Abstract
Myb transcription factors have been implicated in a wide variety of plant-specific processes, including secondary metabolism, cell shape determination, cell differentiation, and stress responses. Very recently, AtMyb41 from Arabidopsis (Arabidopsis thaliana) was described as a gene transcriptionally regulated in response to salinity, desiccation, cold, and abscisic acid. The corresponding transcription factor was suggested to control stress responses linked to cell wall modifications. In this work, we have characterized AtMyb41 further by subjecting independent AtMyb41-overexpressing lines to detailed transcriptome and metabolome analysis. Our molecular data indicate that AtMyb41 is involved in distinct cellular processes, including control of primary metabolism and negative regulation of short-term transcriptional responses to osmotic stress.
Myb transcription factors (TFs) are widespread in animals, plants, and fungi. They were first identified as oncogenes in animals, where their function was linked to control of the cell cycle (Ito et al., 2001). Most animal Myb TFs are characterized by three repeats of the Myb domain, R1R2R3, which is an imperfect helix-turn-helix repeat of 50 to 53 amino acids. In contrast, most plant Myb TFs are of the R2R3 type that features only two repeats of the Myb domain. This basic structural difference between kingdoms raised questions about specific roles of Myb TFs in plants. Over the past decade, characterization of plant Myb TFs has implicated them in a wide variety of plant-specific processes, including secondary metabolism, cell shape determination, cell differentiation, and stress responses. For example, overexpression of AtMyb75 (AtPAP1) and AtMyb90 (AtPAP2) resulted in the accumulation of anthocyanin (Borevitz et al., 2000; Xie et al., 2006), while overexpression of AtMyb28 enhanced Met-derived glucosinolate content of Arabidopsis (Arabidopsis thaliana; Gigolashvili et al., 2007). AtMyb66 (WEREWOLF) promotes cell differentiation to the nonhair fate (Lee and Schiefelbein, 1999). AtMyb23 and AtGl1 both promote initiation of leaf edges, and AtMyb23 also controls trichome branching (Kirik et al., 2005). A mutation in AtMyb26 impairs secondary wall thickening of endothecial cells in the anthers and leads to male sterility (Yang et al., 2007), and AtMyb46 has been linked to secondary cell wall biosynthesis (Zhong et al., 2007). Myb TFs have also been implicated in abiotic stress signaling pathways (Yamaguchi-Shinozaki and Shinozaki, 2005). For instance, AtPHR1 is a critical regulator of phosphorus stress responses (Rubio et al., 2001). AtMyb15 interacts with AtICE1 and negatively regulates the induction of cold-responsive genes (CBF regulon) independently of abscisic acid (ABA; Agarwal et al., 2006). AtMyb8 is required for basal freezing tolerance. Loss of AtMyb8 function also results in hypersensitivity to NaCl and increased expression of stress genes (Zhu et al., 2005). AtMyb2 mediates the ABA-dependent expression of salt- and dehydration-responsive genes and interacts with a calmodulin protein to enhance salt tolerance (Hoeren et al., 1998; Yoo et al., 2005). AtMyb60, AtMyb61, and AtMyb44 regulate stomatal aperture in response to drought stress (Cominelli et al., 2005; Liang et al., 2005; Jung et al., 2008).
Given the established stress elicitation of many Myb TFs (Yanhui et al., 2006), it is likely that other uncharacterized Myb TFs are also involved in plant responses to adverse environmental conditions. Very recently, the AtMyb41 gene was found to be transcriptionally regulated in response to salinity, desiccation, and cold as well as the endogenous plant hormone ABA (Cominelli et al., 2008). Overexpression of AtMyb41 in transgenic plants resulted in a dwarf phenotype due to alterations of cell expansion and cuticle integrity and increased their sensitivity to desiccation, suggesting that AtMyb41 may control stress responses linked to cell wall modifications (Cominelli et al., 2008). Here, we have characterized AtMyb41 further by subjecting independent AtMyb41-overexpressing lines to detailed transcriptome and metabolome analysis. Our molecular data indicate that AtMyb41 is involved in distinct cellular processes, including control of primary metabolism and negative regulation of short-term transcriptional responses to osmotic stress.
RESULTS
AtMyb41 Expression Is Induced by Salt and Osmotic Stress in an ABA-Dependent Manner
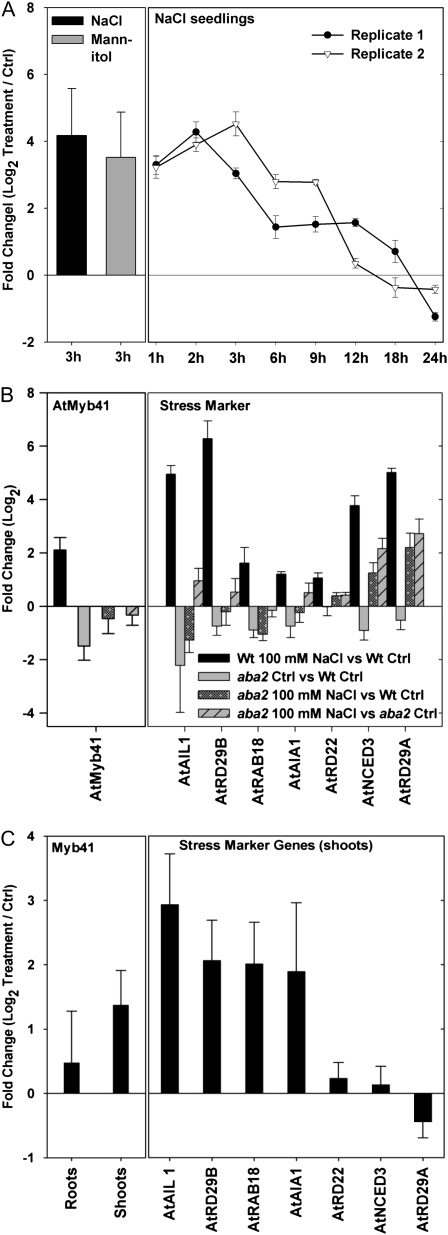
We identified AtMyb41 in a screen for salt shock- and osmotic shock-inducible TFs in 9-d-old Arabidopsis seedlings grown in sterile liquid culture using a high-throughput quantitative real-time reverse transcription (RT)-PCR resource for almost all known Arabidopsis TFs (Czechowski et al., 2004). AtMyb41 transcript levels in seedlings increased reproducibly after 3 h of 100 mm NaCl or 200 mm mannitol treatment (Fig. 1A). Subsequent analysis of the kinetics of AtMyb41 induction revealed a rapid increase in transcript levels within 1 h of NaCl addition, as described previously (Cominelli et al., 2008), but under our experimental conditions AtMyb41 peaked between 2 and 3 h and subsequently declined to basal, nonstressed control levels (Fig. 1A). Induction of AtMyb41 transcription was partially blocked in the aba2 mutant with reduced capacity to synthesize ABA (Fig. 1B). Similar results were obtained for a set of known stress marker genes, including AtRd29b and AtRab18, confirming an ABA-dependent regulation (Fig. 1B). NaCl induction of AtMyb41 expression was also observed in 4-week-old hydroponically grown plants acclimated to 50 mm NaCl (Fig. 1C), implicating AtMyb41 in long-term adjustment of plant cells to moderate osmotic stress in addition to more dramatic, short-term responses to osmotic shock (Fig. 1A). In line with a previous report (Cominelli et al., 2008), we observed that AtMyb41 expression was up-regulated by drought, cold, and ABA treatments (data not shown).
Figure 1.
A, Expression of AtMyb41 in 9-d-old seedlings after 3 h of treatment with 100 mm NaCl or 200 mm mannitol (left panel; data represent means ± sd of three independent replicates) and after 24 h of treatment with 100 mm NaCl (right panel; data from two independent experiments are shown separately, with error bars representing sem from three biological replicates). Ctrl, Control. B, Expression of AtMyb41 (left) and stress marker genes (right) after 3 h of 100 mm NaCl treatment in 9-d-old aba2 mutant and wild-type seedlings. Data represent means ± sd of three independent replicates. Black bars represent salt-stressed wild type (Wt) versus control wild type (Wt Ctrl); gray bars represent control aba2 versus control wild type; dotted black bars represent salt-stressed aba2 versus control wild type; striped gray bars represent salt-stressed aba2 versus control aba2. C, Expression of AtMyb41 (left) and stress marker genes (right) in hydroponically grown 4-week-old plants. Plants were treated with increasing salt concentrations, reaching 50 mm NaCl in the final week. Data represent means ± sd of three independent replicates. The Arabidopsis Genome Initiative codes of the genes are provided in Supplemental Table S4.
Ectopic Overexpression of AtMyb41 Retards Arabidopsis Growth
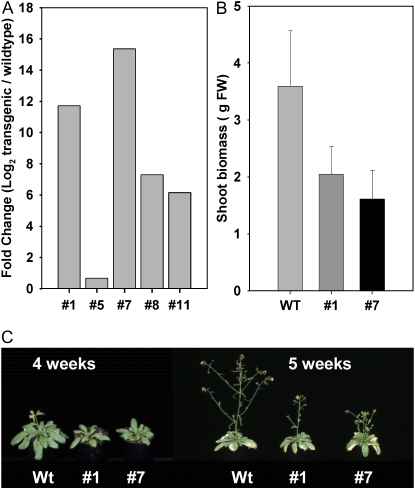
To investigate the role of AtMyb41 in plant response to stress, transgenic Arabidopsis lines were created using the constitutive promoter Cauliflower mosaic virus-35S to drive AtMyb41 expression. In the T3 generation, eight kanamycin-resistant lines were identified and AtMyb41 overexpression was confirmed by quantitative real-time RT-PCR in four of five putative single-insertion lines (Fig. 2A). Growth was not affected significantly in seedlings of AtMyb41 overexpressors (data not shown). However, prolonged overexpression of AtMyb41 inhibited growth (Fig. 2, B and C), albeit to a much lower extent than described previously for independent AtMyb41 transgenic lines (Cominelli et al., 2008). Despite the mild dwarf phenotype, no significant differences in relative growth rate were observed between AtMyb41 overexpressors and wild-type plants in vitro on salt-containing plates, in long-term salt acclimation experiments performed in hydroponic culture, or in soil-grown salt- or drought-stressed plants (data not shown).
Figure 2.
A, Analysis of transgene expression in AtMyb41-overexpressing lines using quantitative real-time RT-PCR. B, Growth of AtMyb41-overexpressing plants compared with wild-type plants, as estimated by shoot biomass. Data represent means ± sd of 45 plants from eight independent replicates, harvested before bolting. FW, Fresh weight; WT, wild type. C, AtMyb41-overexpressing plants from lines 1 and 7 compared with wild-type (Wt) plants at 4 and 5 weeks after sowing.
AtMyb41 Overexpression Deregulates a Subset of Salt-Induced Genes
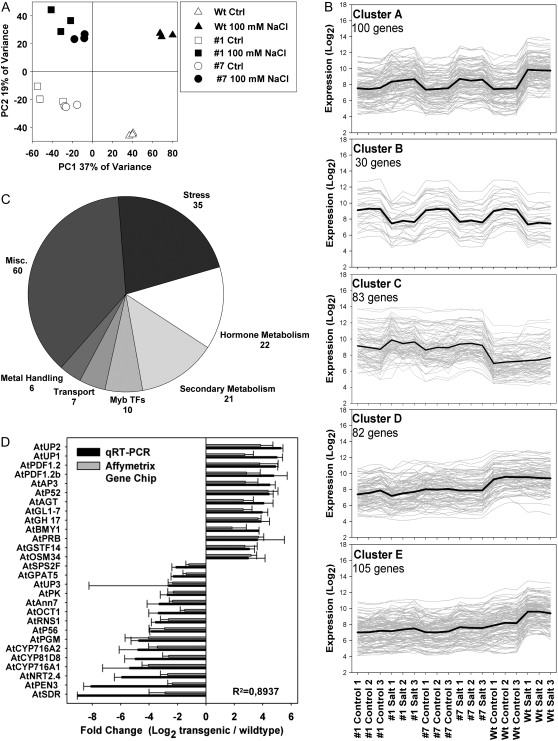
Rapid induction of AtMyb41 expression in response to osmotic and salt stress indicated a possible role for AtMyb41 in immediate/early responses of seedlings to osmotic stress. To identify potential targets of AtMyb41 action as a step toward defining its physiological role, transcriptome analysis of the two most highly overexpressing AtMyb41 lines, lines 1 and 7, was performed using the Affymetrix GeneChip Arabidopsis ATH1 Genome Array. Three independent experiments were performed with seedlings of the transgenic lines and wild-type plants grown in vitro under control conditions and after treatment with 100 mm NaCl for 3 h. For a global overview, gene expression data were analyzed first via unsupervised principal component analysis, which demonstrated good reproducibility between independent experiments (Fig. 3A). Principal component 1 (PC1), representing 37% of the variance within the data set, separated the transgenic lines from wild-type seedlings, while PC2, encompassing 19% of the variance, separated stressed plants from controls (Fig. 3A). To assess differential expression among the genotypes and the treatments, data were further analyzed by a significance-based comparison applying a false discovery rate (FDR)-corrected false positive rate (type I error) of 5% (FDR P < 0.05; Benjamini and Hochberg, 1995) and a threshold >1.5-fold change (log2 scale). Comparisons were made between genotypes for a given treatment or between treatments for a given genotype (Supplemental Table S1). Genes differentially expressed between genotypes and/or treatments were clustered with the K-means method, revealing five groups with broad differential patterns of expression (Fig. 3B). Clusters A and B represented salt-induced and salt-repressed genes, respectively, regardless of the genotype, many of which are known to be stress responsive like AtRd29a, AtCor15a, and AtKin1 (Supplemental Table S1). The other three clusters included genotype-specific genes. We found transcripts induced (cluster C) or repressed (cluster D) in the transgenics that did not respond to salt treatment, indicating stress-unrelated roles for AtMyb41 or reflecting pleiotropic effects of AtMyb41 overexpression. Most interesting, cluster E contained genes that were salt induced in the wild type but not in AtMyb41-overexpressing plants, indicating a possible role for AtMyb41 in feedback repression of these genes. This cluster contained a number of known stress-responsive genes, including some late embryogenesis abundant genes, AtDreb2a and AtNced3.
Figure 3.
A, Principal component analysis of the transcriptomic profiles obtained from three independent biological replicates of each genotype and treatment. The first two principal components are shown, which constitute the major variation of the data set. Ctrl, Control; Wt, wild type. B, K-means clustering of the genes declared to be changed (FDR P < 0.05, >1.5-fold changed in log2 scale) in AtMyb41 overexpressors versus the wild type and treatment versus control. Cluster A, Salt-induced genes; cluster B, salt-repressed genes; cluster C, genes up-regulated in the transgenic lines; cluster D, genes down-regulated in the transgenic lines; cluster E, salt-induced genes in the wild type but not in the transgenic lines. C, PageMan statistical overrepresentation analysis of genes significantly changed in expression due to AtMyb41 overexpression (genes only responding to salt stress were excluded). D, Comparison of microarray and quantitative real-time RT-PCR (qRT-PCR) data (data represent means ± sd of two overexpressing lines under control conditions in three independent experiments) of selected genes found significantly changed in the transcriptomic profiles. The Arabidopsis Genome Initiative codes of the genes are provided in Supplemental Table S4.
To determine the broad functions of genes that may be controlled by AtMyb41, transcripts that were differentially expressed between AtMyb41 overexpressors and the wild type under both stress and nonstress conditions (FDR P < 0.05, >1.5-fold change on a log2 scale) were collectively subjected to statistical overrepresentation analysis using PageMan software (Usadel et al., 2006). Overrepresented functional groups included large enzyme families, stress responses, hormone metabolism, secondary metabolism, Myb TFs, transport, and metal handling (Fig. 3C). This analysis indicated that AtMyb41 controls, directly or indirectly, a panoply of genes involved in multiple cellular processes.
Quantitative real-time RT-PCR was used to confirm the results of the microarray studies for the most strongly up- and down-regulated genes (Fig. 3D). All of the genes tested were confirmed to be either induced or repressed in AtMyb41 overexpressors compared with the wild type. In many cases, the magnitude of changes calculated from quantitative real-time RT-PCR data were greater than from array data, as expected, but the correlation between technologies was remarkably good (linear regression r2 = 0.8937).
AtMyb41 Overexpression Alters Primary Metabolism in Seedlings
Metabolite profiling of seedlings, using gas chromatography coupled to electron impact ionization-time of flight-mass spectrometry (GC/EI-TOF-MS), was carried out to determine the extent to which AtMyb41-induced changes in gene expression altered plant metabolism. A total of 92 mass spectral tags (MSTs; Desbrosses et al., 2005) were recognized, and 60 of these were ascribed to known chemical compounds (Supplemental Table S2). Data were analyzed first using unsupervised independent component analysis (ICA), which revealed marked differences between genotypes (transgenic versus wild type) but not between the treatments (control versus NaCl; Fig. 4A). Further statistical analysis using ANOVA also demonstrated significant changes in metabolite pools between genotypes but only minor changes between treatments. Sugars, amino acids, and some organic acids were changed in the AtMyb41 overexpressors relative to wild-type seedlings (Table I). Differences due to NaCl treatment were small, and only six metabolites, mostly sugars, accumulated in wild-type and transgenic lines in response to salt shock (Table II). Similar results in other plant species implicate glycolysis and Suc metabolism in evolutionarily conserved responses to short-term osmotic stress (Sanchez et al., 2008b). Only Glc and Gal accumulated under short-term osmotic stress in the wild type and constitutively in AtMyb41 overexpressors (Tables I and II). Taken together, these results point to a role of AtMyb41 in the control of primary metabolism.
Figure 4.
Unsupervised ICA of metabolomic profiles. A, Wild-type (Wt) and AtMyb41-overexpressing seedlings under control conditions (Ctrl; black symbols) and after 3 h of 100 mm NaCl treatment (red symbols). Triangles represent the wild type; squares represent line 1; and circles represent line 7. B, Wild-type and AtMyb41-overexpressing adult soil-grown plants under different stress conditions. Triangles represent the wild type; squares represent line 1; circles represent line 7; black represents control; red represents NaCl; orange represents ABA; blue represents cold; and gray represents drought.
Table I.
Significantly changed metabolites due to AtMyb41 overexpression in lines 1 and 7 (ANOVA P < 0.01)
Data represent means from three independent experiments. To aid comprehension, the metabolite pool sizes in the overexpressors were averaged and expressed as ratios of wild-type seedlings under control or 100 mm NaCl conditions. The chemical identities of metabolites shown in brackets could not be accurately ascertained. The unknowns are unidentified metabolites that have been detected before and are denoted by Golm Metabolite Database code (Kopka et al., 2005).
| Type | Metabolite | Lines 1 and 7 Control/Wild-Type Control | Lines 1 and 7 NaCl/Wild-Type NaCl |
|---|---|---|---|
| Amino acids | Pyroglutamic acid | 5.40 | 5.68 |
| Ala | 5.16 | 5.40 | |
| Glu | 3.87 | 4.16 | |
| Asp | 2.97 | 3.06 | |
| Phe | 2.78 | 2.50 | |
| Thr | 2.22 | 2.53 | |
| Gly | 2.08 | 2.58 | |
| Se | 1.93 | 2.18 | |
| [Orn lactam] | 0.57 | 0.75 | |
| Sugars | Maltose | 1.98 | 2.92 |
| Xyl | 1.97 | 2.12 | |
| Rha | 1.81 | 3.68 | |
| 1,6-Anhydro-β-d-Glc | 1.57 | 2.47 | |
| Glc-6-P | 1.46 | 1.77 | |
| Gal | 1.40 | 1.83 | |
| Glc | 1.32 | 1.77 | |
| Myoinositol | 0.63 | 0.84 | |
| [2-NH 2-deoxy Glc] | 0.48 | 0.74 | |
| Organic acids | Gluconic acid | 3.79 | 2.32 |
| Citric acid | 2.15 | 2.41 | |
| Sinapic acid | 1.65 | 1.86 | |
| Fumaric acid | 1.56 | 2.94 | |
| Malic acid | 0.57 | 0.85 | |
| Glyceric acid | 0.09 | 0.09 | |
| Miscellaneous | Putrescine | 9.17 | 7.81 |
| [Glycerophosphoglycerol] | 1.67 | 1.84 | |
| [Glycerol-3-P] | 1.25 | 1.51 | |
| Unknown | A139006 | 10.12 | 7.01 |
| A151009 | 5.94 | 5.53 | |
| A144007 | 3.09 | 3.05 | |
| A302001 | 2.39 | 2.17 | |
| A199004 | 1.76 | 1.92 | |
| A203003 | 1.50 | 1.62 | |
| A147001 | 0.64 | 0.83 | |
| A1990904 | 0.48 | 0.75 | |
| A126003 | 0.19 | 0.23 | |
| A237006 | 0.13 | 0.31 |
Table II.
Significantly changed metabolites in seedlings under salt shock, regardless of the genotype (ANOVA P < 0.01)
To aid comprehension, metabolite pool sizes were averaged for both overexpressors. Data represent means of three independent experiments and are expressed as ratios of the 100 mm NaCl treatment to controls. The unknown is an unidentified metabolite that has been detected before and is denoted by Golm Metabolite Database code (Kopka et al., 2005).
| Type | Metabolite | Wild-Type NaCl/Control | Lines 1 and 7 NaCl/Control |
|---|---|---|---|
| Sugars | Suc | 1.76 | 1.70 |
| Glc | 1.37 | 1.87 | |
| Gal | 1.40 | 1.87 | |
| Fru | 1.39 | 1.62 | |
| Unknown | A1990904 | 1.19 | 1.89 |
Long-Term Effects of Myb41 Overexpression in Plants
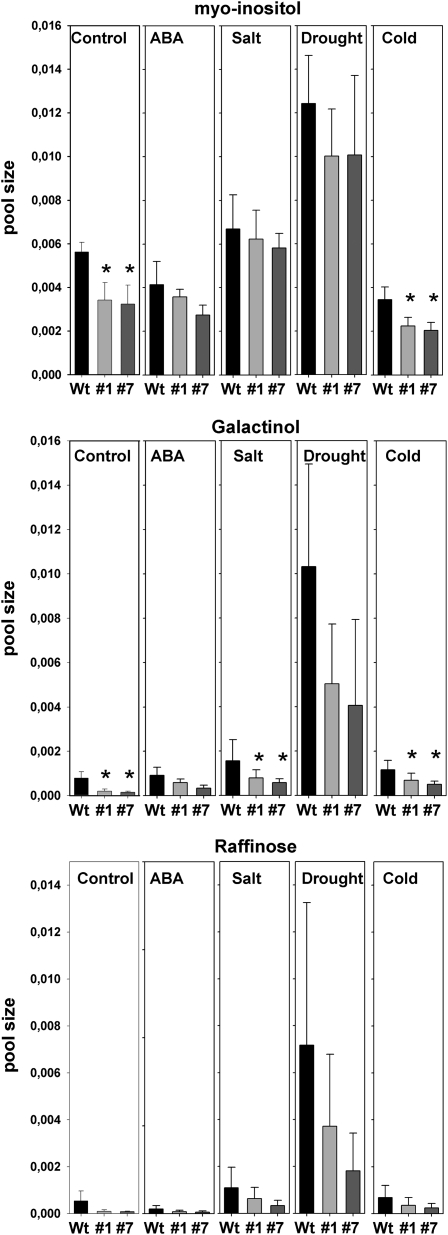
Given the effects of AtMyb41 overexpression on long-term plant growth (Fig. 2, B and C), it was of interest to us to investigate the molecular consequences of AtMyb41 overexpression in older plants. Rosettes from AtMyb41 transgenics and wild-type plants, subjected to control and various stress treatments, were profiled at the metabolome level. A total of 91 MSTs were recognized, but only 54 were found in both seedlings and mature plants. Seventy MSTs of rosettes of mature plants were ascribed to known chemical compounds (Supplemental Table S3). Changes in metabolite profiles following salt, drought, cold, and ABA treatments were revealed by unsupervised ICA, but surprisingly, no clear separation was observed between genotypes for these treatments (Fig. 4B). A significance-based analysis (ANOVA) demonstrated that the pool sizes of only two metabolites were statistically different in the shoots of transgenic compared with wild-type plants: myoinositol and galactinol (Fig. 5). Myoinositol levels were significantly lower in control and cold-treated AtMyb41 overexpressors than in wild-type plants, and galactinol was lower in the transgenic lines not only under these two conditions but also under salt-stress conditions. Raffinose levels also tended to be lower in the transgenic lines than in the wild type under control and stress conditions, although differences were not significant at P < 0.01 (Fig. 5). Note that myoinositol levels were also lower in seedlings of AtMyb41 overexpressors than in wild-type controls (Table I).
Figure 5.
Metabolites significantly changed in shoots of mature AtMyb41-overexpressing plants grown in soil under different stress conditions. Data represent means ± sd of eight independent biological replicates. Asterisks highlight statistically significant difference between the wild type (Wt) and transgenics within each treatment at P < 0.01 (ANOVA).
To test if the genes that were differentially expressed in Myb41 overexpressor compared with wild-type seedlings were also differentially expressed in adult plants of these genetic backgrounds, we measured expression of AtMyb41-responsive genes (Fig. 3D) in greenhouse-grown plants. A comparison between seedlings and adult plants grown under control, nonstress conditions revealed that only AtAp3 was differentially expressed between rosettes of AtMyb41 overexpressors and those of the wild type (Fig. 6A). Transcripts of several genes were barely detected, and subsequent analysis of seedlings revealed that some of these genes were predominantly expressed in roots, explaining the lack of expression in mature rosettes (Supplemental Fig. S1). However, differences between transgenic and wild-type plants in the expression of some of these genes were observed when adult plants were exposed to abiotic stress, suggesting that AtMyb41 overexpression alters stress signaling pathways not only in seedlings but also in mature rosettes (Fig. 6, B–E).
Figure 6.
Expression analysis of the most highly regulated transcripts in AtMyb41-overexpressing rosettes. A, Expression comparison of AtMyb41-overexpressing plants grown under greenhouse control conditions with in vitro-grown seedlings. Ctrl, Control; WT, wild type. B to E, The expression of these genes under salt (B), drought (C), or cold (D) stress and after ABA treatment (E) in rosettes from greenhouse-grown plants. Data represent means ± sd of four biological replicates, each a pool of eight plants. The arrow in A highlights the similar expression pattern of AtAp3 between rosettes and seedlings, while the asterisks in B to E indicate differentially expressed genes between adult wild-type and overexpressor plants, according to t test (* P < 0.05, ** P < 0.01). nd, Not detected in rosettes. The Arabidopsis Genome Initiative codes of the genes are provided in Supplemental Table S4.
DISCUSSION
Although many TFs of the Myb family are expected to be involved in stress responses, few have been characterized functionally. Recently, AtMyb41 was implicated in stress-induced cell wall expansion and modification in Arabidopsis, based on the dual observations that AtMyb41 transcript levels could not be detected under normal growth conditions but accumulated under stress and that overexpression of AtMyb41 resulted in abnormal morphology with severely inhibited cell and whole plant growth (Cominelli et al., 2008). Using quantitative real-time RT-PCR, we were able to detect AtMyb41 transcripts under nonstress conditions not only in seedlings but also in shoots and roots of mature plants. We showed that AtMyb41 was induced by osmotic stress in an ABA-dependent manner, similar to known ABA-responsive genes like AtAil1, AtRab18, AtRd29b, and AtAia1, suggesting that AtMyb41 may be part of the AREB regulon (Fujita et al., 2005). Consistent with this idea, putative ABRE cis elements are present in the promoter region of AtMyb41, together with myc recognition sites (Supplemental Fig. S2). A gain-of-function approach was used to shed light on possible molecular and physiological roles of AtMyb41. Constitutive overexpression of AtMyb41 in Arabidopsis inhibited plant growth, but to a much lower extent than described previously for similar transgenic plants (compare figure 2C with figure 2F in Cominelli et al., 2008).
Transcriptome analysis revealed a subset of genes that were induced by salt treatment in wild-type seedlings that failed to be induced in AtMyb41 overexpressors (Fig. 3B). Clearly, AtMyb41 overexpression interfered with the response of these genes, suggesting that this Myb TF may act as a transcriptional repressor. Among these transcripts were a number of known stress-responsive genes, including six late embryogenesis abundant-like proteins that are crucial in seed development and stress responses (Hundertmark and Hincha, 2008), AtDreb2a encoding a key TF involved in stress signaling (Sakuma et al., 2006), and AtNced3 encoding a key enzyme of ABA biosynthesis with a major role under stress (Iuchi et al., 2001). Noteworthy, AtBgl1, which encodes a β-glucosidase involved in the activation of conjugated ABA pools (Lee et al., 2006), was more expressed in the AtMyb41 overexpressors than in the wild type under both stress and nonstress conditions (Supplemental Table S1). Transcriptional changes in AtNced3 and AtBgl1 indicate that a disturbance in ABA metabolism may have been responsible for the deregulation of stress-related genes in the AtMyb41 overexpressors, but further work will be required to test this hypothesis.
Interestingly, 10 Myb TFs were differentially expressed in AtMyb41 overexpressors compared with the wild type (Fig. 3C). Eight of these, AtMyb71, AtMyb121, AtMyb9, AtMyb52, AtMyb49, AtMyb64, AtMyb102, and AtMyb93, were responsive to salt treatment in the wild type but not in the transgenic lines, while expression of AtMyb40 and AtMyb74 was repressed in the AtMyb41 overexpressors under nonstress conditions (Supplemental Table S1). Phylogenetic analysis of the protein sequences revealed that the majority of these TFs are closely related (Kranz et al., 1998), and a survey of publicly available transcriptomic data suggested that they are similarly transcriptionally regulated, particularly in the case of osmotic and salt stresses (Supplemental Fig. S3). Of the 10 Myb TFs, only AtMyb102 has been characterized in any detail: AtMyb102 transcripts accumulated rapidly following osmotic stress and wounding and contain a 5′ untranslated region with a crucial role in posttranscriptional regulation (Denekamp and Smeekens, 2003). Taken together, these results point to the existence of a network of interregulated Myb TFs that orchestrates transcriptional reprogramming in Arabidopsis to mount appropriate responses to abiotic stress.
Transcriptomic analysis identified a set of well-known stress-responsive genes that are part of the overall response to stress, such as AtRd29a, AtCor15a, and AtKin1 (Ma et al., 2006), which were apparently not deregulated by AtMyb41 overexpression. Thus, AtMyb41 controls some but not all of the signaling pathways responding to salt shock in seedlings. In addition, transcriptome analysis also revealed a subset of transcripts that were either up- or down-regulated in AtMyb41 overexpressors relative to the wild type, which were not responsive to salt treatment in any genotype (Fig. 3B; Supplemental Table S1). Altered expression of these genes in transgenic lines may have been a pleiotropic effect of prolonged, high-level AtMyb41 expression. However, it may also reflect physiological roles of AtMyb41 not necessarily linked to osmotic stress. For example, AtMyb41 may be involved in sulfur homeostasis, because its expression increased under low sulfur and in the sulfate transporter sultr1;2 mutant (sel1-10; Maruyama-Nakashita et al., 2003). In our data set, this transporter was up-regulated in the AtMyb41-overexpressing seedlings, while AtSultr3;4 was down-regulated under salt treatment in the transgenic lines but not in the wild type (Supplemental Table S1). These results support the hypothesis that AtMyb41 may be involved in the regulation of nutrient homeostasis.
Metabolomic analysis revealed strong differences between seedlings of AtMyb41 overexpressors and the wild type, indicated by constitutive changes in the level of primary metabolites including amino acids, organic acids, and sugars (Tables I and II). Some of the AtMyb41-induced metabolic changes, such as the increase in amino acids and organic acids, have been observed under various abiotic stress conditions (Guy et al., 2008; Sanchez et al., 2008b). It is unlikely that the altered metabolic phenotype of AtMyb41 overexpressor seedlings was a result of changes in growth rate, as reduced plant size was evident only later in plant development. Almost all of the major metabolic changes were observed in seedlings but not in adult plants of the AtMyb41 transgenic lines, which indicates some form of developmental control of AtMyb41 function. The potential involvement of Myb TFs in the control of primary metabolism is an interesting area for further research, since, to the best of our knowledge, this family of TFs has so far only been shown to be involved in the control of secondary metabolism (Borevitz et al., 2000; Xie et al., 2006). Consistent with the changes in metabolite levels, transcript levels of genes encoding a variety of enzymes involved in central metabolism were affected in the AtMyb41 overexpressors, including β-amylase (AtBmy1), pyruvate kinase (AtPk), phosphoglycerate/biphosphoglycerate mutase (AtBpm), Suc-P synthase (AtSps2f), Ala-glyoxylate aminotransferase (AtAgt), Gln synthetase (AtGln-1;4), and Tyr aminotransferase (AtTat3). Furthermore, transcripts for several transporters, including sulfate (AtSultr1;2 and AtSultr3;4), nitrate (AtNrt2;4), sugar, and amino acid transporters were differentially regulated (Supplemental Table S1).
Despite the negative impact of AtMyb41 overexpression on the growth of adult plants (Fig. 2), these lines showed few differences in metabolite levels compared with wild-type controls under nonstressed or stressed conditions at this developmental stage. Only myoinositol and galactinol levels were significantly lower in adult AtMyb41 overexpressors compared with wild-type controls, and raffinose levels showed a similar trend (Fig. 5). Note that the raffinose biosynthetic pathway, which includes all of these three compounds, has been linked to osmotic and cold stresses in Arabidopsis (Taji et al., 2002; Zuther et al., 2004). Two galactinol synthase genes (AtGols2 and AtGols4) were deregulated in the AtMyb41-overexpressing seedlings (Supplemental Table S1), suggesting the involvement of AtMyb41 in transcriptional control of this metabolic pathway. Taken together, this indicates that at least some of the stress-related roles of AtMyb41 in seedlings are conserved at later stages of plant development. Consistent with this idea, some of the transcripts that were differentially expressed between wild-type and transgenic seedlings were also differentially regulated under stress conditions in adult plants (Fig. 6).
Cominelli et al. (2008) identified numerous genes that were either up- or down-regulated in AtMyb41 overexpressors compared with the wild type under nonstress conditions. We retested 20 of these by real-time quantitative RT-PCR but found few that were regulated in the same way in our AtMyb41 overexpressors (Supplemental Fig. S4A). Although some of these genes were regulated in the wild type by abiotic stress (salt, drought, cold) or ABA, most were regulated in the same way in our AtMyb41 overexpressors (Supplemental Fig. S4, B–E). Therefore, few if any of these genes appear to be controlled by AtMyb41. The discrepancies between our results and those of Cominelli et al. (2008) may be explained, in part, by differences in the magnitude of the effects of AtMyb41 overexpression in the different transgenic lines. The AtMyb41 overexpressors of Cominelli et al. (2008) were highly stunted, while those in this study were only moderately affected in growth, and only at later stages of development. Differences in plant culture and treatment conditions may also account for some of the differences observed between the two studies. It should also be pointed out that the results of Cominelli et al. (2008) lack internal consistency: some of their results on cell wall-related gene transcripts obtained by RT-PCR were not confirmed by their microarray experiments, undermining their conclusion that changes in the expression of such genes in the AtMyb41 overexpressors may account for the growth defects of their transgenic lines. In fact, no causal relationship between cell wall-related genes and the dwarf phenotype of their AtMyb41 overexpressors was proven (Cominelli et al., 2008).
Whatever the explanation for the growth defect in AtMyb41 overexpressors, it seems unlikely that it was caused by constitutive activation of stress pathways by AtMyb41, as none of the stress-related metabolites identified in previous studies, including amino acids, organic acids, sugars, and polyols (Sanchez et al., 2008b), were altered in mature, nonstressed transgenic plants compared with the wild-type controls (Fig. 4). In contrast to previous results (Cominelli et al., 2008), we observed no increase in the sensitivity of growth to salt and drought stresses of AtMyb41-overexpressing plants compared with wild-type controls, which is perhaps not surprising given the multigenic nature of abiotic stress tolerance. Again, differences between these two studies in this regard may be related to differences in the severity of the growth defects exhibited by the two sets of AtMyb41 overexpressors, with the dramatic dwarf phenotype observed by Cominelli et al. (2008) reflecting marked pleiotropic effects that would confound any interpretation of the physiological roles of AtMyb41.
In summary, we demonstrated the involvement of the ABA-regulated AtMyb41 TF in the control of distinct cellular processes in Arabidopsis seedlings, including negative regulation of short-term transcriptional responses to osmotic stress and associated changes in primary metabolism.
MATERIALS AND METHODS
Plant Material, Growth, and Experimental Conditions
Arabidopsis (Arabidopsis thaliana) lines overexpressing AtMyb41 were obtained using the PyAt4g28110 cDNA clone from the ORFeome collection (Gong et al., 2004). Through the Gateway system (Invitrogen), the At4g28110 open reading frame was cloned into PMDC 32 (Curtis and Grossniklaus, 2003), where the expression is driven by the constitutive Cauliflower mosaic virus-35S promoter. Plants were transformed and selected with kanamycin for at least three generations.
For in vitro culture, approximately 100 seedlings were grown at 22°C in sterile liquid medium in Erlenmeyer flasks on a shaker under constant fluorescent light (approximately 50 μE m−2 s−1 in the flask; Scheible et al., 2004). Seven days after sowing, the medium was changed, and on day 9, NaCl or mannitol was added at a final concentration of 100 or 200 mm, respectively. Whole seedlings were harvested after 1, 2, 3, 6, 12, 18, and 24 h. Each experiment consisted of sample sets with at least three biological replicates (i.e. independent flasks). For hydroponic culture, seeds were sown in 0.5-mL plastic tubes filled with half-strength Hoagland medium (Hoagland and Arnon, 1938) including 0.8% agar. After 1 week, tubes were cut open at the bottom and transferred to a 2-L box filled with nutrient solution without agar and grown at 20°C and 12 h of light/12 h of darkness. The salt acclimation experiment was performed by increasing NaCl concentration weekly in three steps, from 0 to 10 mm, 25 mm, and 50 mm NaCl, with fresh nutrient solution each week. Medium was also changed every week for control plants. For greenhouse culture, plants were grown in soil (type GS90 + vermiculite; Einheitserde) using 10-cm pots under 20°C/18°C day/night temperatures with a long-day (16 h) photoperiod (minimum 200 μE m−2 s−1). Salt and drought stress treatments were performed on 1-week-old plants, watering with a 50 mm NaCl solution or withholding irrigation for 2 weeks, respectively. For cold stress, 3-week-old plants were transferred to a 4°C growth chamber with a 16-h photoperiod at 90 μE m−2 s−1 for 24 h. ABA-treated plants were sprayed with 100 μm ABA solution 24 h before harvest. For each condition, rosette leaves were harvested before bolting in eight biological replicates consisting of pools of five plants each.
Gene Expression Analysis
For the Affymetrix GeneChip Arabidopsis ATH1 Genome Array experiments, RNA extraction was performed from seedling material grown in vitro from three independent experiments in pools of three biological replicates each (i.e. independent flasks). Total RNA was isolated using the hot borate method (Wan and Wilkins, 1994), and quality and quantity were assessed with a Bioanalyzer- 2100 using RNA 6000 NanoChips (Agilent Technologies) and a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), respectively. RNA was labeled with the Message AmpII-Biotin Enhanced kit following the manufacturer's instructions (Ambion). Hybridization and scanning were performed at ATLAS Biolabs. For quantitative real-time RT-PCR analysis, total RNA was treated with TURBO DNAseI (Ambion) and first-strand cDNA was synthesized with an oligo(dT) primer and SuperScript III reverse transcriptase (Invitrogen). Real-time PCR was performed using the 2xSYBR GreenI PCR Mastermix and an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Primer design, reaction conditions, cycling and dissociation curve parameters, DNA contamination, and 3′ to 5′ ratio checks were performed as described by Czechowski et al. (2005) without spike-in controls in a reaction volume of 10 μL. Amplification efficiency was assessed with the LinRegPCR program (Ramakers et al., 2003). Analysis of expression data was performed as described previously (Czechowski et al., 2004, 2005) using the geometric mean of three housekeeping genes for normalization (Vandesompele et al., 2002). The housekeeping genes were AtSand (At2g28390), AtGapdh (At1g13440), and AtPdf2 (At1g13320), selected among the most stably expressed genes in Arabidopsis (Czechowski et al., 2005). A list of the primers used is available in Supplemental Table S5.
Metabolite Analysis
Metabolomic analysis was performed essentially as described by Sanchez et al. (2008a). In brief, plant material (60 mg) was extracted with chloroform/methanol, and the polar fraction was prepared by liquid partitioning into water, derivatized, and injected in a GC/EI-TOF-MS system (Wagner et al., 2003). Peak height representing arbitrary mass spectral ion currents was normalized with the sample fresh weight and ribitol content for internal standardization, and the resulting metabolomic profiles were analyzed using the TagFinder software (Luedemann et al., 2008). A series of discrete metabolic features were obtained after filtering for those represented by three or more intercorrelated mass fragments within the whole data set. Metabolites were identified using the NIST05 software (www.nist.gov/srd/mslist.htm) and the mass spectral and retention time index collection of the Golm Metabolome Database (Kopka et al., 2005). Mass spectral matching was manually supervised and accepted with thresholds of match > 650 (maximum 1,000) and retention time index < 1.0%.
Data Analysis and Statistics
K-means clustering, principal component analysis, and ICA were used as clustering algorithms to analyze profiling data in a nonsupervised approach, using The Institute for Genomic Research multiple experiment viewer software (TMEV_3.1), the pcaMethods package for the R programming language (www.bioconductor.org/packages/2.1/bioc/html/pcaMethods.html), and the MetaGeneAlyse Web page (http://metagenealyse.mpimp-golm.mpg.de).
Microarray results were analyzed using the bioconductor software package for R (Gentleman et al., 2004). Data quality was assessed with the affy (Gautier et al., 2004) and AffyPLM packages, and expression estimates were obtained with the Robust Multi-Array algorithm (Irizarry et al., 2003). Genes assigned as present (P < 0.05) using the MAS5 present/absent algorithm were statistically tested for differential expression using mixed models with the LIMMA bioconductor package (Smyth, 2004). P values describing control versus treatment and genotype comparisons were corrected for multiple testing using the FDR correction (Benjamini and Hochberg, 1995). Metabolomic data were log10 transformed prior to statistical analysis, performed with ANOVA using the TMEV_3.1 software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Root-shoot expression ratio of selected genes identified in the microarray analysis as differentially expressed in AtMyb41-overexpressing seedlings.
Supplemental Figure S2. cis-Regulatory elements in the putative promoter region of AtMyb41 identified from the PLACE database (www.dna.affrc.go.jp/PLACE).
Supplemental Figure S3. Coexpression analysis of the 10 Myb TFs using the Genevestigator database (www.genevestigator.ethz.ch).
Supplemental Figure S4. Comparison of the expression of the genes most highly regulated by AtMyb41 overexpression in the study of Cominelli et al. (2008) with the expression in the independent transgenic lines (lines 1 and 7) described in our study.
Supplemental Table S1. Transcriptomic profile data from seedlings.
Supplemental Table S2. Metabolic profile data from seedlings.
Supplemental Table S3. Metabolic profile data from rosettes.
Supplemental Table S4. Arabidopsis Genome Initiative codes of all of the genes described.
Supplemental Table S5. List of primers used for quantitative real-time RT-PCR.
Supplementary Material
Acknowledgments
We gratefully acknowledge the long-standing support of all directors at the Max Planck Institute for Molecular Plant Physiology (MPIMP), particularly Prof. Dr. Lothar Willmitzer. We are grateful to Dr. Jochim Kopka, Alexander Erban, and Ines Fehrle for assistance and outstanding technical support concerning the GC/EI-TOF-MS profiling. We thank Franziska Schwabe for her day-to-day assistance, the MPIMP “greenteam” for support in the greenhouse experiments, and Dr. Matthew Hannah for help with microarray analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael K. Udvardi (mudvardi@noble.org).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25 1263–1274 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 57 289–300 [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15 1196–1200 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53 53–64 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari R, Stitt M, Scheible WR, Udvardi M (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses GG, Kopka J, Udvardi MK (2005) Lotus japonicus metabolic profiling: development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol 137 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Muller C, Flugge UI (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51 247–261 [DOI] [PubMed] [Google Scholar]
- Gong W, Shen YP, Ma LG, Pan Y, Du YL, Wang DH, Yang JY, Hu LD, Liu XF, Dong CX, et al (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK (2008) Metabolomics of temperature stress. Physiol Plant 132 220–235 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1938) The water culture method for growing plants without soil. Calif Agric Exp Stn Circ 357 1–39 [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BCM Genomics 9 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad B, Collin F, Cope L, Hobbs B, Speed T (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 325–333 [DOI] [PubMed] [Google Scholar]
- Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Do Choi Y, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng ZG, Oppenheimer D, Schiefelbein J, Hulskamp M (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al (2005) GMD@CSBDB: the Golm Metabolome Database. Bioinformatics 21 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16 263–276 [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jian F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J (1999) WEREWOLF, a Myb related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 1539–1546 [DOI] [PubMed] [Google Scholar]
- Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM (2005) AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol 15 1201–1206 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS) based metabolite profiling experiments. Bioinformatics 24 732–737 [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57 1097–1107 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DH, Lippold F, Redestig H, Hannah MA, Erban A, Krämer U, Kopka J, Udvardi MK (2008. a) Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J 53 973–987 [DOI] [PubMed] [Google Scholar]
- Sanchez DH, Siahpoosh MR, Roessner U, Udvardi MK, Kopka J (2008. b) Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol Plant 132 209–219 [DOI] [PubMed] [Google Scholar]
- Scheible W, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi M, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 3. [DOI] [PubMed] [Google Scholar]
- Taji T, Oshumi C, Iuchi S, Seki M, Kasuga M, Kabayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold- inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29 417–426 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bäsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 research0034I–research0034II [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J (2003) Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62 887–900 [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA (1994) A modified hot-borate method significantly enhances the yield of high-quality RNA from cotton Gossypium hirsutum L. Anal Biochem 223 7–12 [DOI] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Wright E, Wang ZY, Dixon RA (2006) Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J 45 895–907 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Yang CY, Xu ZY, Song J, Conner K, Barrena GV, Wilson ZA (2007) Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60 107–124 [DOI] [PubMed] [Google Scholar]
- Yoo JH, Park CY, Kim JC, Heo WD, Cheong MS, Park HC, Kim MC, Moon BC, Choi MS, Kang YH, et al (2005) Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J Biol Chem 280 3697–3706 [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Zheng-Hua Y (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary cell wall biosynthesis in Arabidopsis. Plant Cell 19 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee BH, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102 9966–9971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zuther E, Buchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG (2004) The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett 576 169–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.