-dimethyl-cholesta-8,24-dien-3-ol (tR = 1.421) and 4α-carboxy-4-methyl-cholesta-8,24-dien-3-ol (tR = 1.457).
Other sterols were 4α-methyl-cholesta-8,24-dien-3-ol (tR = 1.112), 4α-methyl-cholesta-8,24-dien-3-one (tR = 1.134), 4α,14α-dimethyl-ergosta-8,24(28)-dien-3-ol (tR = 1.165), 4α-methyl-ergosta-8,24(28)-dien-3-ol (tR = 1.176), and 4α-methyl-ergosta-7,24(28)-dien-3-one (tR = 1.245).
Percentage of total sterols.
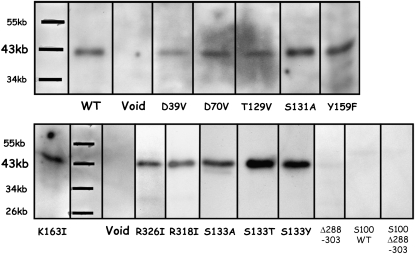
To corroborate the in vivo sterol profile data, we next performed an in vitro enzymatic assay with the recombinant wild-type and mutated 3βHSD/Ds. In the case of an enzyme that is part of a membrane-bound multienzyme complex, interactions with other components of the complex are probably crucial for optimum enzymatic activity. Thus, 3βHSD/D activity was assayed in the microsomal extracts and the corresponding cytosolic fractions were prepared from the different mutants described below, using the standard assay conditions for recombinant 3βHSD/D (as described in “Materials and Methods” and in Rahier et al., 2006). The immunoblotting analysis indicated that the wild-type 3βHSD/D produced a discrete band with the expected Mr in the microsomal extracts (Fig. 6, WT), but this band was absent from the corresponding supernatant fraction (Fig. 6, S100 WT).
Figure 6.
Western blot analysis. Forty micrograms of microsomal protein from erg26 yeast strain overexpressing the FLAG-tagged wild-type At3βHSD/D cDNA (WT) and cDNAs with the shown mutations and of yeast transformed with vector without cDNA (Void) were analyzed. Analyses of 100 μg of concentrated 100,000g supernatant protein from erg26 yeast strain overexpressing the FLAG-tagged wild-type At3βHSD/D cDNA (S100 WT) and concentrated 100,000g supernatant from mutant Δ[288-303] (S100 Δ288–303) are also shown. Proteins were submitted to immunoblotting with an anti-FLAG serum.
Analysis of Mutants D39V, D70V, D70A, and T129V
The model of 3βHSD/D predicts that Asp-39, Asp-70, and Thr-129 interact with different Rib hydroxyl groups of NAD+. In addition, analysis of the conserved residues of the fingerprint of DesIV identifies the Asp-39 residue in 3βHSD/D that may be critical to specific recognition of NAD+. Thus, we selected for further studies the D39V, D70V, D70A, and T129V mutants of 3βHSD/D, as suggested by the model. The immunoblotting analysis confirmed that each of the mutant proteins produced a discrete band with the expected Mr (Fig. 6). Substitution of Asp-39, Asp-70, and Thr-129 for Val or Ala rendered the enzyme totally inactive in vivo, since these mutants could not grow in the absence of added cholesterol in the medium and no traces of C4-demethylated sterols could be detected (Table I). Moreover, no 3βHSD/D activity could be detected in vitro (Table II). These results indicate that these residues are essential for enzyme activity.
Table II.
Apparent kinetics parameters for reactions of wild-type Arabidopsis 3βHSD/D and mutants
Data shown are means ± sd of two or three experiments. The parameters for the [2] dehydrogenation/decarboxylation reaction were determined in the corresponding microsomal extracts under the standard assay conditions as described in “Materials and Methods.” Apparent Vmax and Km values were determined by fitting the data to the Michaelis-Menten equation. ND, Not determined.
| Enzyme Used to Transform the erg26 Null Mutant | Km, [2] | Vmaxa | Vmax/Km, [2] |
|---|---|---|---|
| mm | nmol mg−1 h−1 | mL h−1 mg−1 | |
| erg26 null mutant | No activityb | ||
| Wild type | 0.13 ± 0.01 | 88 ± 7 | 0.68 |
| D39V | ND | No activity | |
| D70V | ND | No activity | |
| D70A | ND | No activity | |
| T129V | ND | No activity | |
| Y159F | ND | No activity | |
| K163I | ND | No activity | |
| R326I | ND | No activity | |
| R318I | 0.14 ± 0.01 | 83 ± 5 | 0.59 |
| Δ[288–303] | ND | No activity | |
| S131A | 0.17 ± 0.01 | 74 ± 4 | 0.44 |
| S133A | 0.11 ± 0.01 | 50 ± 7 | 0.45 |
| S133T | 0.51 ± 0.09 | 31 ± 2 | 0.061 |
| S133Y | ND | No activity |
The Vmax for each enzyme was normalized for differences in expression by comparison with the wild type using immunoblots as shown in Figure 6.
No activity detectable. The estimated limit of detection of the 3βHSD/D activity is 0.1 nmol h−1 mg−1.
Analysis of Mutants Y159F and K163I
On the basis of amino acid sequence analysis, 3βHSD/D is a member of the SDR superfamily, containing the highly conserved YX3K catalytic couple (Jörnvall et al., 1995; Kallberg et al., 2002). The model suggests that Tyr-159 and Lys-163 are well positioned for deprotonation of the 3β-hydroxyl group of the carboxysterol substrate. There is a single 159YX3K163 motif in 3βHSD/D, and we targeted Y159F and K163I mutants. Both mutants were successfully expressed in the erg26 strain microsomal extract (Fig. 6), but the substitution of Tyr-159 for Phe and Lys-163 for Ile rendered the enzyme totally inactive both in vivo and in vitro (Tables I and II), indicating that these residues were essential for the enzyme activity.
Analysis of Mutants R326I and R318I
3βHSD/D is a unique HSD of the SDR family that catalyzes both the dehydrogenation of a 3β-hydroxyl group and decarboxylation at C4 of a carboxysterol by a single membrane-bound protein. 3βHSD/D is significantly larger than the typical HSD (380 versus 250 amino acid residues). It is conceivable that the dual action could be linked to the 130 additional residues on the C-terminal end of the enzyme. The model localizes Arg-326 as the only positively charged residue in the carboxysterol-binding cavity of 3βHSD/D. Arg-326 is positioned in close proximity to the carboxylate group of the substrate, which is at a distance to form a salt bridge with the amino groups of Arg-326. Thus, we targeted R326I to examine this interaction. As a comparison, we also substituted Arg-318, which the model localizes as distant from the substrate and cofactor pocket, near the protein external surface, for an Ile residue. The immunoblotting analysis confirmed that each of the mutant proteins produced a discrete band with the expected Mr (Fig. 6). Data from Tables I and II clearly show that substitution of Arg-326 for Ile resulted in an inactive enzyme, as shown by (1) the absence of growth of the transformant in the absence of added cholesterol, (2) a sterol profile comprising exclusively C4-methylated sterols and C4-carboxy-3β-hydroxy-sterols, and (3) no detectable 3βHSD/D activity in the corresponding microsomal extracts. In addition, comparison of the incubations of carboxysterol substrate [2] and corresponding controls performed with inactivated microsomes revealed the absence of dehydrogenated product, including a putative 3-oxo-C4-carboxyl-steroid intermediate, and complete recovery of [2] as confirmed by selective ion monitoring. These data clearly show that Arg-326 is essential for the 3βHSD/D activity.
In contrast, mutant R318I exhibited a similar sterol profile as the wild type. Furthermore, mutant R318I had sufficient activity to allow measurement of a variety of 3βHSD/D kinetic parameters, attesting that the microsomal extracts obtained with such a mutated enzyme were functional (Table II). The R318I mutant has Km and Vmax/Km values similar to those of the wild-type enzyme, indicating that Arg-318 is not involved in the function of the enzyme.
Analysis of a 290 to 306 Deletion Mutant of 3βHSD/D
In contrast to most SDR proteins, which are cytosolic, including HSDs, biochemical studies performed in plants (Rondet et al., 1999) have shown that native 3βHSD/D is membrane bound. A single putative short membrane-binding domain, including a hydrophobic patch of amino acids 283 to 306, is found in 3βHSD/D, which also possesses an endoplasmic reticulum retrieval signal. Indeed, we observed that when 3βHSD/D is expressed in the yeast erg26 strain, the resulting protein (Fig. 6, WT) and the enzymatic activity were observed only in the microsomal fraction. 3βHSD/D was not present in the supernatant (Fig. 6, S100 WT), which remained enzymatically inactive (data not shown). Our model predicts that this hydrophobic patch is included in the unstructured loop of about 45 amino acids near the C terminus that we could not model. This loop is distant from the active site cavity and protrudes from the external surface of the 3βHSD/D. Hydrophobic patches often constitute membrane association or protein interaction sites, and the loop may well turn out to serve an important function by mediating the attachment of 3βHSD/D to the membrane or to another component of the C4-demethylation multienzyme complex. Because of its shortness and its localization, this hydrophobic domain would interact with only one leaflet of the bilayer, thus classifying 3βHSD/D in the monotopic enzyme group (Blobel, 1980). We deleted 17 amino acids of the hydrophobic patch to produce mutant Δ[290-306], expecting to delocalize the 3βHSD/D to the soluble supernatant fraction. Attempts to substantiate this suggestion experimentally failed, since no detectable 3βHSD/D activity could be measured in both fractions. Moreover, expression of mutant Δ[290-306] without the hydrophobic stretch could be achieved in neither the erg26 microsomal extract (Fig. 6, Δ288-303) nor the corresponding supernatant fraction (Fig. 6, S100 Δ288-303). It is reasonable to consider that gross conformational changes to the native enzyme would lead to its proteolyse in the yeast system. This is probably illustrated by the absence of expression of the Δ[288-303] mutated enzyme possessing a 15-amino acid truncation, while all of the other mutated and wild-type 3βHSD/Ds were substantially and similarly expressed in the microsomal extracts, consistent with the conservation of their gross conformation.
Analysis of Mutant S131A
Sequence alignments indicate that Ser-131 corresponds to Thr-127 in DesIV, which is involved in the formation of a hydrogen bond with the keto group formed during substrate dehydrogenation (Allard et al., 2004). The model predicts that Ser-131 is in proximity to the 3β-hydroxyl group of the substrate and to the ε-NH2 group of Lys-163. Substitution of Ser-131 for Ala led to an in vivo sterol profile of the transformants similar to that of the wild-type enzyme (Table I). Microsomal extracts obtained with this mutated enzyme were functional (Table II). The data clearly show that Ser-131 is not crucial for the 3βHSD/D activity and that it is not a key catalytic or substrate-binding residue for 3βHSD/D. Mutant S131A exhibits a 1.5-fold decrease in efficiency (V/K) compared with the wild type, because it has a slightly higher value of Km for [2] and a slightly lower Vmax. These data indicate a weak participation of Ser-131 in both catalysis and substrate binding and are consistent with a possible weak hydrogen bonding of Ser-131 to the C3 hydroxyl or oxo group of the substrate.
Analysis and Substrate Specificity of Ser-133 Mutants
The model localizes Ser-133 in closer proximity to the C4β-methyl group of the substrate (<3 Å) than to its 3β-hydroxyl group. Substitution of Ser-133 for Ala led to in vivo sterol profiles of the transformants similar to that of the wild-type enzyme (Table I), and the corresponding microsomal extracts were functional (Table II). These data clearly show that Ser-133 is not crucial for the 3βHSD/D activity and that it is not a key catalytic or substrate-binding residue for 3βHSD/D. Mutant S133A has similar Km values for both 4α-carboxysterol [2] and 4β-methyl,4α-carboxysterol [3] as the wild-type enzyme (Tables II and III). In addition, it exhibits a small 1.5-fold decrease in efficiency with [2], which reflects a lower Vmax, and a similar efficiency with [3] compared with the wild type. These data suggest that the hydroxyl function of Ser-133 is not involved in substrate binding and plays a minimal catalytic role.
Table III.
Apparent kinetics parameters for reaction, and substrate preference of wild-type Arabidopsis 3βHSD/D and Ser-133 mutants
Data shown represent means ± sd of two or three measurements. The parameters for the [2] and [3] dehydrogenation/decarboxylation reactions were determined in the corresponding microsomal extracts under the standard assay conditions as described in “Materials and Methods.” Apparent Vmax and Km values were determined by fitting the data to the Michaelis-Menten equation.
| Enzyme Used to Transform the erg26 Null Mutant | Substrate: [2]
|
Substrate: [3]
|
Specificity
|
||||
|---|---|---|---|---|---|---|---|
| Km | Vmaxa | Vmax/Km | Km | Vmaxa | Vmax/Km | V/K [2]b to V/K [3] | |
| mm | nmol mg−1 h−1 | mL h−1 mg−1 | mm | nmol mg−1 h−1 | mL h−1 mg−1 | ||
| erg26 null mutant | No activity detectable | No activity detectable | |||||
| Wild type | 0.13 ± 0.01 | 88 ± 7 | 0.68 | 0.26 ± 0.02 | 111 ± 5 | 0.43 | 1.6 |
| S133A | 0.11 ± 0.01 | 50 ± 7 | 0.45 | 0.20 ± 0.02 | 84 ± 9 | 0.42 | 1.07 |
| S133T | 0.51 ± 0.09 | 31 ± 2 | 0.061 | 0.77 ± 0.12 | 4.5 ± 1.2 | 0.0058 | 10.5 |
| S133Y | No activity detectable | No activity detectable | |||||
The Vmax for each enzyme was normalized for differences in expression by comparison with the wild type using immunoblots as shown in Figure 6.
For each mutant or wild-type enzyme, both substrates were compared in the same enzyme preparation. Specificity for competing substrates is determined by the ratios of specificity constants (kcat/Km): Vmax = kcat × enzyme concentration; thus, for two substrates, A and B, using the same enzyme preparation: (kcat/Km)a/(kcat/Km)B = (Vmax/Km)a/(Vmax/Km)B. The estimated limit of detection of the 3βHSD/D activity is 0.1 nmol h−1 mg−1.
To further examine the predicted proximity between Ser-133 and the substituent at C4β of the substrate, we substituted Ser-133 with larger Thr or Tyr residues. The immunoblotting analysis confirmed that each of the mutant proteins produced a discrete band with the expected Mr (Fig. 6). In contrast to mutant S133A, mutant S133T led to a lower amount of C4-demethylated sterols and higher quantities of 4,4-dimethylated sterol intermediates. Mutant S133Y led to a sterol profile comprising exclusively C4-methylated sterols and 4α-carboxy-3β-hydroxy sterols (Table I). Moreover, the ratio of 4,4-dimethylsterols to 4α-methylsterols is about 8-fold higher in mutant S133T than in mutant S133A or wild-type 3βHSD/D.
The validity of the in vivo data were further examined in vitro by comparing the kinetic parameters of wild-type 3βHSD/D and mutant enzymes S133A, S133T, and S133Y (Table III). The reactions were measured using two substrates: 4α-carboxysterol [2] devoid of substituent at C4β, and 4β-methyl,4α-carboxysterol [3] possessing an additional C4β-methyl substituent. The Km values of [2] and [3] for mutant S133T were increased about 3- to 4-fold, indicating that the binding of [2] and [3] to 3βHSD/D was impaired by replacing Ser-133 with Thr. In addition, the Vmax/Km was decreased 11-fold and 70-fold for [2] and [3], respectively, compared to that of the wild-type enzyme, because the S133T has a much lower Vmax for [3] than for [2]. These kinetic data indicate that increased bulkiness at residue 133 impairs carboxysterol substrate binding and 3βHSD/D catalysis. Furthermore, the more bulky amino acid change in the S133Y mutant totally abolished catalytic activity with both substrates (Tables I–III).
Moreover, data from Table III indicate that specificity between substrates [2] and [3], determined by the ratio of their respective specificity constants, (kcat/Km)[2]/(kcat/Km)[3], increased from 1 for the less bulky Ala residue at position 133 to 10 in favor of [2] for the more bulky Thr residue.
Taken together, the data are consistent with the close proximity of Ser-133 to the C4β domain of the bound substrate, as suggested by the model, and show that increasing the size of the residue at position 133 alters the substrate specificity of the 3βHSD/D in favor of the C4β-demethylated carboxysterol substrate.
DISCUSSION
Homology modeling has been recently used to identify key amino acids in a number of plant enzymes (Thorsøe et al., 2005; Geissler et al., 2007; Osmani et al., 2008). In this work, a homology model of the tertiary structure of the monomeric 3βHSD/D was built and constitutes a novel example of the application of this methodology to a plant membrane-bound enzyme. It revealed several essential or key amino acid residues involved in substrate and cofactor binding and 3βHSD/D catalysis. We could align correctly and model the 3βHSD/D through careful editing of 80% of the residues, allowing us to build its structural fold. Incidentally, models with 2 Å accuracy have been built previously, although the amino acid sequence identity between the target and the template was below 15% (Koehl and Levitt, 1999). The model shows a typical SDR α/β folding pattern highly similar to a Rossmann fold, which consisted of central β-sheets flanked by α-helices. In addition, the quality of the model is reinforced by the conservation of many of the residues of the DesIV template that are involved in NAD+ cofactor binding. The predictions of the 3D model were substantiated by rational mutation studies and biochemical analyses. The obtained in vivo data were totally corroborated in vitro by measuring the corresponding enzymatic activities.
Binding of NAD+
Based on the obtained data, several essential residues, including Asp-39, Asp-70, Thr-129, and Lys-163, appear to take part in the hydrogen bond network around the NAD+ cofactor (Figs. 4, 5, and 7). In particular, the data suggest a close contact between the essential Asp-39 residue and the hydroxyl groups of the adenine Rib that does not leave enough space for the phosphate group in NADP(H). Many members of the SDR family of enzymes that utilize NAD+ as the preferred cofactor have an Asp residue in the first β2α2 loop of the Rossmann fold, while several members that utilize NADP+ have an Arg residue in this position (Huang et al., 2001; Duax et al., 2003). Furthermore, the formation of a pair of hydrogen bonds between the side chain of this Asp residue and the 2′ and 3′ hydroxyl groups on the adenine Rib has been identified as critical to the binding of the NAD+ cofactor (Tanaka et al., 1996; Duax et al., 2003). For example, in the case of NAD+-dependent Drosophila alcohol dehydrogenase, replacement of the corresponding Asp-38 residue by a hydrophobic Leu residue led to an inactive enzyme (Chen et al., 1991), in good agreement with our results. Thus, our study has targeted the essential Asp-39 residue as the amino acid that is probably responsible for the known strict NAD+ specificity of the 3βHSD/D (Rondet et al., 1999).
Figure 7.
Scheme of the predicted interactions in the catalytic pocket of 3βHSD/D with NAD+ and 4α-carboxy-4β,14α-dimethyl-9β,19-cyclo-5α-cholest-24-en-3β-ol. Hydrogen bonds, the salt bridge, and steric interactions between amino acids, NAD+, and carboxysterol substrate are indicated by dashed lines. Hydrophobic residues that are in the immediate surroundings of the carboxysterol in its docking pocket are also shown.
The data clearly show that Asp-70 is essential for 3βHSD/D activity both in vivo and in vitro. As shown in Figures 4, 5, and 7, Asp-70 is located in proximity to form a hydrogen bond with the amino group of the adenine ring of NAD+ and to participate in positioning the adenine ring for productive binding. The location of Asp-70 near the amine group of the adenine ring is analogous to Asp-62 in porcine carbonyl reductase (Ghosh et al., 2001), Asp-60 in 3α,20β-HSD (Ghosh et al., 1994), and Asp-68 in 7αHSD (Tanaka et al., 1996). Moreover, drastic reduction in enzymatic efficiency for the oxidative reaction of the D60A mutant of 3β,17βHSD has been measured (Filling et al., 2002).
Residues Thr-129 and Lys-163 were both shown to be essential for the 3βHSD/D activity, suggesting their participation in positioning the nicotinamide Rib moiety for productive binding.
Taken together, these data suggest the existence of five hydrogen bonds between NAD+ and its 3βHSD/D binding site (Figs. 4, 5, and 7). Such a hydrogen bond pattern was also found in a number of HSDs (Tanaka et al., 1996) or other members of the SDR family, including DesIV.
Interactions of 4α-Carboxysterol with 3βHSD/D
A common characteristic feature of several members of the SDR family is to involve a catalytically important YX3K motif in the middle of the chain. There is a single 159YX3K163 motif in 3βHSD/D, and our model shows that the essential Tyr-159 residue interacts directly with the 3β-hydroxyl of the steroid substrate (Figs. 4, 5, and 7). These data are consistent with the conclusion that Tyr-159 acts as a general base to abstract the proton from the hydroxyl group of the steroid substrate in order to facilitate the hydride transfer that produces the 3-keto-C-4-carboxysteroid intermediate, as generally proposed for most of the SDRs characterized to date (Jörnvall et al., 1995; Oppermann et al., 2003; Wu et al., 2007). In addition, the model places the ε-NH2 group of the essential Lys-163 residue in close proximity to Tyr-159. One possible critical role of Lys-163 would be to lower the pKa of Tyr-159, as suggested before for various SDRs (Chen et al., 1993; Wu et al., 2007), in addition to its interaction with the 2′-hydroxyl group of the nicotinamide Rib.
3βHSD/D comprises 130 additional residues on the C-terminal end of the enzyme compared with most of other monofunctional HSD members of the SDR family, and it is reasonable to postulate that its dual action is linked to these additional residues. Ser-131 is a homolog to the essential Ser-124 of the mammalian 3βHSD (Thomas et al., 2004) and Ser-138 is a homolog of 3β,17βHSD (Filling et al., 2002), which have been shown to bind the targeted hydroxyl or oxo group of the substrate (Kallberg et al., 2002; Oppermann et al., 2003; Wu et al., 2007). The residual activity of the S131A mutation, which led to a complete loss of activity in several other SDRs, suggests that formation of the Arg-326-substrate carboxylate salt bridge can fulfill the stabilization of the substrate docking in the case of 3βHSD/D, in accord with the strict substrate specificity of 3βHSD/D that required a free C-4 carboxyl group (Rahier et al., 2006). Thus, Arg-326 is one of the crucial residues that gives 3βHSD/D its unique features. Moreover, by stabilizing the C-4 carboxyl group of the 3-keto C4-carboxysteroid intermediate in a deprotonated state, Arg-326 could facilitate its decarboxylation to produce the enol intermediate. Incidentally, the Arg-substrate carboxylate salt bridge is a critical constituent in several key enzymatic reactions and is involved, for example, in the binding of fatty acid substrate of peroxygenase P450 (Matsunaga et al., 2001) and malate substrate of malate dehydrogenase (Wright and Viola, 2001).
In this decarboxylation reaction, with the assistance of protonated Lys-163, Tyr-159 functions now as a general acid and donates a proton to the 3-keto group to produce the neutral enol. Taken together, these data suggest that Tyr-159 activated by Lys-163 acts as a general base in the dehydrogenation step and as a general acid during the subsequent decarboxylation step, thus acting as a proton shuttle between the C3 hydroxyl and itself.
The data show that an increased size of residue 133 alters the substrate specificity of the 3βHSD/D in favor of substrates devoid of the C4β-methyl group, in accord with the close steric interaction between Ser-133 and this group predicted by the model. A bulkier residue at position 133 probably prevents the C4β-methyl substituted substrate [3] from occupying the correct position for its reaction and leads to a low efficiency. Particularly, one can presume that placing a bigger amino acid at residue 133 will widen the distance between the catalytic Tyr-159 and the 3β-hydroxyl or 3-keto group of the substrate.
Although the catalytic efficiency of mutant S133T for 4α-carboxy,4β-methylsterol substrate [3] (0.0058 mL h−1 mg−1) was about 70-fold lower than that of the wild-type enzyme (0.43 mL h−1 mg−1; Table III), it was sufficient to partially complement the erg26 null mutant in vivo (Table I). These data are consistent with previous kinetic data (Rahier et al., 2006) showing that plant and yeast 3βHSD/Ds have higher apparent rates than other enzymes involved in the postsqualene sterol biosynthetic pathway (Bouvier et al., 2005).
In the final tautomerization step of the reaction, a proton is added to the C4β position of the enol intermediate and the enol hydroxyl is deprotonated. This enol intermediate could undergo a spontaneous keto-enol-tautomerization to yield the product. However, in accord with the model, we suggest that Tyr-159 could function with the assistance of Lys-163 as a general base for enol deprotonation. In addition, substitution of Ser-133, in proximity to the C4β domain of the enol, by an aprotic Ala residue had minimal effects on 3βHSD/D kinetic parameters, suggesting that a water molecule is probably recruited into the active site of 3βHSD/D and involved in enol protonation at C4β.
Conservation of Active Site Residues across 3βHSD/D Phylogeny
Alignment of the Arabidopsis isoform 1 of 3βHSD/D studied herein with the previously identified Arabidopsis 3βHSD/D isoform 2 (3βHSD/D2; DQ302749) and with 3βHSD/Ds from animals and yeast (Rahier et al., 2006) indicated that residues forming the hydrogen bond pattern between the NAD+ cofactor and the 3βHSD/D surface, as well as catalytic residues Tyr-159 and Lys-163, were conserved across 3βHSD/D phylogeny. These data fit with the rather conserved NAD+-binding mode and dehydrogenation process in SDRs (Jörnvall et al., 1995). In addition, amino acid residues that interact with the carboxysterol substrate nucleus inside the active site cavity also generally remain conserved or similar across the 3βHSD/D phylogeny. In particular, Arg-326 is conserved across all 3βHSD/Ds from plants and fungi and is replaced by a His residue in all animal NSDHL orthologs. The pKa of His is likely to rise upon interaction with the substrate carboxyl group during binding (Anderson et al., 1990), and the acid form, the imidazolium ion, is able to interact with the substrate by forming a H-bonded ion pair with its carboxyl group. Stabilizing hydrogen-bonded ion pairing between His and Asp residues has been shown to be critical in several enzymatic reactions (Sundaramoorthy et al., 1998; Chen et al., 2004; Tu et al., 2006). Finally, sequence alignments show that aromatic and hydrophobic amino acid residues that are confining the steroid nucleus and side chain in the binding hydrophobic cavity remain conserved or similar across the 3βHSD/D phylogeny (Rahier et al., 2006).
Catalytic Mechanism
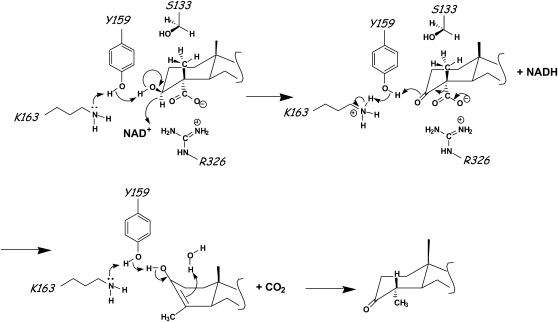
A mechanism consistent with our data is suggested in Figure 8. In the first step catalyzed by 3βHSD/D, Arg-326 stabilizes the substrate while Tyr-159 functions as the catalytic base, and Lys-163 lowers the pKa of the Tyr-OH to promote proton transfer during the dehydrogenation of the carboxysterol to produce the corresponding ketone. In the decarboxylation of the 3-oxo acid to produce the enol, Tyr-159 donates a proton to the 3-keto group while Arg-326 maintains the carboxyl group in a deprotonated state. Tautomerization of the enol to produce the 3-keto-C4-decarboxylated product is possibly helped by extraction of the proton from the C3-hydroxyl by Tyr-159 and addition of a proton on the β-face at C4 from a water molecule. During the course of C4-demethylation of 4,4-dimethysterols, the 4α-methyl group is first oxidized by the SMO to yield a carboxyl; this is decarboxylated, and the intermediate enol is protonated on the β-face, presenting to the oxygenase another 4α-methyl for the same treatment.
Figure 8.
Proposed reaction mechanism of 3βHSD/D. Tyr-159, Lys-163, and Arg-326 are proposed to be involved in the dehydrogenation and decarboxylation of 4α-carboxysterol with NAD+. 4α-Carboxysterol binding is stabilized by Arg-326. Tyr-159 acts as the general base to abstract the proton of the hydroxyl group of steroid, facilitates the hydride transfer to NAD+, and acts as the general acid to deliver a proton to the 3-keto group, and together with Arg-326 it facilitates the decarboxylation of the carboxysteroid. Tyr-159 and a water molecule probably help in the final tautomerization step.
Outlook on the Essential Role of the Sterol C4-Demethylation Step in Plants
Phytosterols play important roles in plants: as structural molecules in membranes (Hartmann, 1998), as plant growth and development factors (Clouse, 2002), and as hormone signaling molecules (Lindsey et al., 2003; Men et al., 2008). For all of these roles, the elimination of the two methyl groups at C4 is a central feature of the biosynthesis and biological function of sterols. The C4-demethylation involves, sequentially, the tight cooperation of three individual enzymes, including 3βHSD/D, which proceeds without accumulation of the transient intermediate products. Several enzymes of the C4-demethylation step control carbon flow to sterol end products (Maillot-Vernier et al., 1991; Holmberg et al., 2003).
It has been suggested that sterols other than brassinolides may serve as novel signals controlling cell fate during plant embryogenesis (Clouse, 2002; Lindsey et al., 2003). Indeed, in plants, sterol biosynthetic mutants upstream of the last biosynthetic intermediate possessing one C4-methyl group (i.e. 24-methylenelophenol) show embryonic defects but those downstream do not. This suggests that sterol C4-demethylation is a crucial process in brassinosteroid-independent plant development. Therefore, knowledge of the active site of 3βHSD/D may help to investigate specific roles of 4,4-dialkylated and 4α-monoalkylated sterol precursors. In animals, a sterol-binding transcription factor (so-called START domain proteins) that controls stage-specific events in embryogenesis has been characterized (Kallen et al., 1998; Ponting and Aravind, 1999). In plants, recent data suggest that the majority of START domain proteins belong to a novel class of putative lipid/sterol-binding transcription factors designated homeodomain-START proteins (Schrick et al., 2004). As yet, plant START proteins have not been shown to bind sterols. It is imaginable that upstream-specific plant C4-methylated sterols, including cycloartenol and its derivatives and transient 4α-carboxysterol and 3-ketosteroid intermediates of the C4-demethylation process, might serve as possible ligands for such proteins and be involved in plant development. At present, we can only speculate.
The present study based on Arabidopsis 3βHSD/D provides the first data, to our knowledge, on the 3D molecular interactions of a sterol biosynthetic enzyme with its substrate in the steps downstream from oxidosqualene cyclase. The homology model can be used to target other key amino acids responsible for coenzyme and steroid binding, membrane interaction, and interactions with other components of the C4-demethylation complex. These data could give clues for the design of novel and selective structure-based inhibitors. In vivo, such inhibitors should offer the possibility to phenocopy not yet identified plant 3βHSD/D mutants.
MATERIALS AND METHODS
Materials
All materials were purchased from Sigma unless otherwise specified.
Strains and Plasmids
The erg26 SDG200 strain of Saccharomyces cerevisiae, deficient in 3βHSD/D activity (Mat a ade5, his3, leu2-3, ura3-52, erg26Δ∷TRP1 trp1∷hisG Δhem1), used in the present study has been described previously (Gachotte et al., 1998). Sterol auxotrophs were grown aerobically at 30°C on solid enriched medium (1% yeast extract, 2% peptone, and 2% Glc) supplemented with 2% ergosterol or cholesterol dissolved in ethanol-Tween 80 (1:1, v/v) or on minimal medium (yeast nitrogen base [YNB]; 0.67% YNB and 2% Glc) containing suitable supplements (50 mg L−1 each), casamino acids (1 g L−1), and 2% ergosterol or cholesterol. In the case of liquid medium, the concentration of sterol used was 0.5%. Sterol prototrophic strains were grown aerobically at 30°C on solid or liquid YNB medium containing suitable supplements (50 mg L−1 each) or enriched medium (yeast extract, peptone, Glc) in the presence of δ-ala (50 mg L−1).
The pVT102U (Vernet et al., 1987) S. cerevisiae shuttle vector optimized for expressing recombinant proteins in yeast was used for cloning, sequencing, and transformation of the erg26 strain. This plasmid contains an Escherichia coli origin of replication, a yeast 2μ origin of replication, an E. coli ampicillin resistance gene, and the yeast URA3 gene. It contains an expression cassette including the alcohol dehydrogenase promoter and terminator.
Arabidopsis FLAG 3βHSD/D
The 3βHSD/D cDNA (AY957470) used in the present work has been described previously (Rahier et al., 2006). In order to check the expression of wild-type and mutated Arabidopsis (Arabidopsis thaliana) 3βHSD/D in the erg26 yeast strain, an N-terminal FLAG epitope (MDYKDDDDK) was fused to the 3βHSD/D protein. For this purpose, a DNA molecule containing at the 5′ end the corresponding nucleotide sequence was synthesized by PCR using the following forward primer (P1, 5′-ATAATATCTAGAATGGACTACAAGGACGACGATGACAAGGAAGTTACAGAGACTGAGCATG-3′) containing an XbaI site and the reverse primer (P2, 5′-ATAATACTCGAGTTAGTCGATCTTCTTGCTCCCGAACACTTTC-3′) containing an XhoI site using the 3βHSD/D cDNA as a template. The AtFLAG3βHSD/D was further cloned into the XbaI and XhoI sites of the pVT102U vector to generate pVT-AtFLAG3βHSD/D.
Homology Modeling and Sequence Alignment
BLAST was used to search for the structure of the closest 3βHSD/D paralog, with default settings against the National Center for Biotechnology Information database of nonredundant protein sequences (Berman et al., 2000). Only two protein data bank (pdb) sequences appeared among the 1,000 first hits: 1R66 and 1R6D. They correspond to crystal structures of DesIV from Streptomyces venezuelae with bound NAD+ and thymidine-5′-diphosphate for the first one and to a corresponding double mutant with NAD+ and 2′-deoxy-thymidine-5′-diphospho-α-d-Glc (DAU) bound for the second (Allard et al., 2004). The quality of the 1R66 hit is indicated by a score of 82 bits, an E-value of 7e-14, 24% identity, and 40% similarity out of 376 positions in the alignment. A control BLAST against the National Center for Biotechnology Information database pdb sequences identifies 2HUN, the crystal structure of hypothetical protein Ph0414 from Pyrococcus horikoshii Ot3, as the next hit to 1R6D, but with an E-value greater by 5 orders of magnitude. Moreover 3βHSD/D (42 kD) and DesIV (37 kD) share a rather bigger size than other HSDs and have strict NAD+ cofactor specificity (Rondet et al., 1999). 1R66 was thus chosen as the target for homology modeling, although it lacks a small C-terminal fragment of 15 residues (323ATAPQLPATAVEVSA337).
A sequence alignment was calculated with ClustalX and manually modified before 3D modeling with Modeller version 8.2 (Sali and Blundell, 1993). This alignment is shown in Figure 3 annotated with conserved residues and with the secondary elements defined in the 1R66 pdb file. A 3D model of the 3βHSD/D protein was built with Modeller using standard settings. NAD+ was modeled as a rigid structure, while [1] (Fig. 2) was first constructed with O (Jones et al., 1991), starting from cycloartenol coordinates provided by the Cambridge Crystallographic Data Center (entry GAGQAG; Nes et al., 1998), superimposed onto DAU in the target structure, and finally used as a rigid body in Modeller. The superposition was done in order to overlap the 4′-hydroxyl group of DAU with the 3β-hydroxyl group of the sterol substrate [1], the d-Glc cycle of DAU with the A cycle of [1], and the 6′-hydroxyl group of DAU with the 4α-carboxyl group of [1].
Visual analysis and manipulation of the model were done with O and PyMOL (DeLano, 2002), which was also used for illustrations.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed using the Quickchange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. PVT-AtFLAG3βHSD/D was used as a template, and the synthetic oligonucleotide primers are listed in Supplemental Table S1. Putative positive clones were picked, and plasmids were isolated and sequenced.
Transformations
S. cerevisiae transformations were performed using the lithium acetate procedure as described previously (Gietz et al., 1992). The transformed erg26 yeast strain was plated on minimal YNB medium containing suitable supplements (adenine, 50 μg mL−1) without uracil and 2% ergosterol as well as on the same minimal YNB medium without uracil but containing δ-ala (50 mg L−1). Cells were grown aerobically at 30°C.
Sterol Analysis
Lyophilized yeast cells (10–30 mg) were extracted, and sterols were analyzed as described previously (Rahier et al., 2006). Sterols were unequivocally identified by retention times and an electron-impact spectrum identical to that of authentic standards (Rahier and Benveniste, 1989).
Substrates
4α-Carboxy-cholest-7-en-3β-ol [2] was synthesized as described previously (Rondet et al., 1999). Melting point 231°C to 233°C. MS mass-to-charge ratio (relative intensity) M+ = 430 (36), 412 (10), 397 (5), 386 (100), 317 (2), 299 (19), 273 (19), 271 (19), 255 (63), 229 (22), 213 (18). 1H NMR: δ: 0.533 (3H, s, H18), 0.844 (3H, s, H19),0.862 (3H, d, J = 6.6, H26 or 27), 0.867 (3H, d, J = 6.6, H26 or 27), 0.918 (3H, d, J = 6.5, H21), 2.030 (3H, dt, J = 11, J = 4, H6β), 2.364 (1H, dd, J = 11, H4β), 3.809 (1H, m, H3α), 5.131 (1H, S, ω1/2 = 10, H7).
4α-Carboxy-4β-methyl-cholest-8,24-dien-3β-ol [3] was synthesized as described previously (Rahier et al., 2006). Melting point 227°C to 229°C. MS of the 4α-carbomethoxy derivative of [3]: mass-to-charge ratio (relative intensity) M+ = 456 (100), 441 (39), 438 (17), 423 (17), 396 (17), 363 (22), 285 (22), 225 (15). 1H NMR of [3]: δ: 0.588 (3H, s, H18), 0.939 (3H, d, J = 6.4, H21), 1.008 (3H, s, H19), 1.176 (3H, s, H4β), 1.600 (3H, s, H26), 1.680 (3H, s, H27), 4.021 (1H, dd, J = 16, J = 7.5, H3α), 5.090 (1H, tt, J = 7.1, J = 1.3, H24).
Standard Assay for Recombinant 3βHSD/Ds
Yeast microsomes were prepared as described previously (Darnet and Rahier, 2003). The corresponding 100,000g cytosolic supernatants were concentrated 5- to 8-fold by dialysis over carboxymethylcellulose sodium salt (Fluka) for 16 h at 4°C. Yeast microsomes (0.8 mg of protein) were incubated in the presence of exogenous synthetic [2] and NAD+ as described previously (Rahier et al., 2006). The incubations were extracted as described in detail previously (Rahier et al., 2006). Briefly, the extract was analyzed by thin-layer chromatography on silica gel eluted with CH2Cl2 (developed twice). The fraction migrating as authentic standards of coprostanone and cholest-7-en-3-one and containing the enzymatically produced cholest-7-en-3-one (RF = 0.50) was eluted and analyzed by GC (DB1). The amount of cholest-7-en-3-one enzymatically produced (relative retention time to cholesterol in GC [tR] = 1.074) was calculated by comparison of the integrated peak areas with a known amount (2–6 μg) of coprostanone (tR = 1.000) added before extraction and allowed the rate of transformation of [2] to be determined. No endogenous component having the same tR as cholest-7-en-3-one was present in the blank (zero substrate). Under these conditions, the estimated limit of detection of the 3βHSD/D activity was 0.1 nmol h−1 mg−1. Incubation of [3] was performed using the same standard conditions and analyzed by GC as described previously (Rahier et al., 2006). Briefly, after addition of coprostanone (2–6 μg) as an internal standard, extraction, and thin-layer chromatography analysis, the fraction migrating between a standard of 4-desmethyl-sterone (RF = 0.50) and 4,4-dimethyl-sterone (RF = 0.70) was eluted and analyzed by GC and GC-MS. The areas of the GC peaks of coprostanone and of the produced 4α-methyl-cholest-8,24-dien-3-one, corrected from an endogenous component of the same tR (1.141) determined in the blank (zero substrate), allowed the rate of transformation of [3] to be measured.
For incubation of [2] and [3] with inactive 3βHSD/D mutants, comparison with the corresponding controls performed with inactivated microsomes revealed the absence of C3-dehydrogenated product and complete recovery of the untransformed substrates, which was confirmed by ion monitoring that corresponded to the masses of the substrate and expected dehydrogenated product.
Apparent Michaelis-Menten kinetic constants for the 3βHSD/D substrates were determined for the mutated and wild-type enzymes by incubating them under standard assay conditions described above and previously (Rahier et al., 2006). For double reciprocal plots (Supplemental Fig. S1), computer-assisted linear regression analysis gave correlation coefficients greater than 0.89 (n = 5–6). Apparent Vmax and Km values were determined by fitting the data to the Michaelis-Menten equation using the nonlinear regression program DNRP-EASY derived by Duggleby (1984) from DNRP53.
Expression and Accumulation of AtFLAG3βHSD/Ds in Microsomal Extracts
Membrane and soluble proteins were quantified using the Bio-Rad protein assay according to Bradford (1976). We checked protein expression in the microsomal extracts from the wild-type and 3βHSD/D mutated cDNAs using the corresponding 3βHSD/D proteins fused to an N-terminal FLAG epitope (AtFLAG3βHSD/D).
Western blots of microsomes (40 μg of protein) or of the 100,000g supernatants from AtFLAG3βHSD/D transformed yeast were achieved after separation of proteins on SDS-14% (w/v) polyacrylamide gels. After electrophoretic transfer to a polyvinylidene difluoride Immobilon P membrane (Millipore), AtFLAG3βHSD/Ds were immunoblotted with affinity-purified murine monoclonal anti-FLAG M2 antibodies from Sigma (1:6,000 dilution) according to the manufacturer's instructions. Goat anti-mouse IgG-alkaline phosphatase conjugate (Bio-Rad) was used as a secondary antibody (1:10,000 dilution). The membrane was then treated with the chemiluminescent AP substrate, and the blot was further used to expose an instant film for detection.
As shown by the immunoblots in Figure 6, there is evidence that all mutated and wild-type AtFLAG3βHSD/D proteins did accumulate in yeast microsomal extracts, producing a discrete band with the expected Mr. In addition, the expression of the wild-type and mutated 3βHSD/Ds was not detectable in the corresponding soluble fractions (data not shown). Immunoblot band relative intensities were quantified by scanning digital camera images of the gels and assigning to individual bands an intensity value relative to the wild type using Un-Scan-It software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY957470.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Double-reciprocal plots for recombinant At3βHSD/D activities.
Supplemental Table S1. Sequence of the synthetic oligonucleotides used for site-directed mutagenesis of cDNA of the Arabidopsis 3βHSD/D gene.
Supplementary Material
Acknowledgments
We are indebted to Dr. Martin Bard (Indiana University) for providing the yeast mutant erg26. We acknowledge Pr. Peter Buist (Carleton University, Ottawa, Canada) for help in editing the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alain Rahier (alain.rahier@ibmp-ulp.u-strasbg.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Allard ST, Cleland WW, Holden HM (2004) High resolution X-ray structure of dTDP-glucose 4,6-dehydratase from Streptomyces venezuelae. J Biol Chem 279 2211–2220 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Becktel WJ, Dahlquist FW (1990) pH-induced denaturation of proteins: a single salt bridge contributes 3-5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry 29 2403–2408 [DOI] [PubMed] [Google Scholar]
- Bard M, Bruner PA, Pierson CA, Lees ND, Biermann B, Frye L, Koegel C, Barbuch R (1996) Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA 93 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Physiol Plant Mol Biol 55 429–457 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci USA 77 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt D, König A, Happle R, Leveleki L, Bittar M, Danarti R, Vahlquist A, Tilgen W, Reinhold U, Poiares Baptisita A, et al (2005) Mutational spectrum of NSDHL in CHILD syndrome. J Med Genet 42 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44 357–429 [DOI] [PubMed] [Google Scholar]
- Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Chen HP, Lung FD, Yeh CC, Chen HL, Wu SH (2004) The role of the conserved histidine-aspartate pair in the ‘base-off’ binding of cobalamins. Bioorg Med Chem 12 577–582 [DOI] [PubMed] [Google Scholar]
- Chen Z, Jiang JC, Lin ZG, Lee WR, Baker ME, Chang SH (1993) Site-specific mutagenesis of Drosophila alcohol dehydrogenase: evidence for involvement of tyrosine-152 and lysine-156 in catalysis. Biochemistry 32 3342–3346 [DOI] [PubMed] [Google Scholar]
- Chen Z, Lee WR, Chang SH (1991) Role of aspartic acid 38 in the cofactor specificity of Drosophila alcohol dehydrogenase. Eur J Biochem 202 263–267 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002) Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2003) Enzymological properties of sterol-C4-methyl-oxidase of yeast sterol biosynthesis. Biochim Biophys Acta 1633 106–117 [DOI] [PubMed] [Google Scholar]
- Darnet S, Rahier A (2004) Plant sterol biosynthesis: identification of two distinct families of sterol-4α-methyl oxidases. Biochem J 378 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA
- Duax WL, Pletvev V, Addlagatta A, Bruenn J, Weeks CM (2003) Rational proteomics. I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family. Proteins 53 931–943 [DOI] [PubMed] [Google Scholar]
- Duggleby RG (1984) Regression analysis of nonlinear plots: an empirical model and a computer program. Comput Biol Med 14 447–455 [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277 25677–25684 [DOI] [PubMed] [Google Scholar]
- Gachotte D, Barbuch R, Gaylor J, Nickels E, Bard M (1998) Characterization of the Saccharomyces cerevisiae ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) involved in sterol biosynthesis. Proc Natl Acad Sci USA 95 13794–13799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Brandt W, Ziegler J (2007) Molecular modeling and site-directed mutagenesis reveal the benzylisoquiloline binding site of the short-chain dehydrogenase/reductase salutaridine reductase. Plant Physiol 143 1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Sawicki M, Pletnev V, Erman M, Ohno S, Nakajin S, Duax WL (2001) Porcine carbonyl reductase: structural basis for a functional monomer in short chain dehydrogenases/reductases. J Biol Chem 276 18457–18463 [DOI] [PubMed] [Google Scholar]
- Ghosh D, Wawrzak Z, Weeks CM, Duax WL, Erman M (1994) The refined three-dimensional structure of 3α,20β-hydroxysteroid dehydrogenase and possible roles of the residues conserved in short-chain dehydrogenases. Structure 2 629–640 [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann MA (1998) Plant sterols and the membrane environment. Trends Plant Sci 3 170–175 [Google Scholar]
- Holmberg N, Harker M, Wallace AD, Clayton JC, Gibbard CL, Safford R (2003) Co-expression of N-terminal truncated 3-hydroxy-3-methylglutaryl CoA reductase and C24-sterol methyltransferase type 1 in transgenic tobacco enhances carbon flux towards end-product sterols. Plant J 36 12–20 [DOI] [PubMed] [Google Scholar]
- Huang YW, Pineau I, Chang HJ, Azzi A, Bellemare V, Laberge S, Lin SX (2001) Critical residues for the specificity of cofactors and substrates in human estrogenic 17β-hydroxysteroid dehydrogenase 1: variants designed from the three-dimensional structure of the enzyme. Mol Endocrinol 15 2010–2020 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 110–119 [DOI] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàles-Duarte R, Jeffery J, Ghosh D (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34 6003–6013 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B (2002) Short-chain dehydrogenases/reductases (SDRs). Eur J Biochem 269 4409–4417 [DOI] [PubMed] [Google Scholar]
- Kallen CB, Billheimer JT, Summers SA, Stayrook SE, Lewis M, Strauss JF III (1998) Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein. J Biol Chem 273 26285–26288 [DOI] [PubMed] [Google Scholar]
- Koehl P, Levitt M (1999) A brighter future for protein structure prediction. Nat Struct Biol 6 108–111 [DOI] [PubMed] [Google Scholar]
- König A, Happle R, Bornholdt D, Engel H, Grzeschik KH (2000) Mutations in the NSDHL gene, encoding a 3β-hydroxysteroid dehydrogenase, cause CHILD syndrome. Am J Med Genet 90 339–346 [PubMed] [Google Scholar]
- Lees ND, Skaggs B, Kirsch DR, Bard M (1995) Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae. Lipids 30 221–226 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Pullen ML, Topping JF (2003) Importance of plant sterols in pattern formation and hormone signalling. Trends Plant Sci 8 521–525 [DOI] [PubMed] [Google Scholar]
- Liu XY, Dangel AW, Kelley RI, Zhao W, Denny P, Botcherby M, Cattanach B, Peters J, Hunsicker PP, Mallon AM, et al (1999) The gene mutated in bare patches and striated mice encodes a novel 3β-hydroxysteroid dehydrogenase. Nat Genet 22 182–187 [DOI] [PubMed] [Google Scholar]
- Maillot-Vernier P, Gondet L, Schaller H, Benveniste P, Belliard G (1991) Genetic study and further biochemical characterization of a tobacco mutant that overproduces sterols. Mol Gen Genet 231 33–40 [DOI] [PubMed] [Google Scholar]
- Matsunaga I, Ueda A, Sumimoto T, Ichihara K, Ayata M, Ogura H (2001) Site-directed mutagenesis of the putative distal helix of peroxygenase cytochrome P450. Arch Biochem Biophys 394 45–53 [DOI] [PubMed] [Google Scholar]
- Men S, Boutté Y, Ikeda Y, Li X, Palme K, Stierhof YD, Hartmann MA, Moritz T, Grebe M (2008) Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat Cell Biol 10 237–244 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Kato H, Oda J, Yamada Y, Hashimoto T (1999) Site-directed mutagenesis of putative substrate-binding residues reveals a mechanism controlling the different stereospecificities of two tropinone reductases. J Biol Chem 274 16563–16568 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yamashita A, Akama H, Nakatsu T, Kato H, Hashimoto T, Oda J, Yamada Y (1998) Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold. Proc Natl Acad Sci USA 95 4876–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WD, Koike K, Jia Z, Sakamoto Y, Satou T, Nikaido T, Griffin JF (1998) 9,19-Cyclosterol analysis by 1H and 13C NMR, crystallographic observations, and molecular mechanics calculations. J Am Chem Soc 120 5970–5980 [Google Scholar]
- Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordlin E, Kallberg Y, Persson B, et al (2003) Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem Biol Interact 143-144 247–253 [DOI] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Imberty A, Olsen CE, Moller BL (2008) Catalytic key amino acids and UDP sugar donor specificity of a plant glucuronosyl transferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol 148 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal S, Taton M, Rahier A (1993) Plant sterol biosynthesis: identification and characterization of two distinct microsomal oxidative enzymatic systems involved in sterol C4-demethylation. J Biol Chem 268 11639–11654 [PubMed] [Google Scholar]
- Pascal S, Taton M, Rahier A (1994) Plant sterol biosynthesis: identification of a NADPH dependent sterone reductase involved in sterol-4 demethylation. Arch Biochem Biophys 312 260–271 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 24 130–132 [DOI] [PubMed] [Google Scholar]
- Rahier A, Benveniste P (1989) Mass spectral identification of phytosterols. In WD Nes, E Parish, eds, Analysis of Sterols and Other Significant Steroids. Academic Press, New York, pp 223–250
- Rahier A, Darnet S, Bouvier F, Camara B, Bard M (2006) Molecular and enzymatic characterizations of novel bifunctional 3β-hydroxysteroid-dehydrogenases/C4-decarboxylases from Arabidopsis thaliana. J Biol Chem 281 27264–27277 [DOI] [PubMed] [Google Scholar]
- Rondet S, Taton M, Rahier A (1999) Identification, characterization, and partial purification of 4α-carboxysterol-C3-dehydrogenase/C4-decarboxylase from Zea mays. Arch Biochem Biophys 366 249–260 [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234 779–815 [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF (2004) START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 5 R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy M, Terner J, Poulos TL (1998) Stereochemistry of the chloroperoxidase active site: crystallographic and molecular-modeling studies. Chem Biol 5 461–473 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Nonaka T, Tanabe T, Yoshimoto T, Tsuru D, Mitsui Y (1996) Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35 7715–7730 [DOI] [PubMed] [Google Scholar]
- Thomas JL, Duax WL, Addlagatta A, Scaccia LA, Frizzell KA, Carloni SB (2004) Serine 124 completes the Tyr, Lys and Ser triad responsible for the catalysis of human type 1 3beta-hydroxysteroid dehydrogenase. J Mol Endocrinol 33 253–261 [DOI] [PubMed] [Google Scholar]
- Thorn A, Egerer-Sieber C, Jäger CM, Herl V, Müller-Uri F, Kreis W, Muller YA (2008) The crystal structure of progesterone 5β-reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. J Biol Chem 283 17260–17269 [DOI] [PubMed] [Google Scholar]
- Thorsøe KS, Bak S, Olsen CE, Imberty A, Breton C, Lindberg Møller B (2005) Determination of catalytic key amino acids and UDP sugar donor specificity of the cyanohydrin glycosyltransferase UGT85B1 from Sorghum bicolor: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol 139 664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu SL, Sughrue W, Britt RD, Lagarias JC (2006) A conserved histidine-aspartate pair is required for exovinyl reduction of biliverdin by a cyanobacterial phycocyanobilin:ferredoxin oxidoreductase. J Biol Chem 281 3127–3136 [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52 225–233 [DOI] [PubMed] [Google Scholar]
- Wright SK, Viola RE (2001) Alteration of the specificity of malate dehydrogenase by chemical modulation of an active site arginine. J Biol Chem 276 31151–31155 [DOI] [PubMed] [Google Scholar]
- Wu X, Lukacik P, Kavanagh KL, Oppermann U (2007) SDR-type human hydroxysteroid dehydrogenases involved in steroid hormone activation. Mol Cell Endocrinol 265–266 71–76 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.