Abstract
Recent evidence has shown that alternative splicing (AS) is widely involved in the regulation of gene expression, substantially extending the diversity of numerous proteins. In this study, a subset of expressed sequence tags representing members of the reactive oxygen species gene network was selected from the PopulusDB database to investigate AS mechanisms in Populus. Examples of all known types of AS were detected, but intron retention was the most common. Interestingly, the closest Arabidopsis (Arabidopsis thaliana) homologs of half of the AS genes identified in Populus are not reportedly alternatively spliced. Two genes encoding the protein of most interest in our study (high-isoelectric-point superoxide dismutase [hipI-SOD]) have been found in black cottonwood (Populus trichocarpa), designated PthipI-SODC1 and PthipI-SODC2. Analysis of the expressed sequence tag libraries has indicated the presence of two transcripts of PthipI-SODC1 (hipI-SODC1b and hipI-SODC1s). Alignment of these sequences with the PthipI-SODC1 gene showed that hipI-SODC1b was 69 bp longer than hipI-SODC1s due to an AS event involving the use of an alternative donor splice site in the sixth intron. Transcript analysis showed that the splice variant hipI-SODC1b was differentially expressed, being clearly expressed in cambial and xylem, but not phloem, regions. In addition, immunolocalization and mass spectrometric data confirmed the presence of hipI-SOD proteins in vascular tissue. The functionalities of the spliced gene products were assessed by expressing recombinant hipI-SOD proteins and in vitro SOD activity assays.
Pre-mRNA splicing is an important regulatory step in the expression of many eukaryotic genes. Alternative splicing (AS) of pre-mRNA results in the generation of multiple transcripts from a single gene, thereby increasing the coding capacity and proteome diversity of eukaryotic genomes (Ast, 2004; Stamm et al., 2005). The number of AS genes identified (using various bioinformatic, transcriptomic, and other functional genomic tools) has increased tremendously in recent years from a few hundred to many thousands (Reddy, 2007). It has recently been established that AS is a further frequent, important regulatory mechanism of gene expression in Caenorhabditis elegans, human, mouse, and Drosophila, and analyses of the developmental stage- and/or tissue-specific expression patterns of AS variants are likely to become important components of biological research (Lareau et al., 2004; Pajares et al., 2007). In humans, it is estimated that about 60% of genes undergo AS (Modrek and Lee, 2002), exon skipping being the most common form and intron retention (IntronR) the least common (Kim et al., 2007). Most of the AS genes in humans are involved in signaling and regulation (Valdivia, 2007).
Lower proportions of genes in plants are reported to undergo AS (Wang and Brendel, 2006). In an extensive study, Ner Gaon et al. (2007) analyzed 11 plant species using EST and genomic sequences and found indications that AS occurs in all tested species. However, the proportions of AS genes found in these plants varied substantially. In both Arabidopsis and rice (Oryza sativa), AS was found to affect approximately 21% of the genes and IntronR was found to be the most prevalent form (Wang and Brendel, 2006). Furthermore, the AS events were found to be conserved in these plants, indicating that AS is a mechanism (in addition to those previously recognized) that is widely involved in the qualitative and/or quantitative control of gene expression by regulating mRNA stability, making prematurely truncated open reading frames and introducing functional diversity into the products of particular genes.
Reactive oxygen species (ROS) control many processes in plants and are widely acknowledged to play dual roles in their physiology, both causing oxidative stress and acting as signaling molecules in various developmental processes in plants (Karpinski et al., 1999; Foreman et al., 2003; Overmyer et al., 2003). Imbalances in the levels of hydrogen peroxide (H2O2) and superoxide radical (O2−) can cause oxidative stress in plants, which in turn can induce the activation or silencing of genes encoding defensive enzymes, transcription regulators, and structural proteins (Mittler, 2002; Scandalios, 2005). In order to maintain their redox equilibrium, cells must maintain a tight balance between the production of oxidants and antioxidants (Scandalios, 2005). Consequently, in Arabidopsis, a large gene network has been shown to be involved in mechanisms that control ROS toxicity while enabling ROS to act as signaling molecules (Mittler et al., 2004).
Superoxide dismutases (SODs; EC 1.15.1.1), which are key components of the ROS gene network in plants, occur in three different forms (Cu/Zn-SOD, Fe-SOD, and Mn-SOD), all of which catalyze the dismutation of O2− to molecular oxygen (O2) and H2O2, thereby protecting cells against oxidative stress (Liochev and Fridovich, 1994). Isoforms of Cu/Zn-SOD named high-isoelectric-point superoxide dismutases (hipI-SODs) have been detected in pine (Pinus spp.), Zinnia, and Populus in our laboratory and shown to be involved in the regulation of ROS and plant development (Schinkel et al., 1998, 2001; Karpinska et al., 2001; Karlsson et al., 2005; Srivastava et al., 2007). These enzymes have substantially higher pI values than other known SODs (>9.5) and monomeric masses of about 16 kD. We also previously found indications that an isomer of hipI-SOD present in pine may be active as a monomer, unlike other SODs, which are active in dimeric or tetrameric form (Schinkel et al., 1998), strengthening the possibility that various isoforms of hipI-SOD could play different roles in metabolic processes. Furthermore, recent reports indicate that AS generates two different isoforms of various SODs in Drosophila, human, rice, and C. elegans. For instance, two active, alternatively spliced forms of Fe-SOD are produced in rice by IntronR AS (Feng et al., 2006), and in C. elegans a Cu/Zn-SOD gene encodes two isoforms by AS that differ in exons coding for the C-terminal part of the proteins (Fujii et al., 1998).
Attempts to assess the significance of these phenomena, and other contributions of AS to tissue, organ, and developmental stage-specific protein expression, have been greatly facilitated by the increasing availability of genomic, transcript, and protein sequence data for increasing numbers of species, as in the study reported here, in which Populus sequence data were applied. Populus has emerged as a model species for the analyses of tree physiology at cellular and molecular levels because of its rapid growth, the relative ease of genetic transformation, the modest size of its genome, and the availability of its genome sequence (Tuskan et al., 2006; Jansson and Douglas, 2007).
In this study, we investigated and characterized AS events in Populus using a subset of ESTs related to the ROS gene network. The study revealed that all known types of AS events occur in the chosen set of genes in Populus, IntronR being the most frequent type. We also analyzed the sequences around exon-intron boundaries for ROS-related genes. Furthermore, we found that in Populus, AS produces two different forms of hipI-SOD via AS at a donor site in intron 6 of the gene PthipI-SODC1. Expression patterns of both of the spliced forms, together with one other form named hipI-SODC2, were analyzed in vascular tissues. We also report the identification of hipI-SOD proteins in vascular tissues. One of the two alternatively spliced forms of hipI-SODC1 transcript encodes an enzymatically active protein, while the other encodes an inactive protein. All of these forms are suggested to be expressed differentially in different tissues.
RESULTS AND DISCUSSION
Investigation of AS in the Populus ROS Gene Network
A subset of the ESTs related to the ROS gene network (Mittler et al., 2004; Scandalios, 2005) were selected in the PopulusDB database to investigate AS. In this study, 18 different genes in EST clusters examined in PopulusDB (Supplemental Table S1) were identified to undergo different types of AS events (one of which appears to be affected by two such events, resulting in 19 AS events in total; Table I; Supplemental Table S2). The total frequency of alternatively spliced genes identified, as a percentage of the total number of ROS-associated EST clusters analyzed, was low (approximately 8%), compared with the frequencies found in analyses of other plant species (Wang and Brendel, 2006). Since only a few ESTs have been sequenced for most Populus genes, the total numbers of AS events, and the proportion of genes involved, are undoubtedly higher than the numbers reported here. Similarly, the number of known AS events in Arabidopsis has gradually increased with time (The Arabidopsis Information Resource, 2008; http://arabidopsis.org).
Table I.
Genes related to the ROS network undergoing various types of AS in Populus
| No. | Description | EST Cluster | Closest Gene Model | Type of AS | Best ATH Hit |
|---|---|---|---|---|---|
| 1 | bHLH | POPLAR.1596 | grail3.0001043902 | Alternative donor site | At1g68920a |
| 2 | Carbonic anhydrase | POPLAR.1629 | estExt_Genewise1_v1.C_LG_I2426 | Alternative donor site | At3g01500a |
| 3 | Superoxide dismutase | POPLAR.5824 | estExt_fgenesh4_pg.C_LG_XIX0416 | Alternative donor site | At5g18100a |
| 4 | Glutathione S-transferase | POPLAR.6206 | grail3.0273000301 | Alternative position | At2g29420 |
| 5 | Dormancy-associated protein | POPLAR.756 | grail3.0046016501 | Cassette exon | At1g28330a |
| 6 | Carbonic anhydrase | POPLAR.4373 | estExt_fgenesh4_kg.C_LG_X0015 | Cassette exon | At5g14740a |
| 7 | Protein phosphatase 2C | POPLAR.10002 | gw1.XV.329.1 | Exon skipping | At4g27800a |
| 8 | bZIP | POPLAR.2214 | estExt_Genewise1_v1.C_LG_III0940 | Intron retention | At5g06950a |
| 9 | Glutathione peroxidase | POPLAR.4658 | fgenesh4_kg.C_LG_III000037 | Intron retention | At4g11600 |
| 10 | MAP kinase kinase kinase | POPLAR.5177 | estExt_Genewise1_v1.C_LG_VI2808 | Intron retention | At1g08720 |
| 11 | Peroxiredoxin | POPLAR.3209 | estExt_fgenesh4_pm.C_LG_VIII0405 | Intron retention | At1g48130 |
| 12 | Protein kinase | POPLAR.497 | estExt_fgenesh4_pg.C_LG_I2295 | Intron retention | At5g40870 |
| 13 | Protein kinase | POPLAR.5037 | estExt_fgenesh4_pg.C_2600008 | Intron retention | At1g29730 |
| 14 | RNA polymerase II | POPLAR.584 | estExt_fgenesh4_pm.C_LG_VII0230 | Intron retention | At5g09920a |
| 15 | RNA polymerase II | POPLAR.5972 | estExt_Genewise1_v1.C_1370048 | Intron retention | At5g09920a |
| 16 | Thioredoxin | POPLAR.4311 | gw1.XVII.864.1 | Intron retention | At5g39950 |
| 17 | WRKY | POPLAR.57 | estExt_Genewise1_v1.C_LG_II0718 | Intron retention | At2g30590 |
| 18 | MYB transcription factor | POPLAR.3744 | gw1.IX.4341.1 | Intron retention, alternative donor site | At3g46130a |
Alternatively spliced genes reported in Arabidopsis.
DNA sequences are known to be highly conserved among Populus species (Sterky et al., 2004). However, our attempts to identify AS sites could have been confounded or complicated by the fact that the EST databases used in this study include sequences from three taxa (aspen [Populus tremula], hybrid aspen [Populus tremula × Populus tremuloides], and black cottonwood [Populus trichocarpa]), especially since Sterky et al. (2004) reportedly failed to distinguish between black cottonwood and hybrid aspen sequences in a clustering analysis of ESTs. The coding sequences of these ESTs were found to be almost identical (typically greater than 98%) after checking species-specific contigs within the same clusters, probably due to the similarity of orthologous genes from the two species. Therefore, it is plausible that some inferred AS may simply reflect the presence of orthologous sequences originating from different Populus species.
Since we examined a relatively small data set, no definitive conclusions can be made about the overall level of AS in Populus for comparison with the more comprehensively surveyed frequencies of AS genes (approximately 21%) in Arabidopsis and rice (Wang and Brendel, 2006). Nevertheless, interesting indications were found, notably that IntronR was the most common form of AS in the ROS-related data set, accounting for 54% of the AS events detected in the Populus ROS gene network, similar to the percentages reported for other species (e.g. 56% and 53% in Arabidopsis and rice, respectively; Wang and Brendel, 2006).
Conservation of AS events has been suggested to provide a strong indication of functional products and to distinguish splicing errors from functional AS (Wang and Brendel, 2006). In support of this hypothesis, over 40% of the spliced genes in Arabidopsis have been shown to have close AS homologs in rice, and approximately 25% have the same splice types. About 50% (Table I) of the closest homologs in Arabidopsis to the AS genes we identified in Populus were not reported to be alternatively spliced. This may be because the spliced genes we found are more tree specific, and they may play important roles in gene regulation rather than arising from splicing errors, despite their low frequency. True splicing errors occur at a very low frequency and thus constitute a very small proportion of the entries in the EST database (Graveley, 2001).
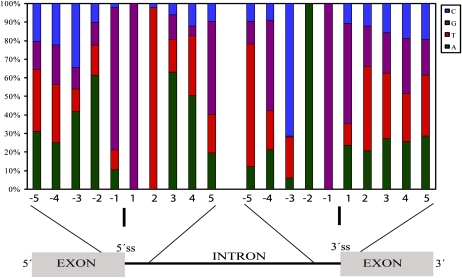
The small Populus data set we analyzed showed very similar types of AS patterns to those reported for Arabidopsis and rice; nevertheless, the closest homologs of many AS genes found in our analysis are not reported as alternatively spliced in Arabidopsis, so we were prompted to check the intron-exon boundaries of the Populus genes and briefly compare them with those of other plant species. For this purpose, sequence patterns around intron-exon junctions of the chosen genes from the ROS network at both the 5′ (acceptor) splice sites and the 3′ (donor) splice sites were examined (Fig. 1). High degrees of sequence similarity around these sites were found to sequences reported for Arabidopsis and rice (Reddy, 2007). These sites are different in human and mouse genes, suggesting that splice site preferences in plants and animals also differ (Wiebauer et al., 1988; Brendel et al., 1998; Reddy, 2007).
Figure 1.
Sequence patterns at the 5′ and 3′ splice sites of introns in Populus. Relative occurrence of different nucleotides at the donor (exon-intron) and acceptor (intron-exon) junctions of ROS-related genes of Populus. The start and end of introns are GT and AG, respectively, which are found to be conserved (at positions 1 and 2 at the exon-intron boundaries and at positions −2 and −1 at the intron-exon boundaries, respectively). This image was produced by extracting 10 nucleotides (five from the exon and five from the intron) at the 5′ and 3′ splice sites of 165 introns from the annotated sequences of selected genes.
ROS can activate gene transcription either via transcription factors that interact directly with specific DNA motifs or via protein kinase cascades that, in turn, activate transcription factors (Scandalios, 2005). In our study, two protein kinases, one of which is a mitogen-activated protein (MAP) kinase kinase kinase, were also found to be alternatively spliced. The expression of several types of transcription factors (including members of the bZIP, Zat, RAV, GRAS, WRKY, and MYB families) is enhanced by ROS (Mittler et al., 2004), and two types of AS were found to affect the expression of transcription regulator genes: IntronR and alternative donor site. The former was found to affect members of the bZIP, WRKY, and MYB families, while the latter affects members of the bHLH and MYB families (Table I), which could have profound significance for the activity of their products. In support of this hypothesis, 18 of 47 SET [for Su(var)3-9, Enhancer-of-zeste, and Trithorax] genes examined by Ng et al. (2007) reportedly undergo various types of AS events, and AS has been proposed to alter the oligomeric assembly of SET gene products in ways that modify their biological roles.
In this study, several genes that are known to be involved in detoxification of both H2O2 and O2− as well as genes involved in oxidative-reductive reactions (Mittler, 2002; Mittler et al., 2004; Scandalios, 2005) were analyzed, and some of them were found to be alternatively spliced (Table I). In spinach (Spinacia oleracea), four isoenzymes of chloroplastic ascorbate peroxidase are known to be encoded by a single gene and the corresponding mRNAs are reportedly generated by AS (Shigeoka et al., 2002). Alternatively spliced forms of the genes whose expression is affected by stress (such as protein phosphatase 2C) and those showing antioxidant activity (such as carbonic anhydrase) have been identified (Slaymaker et al., 2002; Meskiene et al., 2003; Yang et al., 2006). We also observed that most of the ESTs correspond to two or more (in a few cases) gene models, indicating that gene duplication events have occurred in Populus (Jansson and Douglas, 2007). Losses of AS may occur shortly after gene duplication. In humans, duplicate genes have been found to have fewer AS forms than single-copy genes, and a negative correlation has been detected between mean numbers of AS forms and gene family size by Su et al. (2006), who also postulated that AS and gene duplication may not have evolved independently.
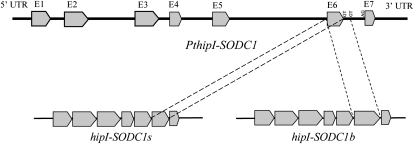
Identification of hipI-SODC1s, hipI-SODC1b, and hipI-SODC2
One of the AS enzymes identified in the ROS network was a recently described hipI-SOD with suggested roles in the development and lignifications of the secondary cell wall (Karpinska et al., 2001; Karlsson et al., 2005; Srivastava et al., 2007). BLAST searches of the Populus genome database for hipI-SOD sequences by Srivastava et al. (2007) showed that the Populus genome contains two hipI-SOD genes, designated PthipI-SODC1 and PthipI-SODC2. Both of these paralogous hipI-SOD genes consist of seven exons separated by six introns (Supplemental Fig. S1), and they share 94% similarity. hipI-SODC2 (GenBank accession no. AJ278671), which represents the contig POPLAR.5824.C2, was cloned and sequenced by Schinkel et al. (2001). Its predicted protein contains 158 amino acids, with an approximate molecular mass of 16 kD. PthipI-SODC2 was found to be the closest gene model for hipI-SODC2. The contig POPLAR.5824.C1 is represented by two EST clones, hereafter termed hipI-SODC1s and hipI-SODC1b (GenBank accession nos. FJ393058 and FJ393059, respectively). Both of these ESTs, corresponding to PthipI-SODC1, were resequenced, analyzed, and found to be quite similar, except in their 3′ regions. hipI-SODC1b is 69 bp larger than hipI-SODC1s and thus encodes an 18.7-kD protein of 181 amino acids (Fig. 2), 23 amino acid longer than the sequence encoded by hipI-SODC1s (158 amino acids, 16 kD). Nevertheless, comparative analysis of the ESTs and PthipI-SODC1 sequences revealed that the C-terminal part of all of the predicted polypeptides is similar. The last eight residues (IIGLKSSV) are encoded entirely by exon 7 (Fig. 2; Supplemental Fig. S1). Analysis of the amino acid sequence indicated that exon 6 in hipI-SODC1b is larger than the corresponding exon in PthipI-SODC1 and that the extra 23 amino acids are encoded by the starting portion of intron 6, thereby making it 69 bp longer. Splicing at this new splice site ligates the seventh exon to the extended exon 6, thereby increasing the size of its transcript (Fig. 3). About 10% of the total AS events detected in Arabidopsis and rice are due to alternative donor site splicing (Wang and Brendel, 2006), similar to the apparent splicing of hipI-SOD of Populus. The generation of multiple isoforms of SOD by AS has also been detected in several other organisms. For instance, in C. elegans, two isoforms of Cu/Zn-SOD, differing in their C termini, are produced by AS (Fujii et al., 1998), and Fe-SOD in rice has been reported to undergo AS, generating two isoforms of different sizes by IntronR (Feng et al., 2006). Ner-Gaon et al. (2004) suggested that most of the introns retained in such events may either be parts of open reading frames in the untranslated regions (UTRs) or located as the last introns in their respective transcripts, supporting the hypothesis that their presence should not contribute to nonsense-mediated decay. They further suggested that gene products with retained introns may be involved in combating stress and other responses to external or internal stimuli.
Figure 2.
Sequence analysis of hipI-SOD proteins. Sequence comparison of all predicted hipI-SOD proteins in Populus. The amino acid sequences were analyzed using the program GeneDoc. Peptides identified by mass spectrometry are highlighted in colors. Residues that differ among the identified peptides are shown in white letters.
Figure 3.
Organization of the PthipI-SODC1 gene. Schematic representation of the PthipI-SODC1 gene and transcripts of hipI-SODC1s and hipI-SODC1b. Pointed gray boxes represent exons, and thick lines between them represent introns. Consensus sequences corresponding to the splice donor and acceptor sites are shown.
Expression of hipI-SOD in Vascular Tissue
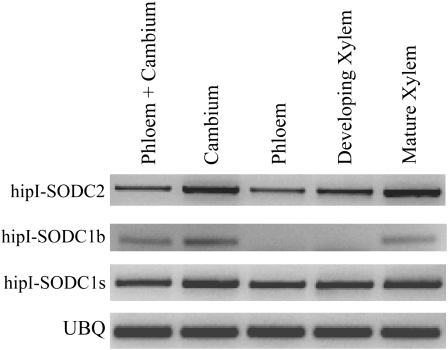
On the basis of microarray expression data (Schrader et al., 2004) and immunolocalization analyses (Srivastava et al., 2007) regarding hipI-SOD, vascular tissue was chosen to check the expression patterns of all three transcripts (hipI-SODC1s, hipI-SODC1b, and hipI-SODC2). Since the hipI-SOD genes differ in their 3′ UTRs, specific PCR primers (see “Materials and Methods”; Table II) for hipI-SODC1s, hipI-SODC1b, and hipI-SODC2 were designed to amplify cDNA from phloem, cambium, and xylem tissues. Pronounced differences in the expression patterns of the hipI-SOD transcripts in the vascular tissues were found. hipI-SODC1s and hipI-SODC2 are amplified as a single band from all samples mentioned above (Fig. 4). hipI-SODC1b transcript was found in cambium, mature xylem, and phloem-cambium but not in phloem and developing xylem samples (Fig. 4). Since the hipI-SODC1b transcript was detected in cambium and phloem-cambium, but not in phloem samples, it may be concluded that in addition to mature xylem hipI-SODC1b is expressed in cambial tissue. To verify the identities of the amplification products, all of the bands representing different transcripts of hipI-SOD were cloned and sequenced. The sequencing results corroborated the presence of hipI-SOD transcripts in all of the tissues in which they had been detected. The expression patterns of the hipI-SOD transcripts indicated that they had tissue-specific distributions. AS-related differences in tissue-specific patterns of transcripts have been detected in several other plants. For instance, Marsh et al. (2003) detected an AS mechanism that generates two transcripts of a phosphoenolpyruate carboxylase (PEPc) kinase gene with differential expression patterns and is involved in tissue-specific regulation of PEPc in tomato (Solanum lycopersicum ). Homeodomain proteins in rice (Ito et al., 2002) and RNA products of a rice homeobox gene (Tamaoki et al., 1995) are also reportedly expressed in a tissue-specific manner. The larger transcript (exons 1 and 3–7) of the rice homeobox gene OSH42 appears to be expressed in all tissues, while the smaller transcript (exons 2–7) is expressed in leaves, stems, and the rachis. In addition, de la Fuente van Bentem et al. (2003) have shown that an additional exon arising from AS of protein phosphatase 5 genes of Arabidopsis appears to be responsible for its transport to integral membranes of the endoplasmic reticulum and the nuclear envelope.
Table II.
Primers used in RT-PCR analysis
| Primer Name | Sequence |
|---|---|
| hipI-SODC1s_fw | 5′-GGTTGGATGTGGCATCATTGGGCTCA-3′ |
| hipI-SODC1s_rev | 5′-GGATCACCCTGCAAAACTTACAATGACA-3′ |
| hipI-SODC1b_fw | 5′-CTCTCATCACCGGAGACTCCAATGTTA-3′ |
| hipI-SODC1b_rev | 5′-CATTCCATCTCATGACTACAAGTTCTTGC-3′ |
| hipI-SODC2_fw | 5′-CTCTCATCACCGGAGACTCCAATGTTA-3′ |
| hipI-SODC2_rev | 5′-AGACCCTGCAAAACTTAAAAGGACGTTC-3′ |
| UBQ_fw | 5′-AGATGTGCTGTTCATGTTGTCC-3′ |
| UBQ_rev | 5′-ACAGCCACTCCAAACAGTACC-3′ |
Figure 4.
RT-PCR analysis of hipI-SODs in Populus. Results of the expression pattern analysis of hipI-SODC2 and alternatively spliced transcripts of hipI-SODC1s and hipI-SODC1b. Specific primers were used for all amplifications (see “Materials and Methods” for details), and UBQ was used as a loading control.
Immunolocalization of hipI-SOD in Vascular Tissue
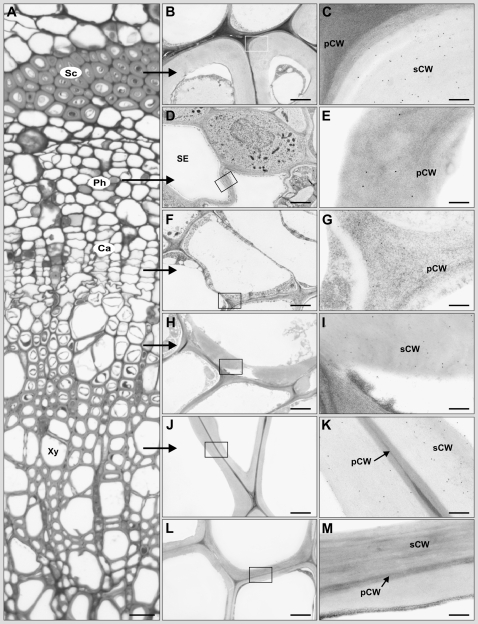
Cross sections of the stem from the 10th internode of hybrid aspen plants were examined by light microscopy and transmission electron microscopy following immunolabeling of hipI-SOD with 10-nm protein A-gold to determine where the protein was expressed at a cellular level. As controls, we used labeling without the primary antibody (data not shown) and preimmune serum as the primary antibody (Fig. 5, L and M). Both did not result in significant labeling of any structures. Significant signals were detected in the secondary walls of phloem fibers and xylem vessels (Fig. 5, B, C, H, I, J, and K), confirming indications obtained in earlier, confocal microscopy studies (Srivastava et al., 2007). In addition, significant labeling was found in the primary wall of sieve element cells (Fig. 5, D and E) and cambial cells (Fig. 5, F and G). The antibody used here was constructed from the N-terminal part of the hipI-SOD (Schinkel et al., 2001) and thus theoretically should not be able to distinguish between the different hipI-SOD forms. Nevertheless, the results show that the transcriptional expression detected in the different tissues results in a protein product localized to the secondary or primary cell wall in phloem fibers, cambium, xylem, and sieve elements in the vascular tissue.
Figure 5.
Immunogold labeling of hipI-SOD in the stem (10th internode) of hybrid aspen plants. A, Light microscopic image of a cross section after histological staining with methylene blue/Azur II. B to K, Transmission electron microscopy images of significant immunogold labeling of hipI-SOD in the secondary cell walls of the sclerenchyma (B and C), developing xylem (H and I), mature xylem (J and K), and the primary cell wall of sieve elements of the phloem (D and E) and cambium cells (F and G). L and M, Control staining for mature xylem with preimmune serum. Boxes in B, D, F, H, J, and L indicate the regions shown at higher magnification in C, E, G, I, K, and M, respectively. Ca, Cambium; Ph, phloem; pCW, primary cell wall; Sc, sclerenchyma; sCW, secondary cell wall; SE, sieve element; Xy, xylem. Bars = 20 μm (A), 2 μm (B, D, F, H, J, and L), and 200 nm (C, E, G, I, K, and M).
Mass Spectrometric Analysis of hipI-SOD Proteins
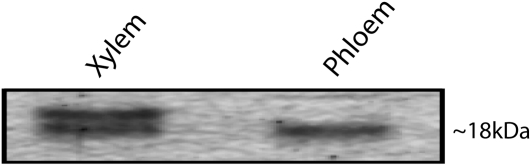
The expression patterns of hipI-SOD transcripts indicated that they have high degrees of tissue specificity. In order to separate the different isoforms, we purified hipI-SOD proteins from phloem (with attached cambium) and xylem samples, separated them by duplicate SDS-PAGE, and used one of the SDS gels for immunoblotting to check the presence of hipI-SOD proteins. In western-blot analysis, one band of approximately 18 kD and two bands close to 18 kD were found in the phloem and xylem tissue, respectively (Fig. 6). These results are similar to those presented previously by Srivastava et al. (2007). All of the bands identified as hipI-SODs by western blotting were extracted from the other stained gel, trypsin digested, and analyzed by mass spectrometry. Several peptides belonging to hipI-SODs were identified from both xylem and phloem samples (Fig. 2, where each of these peptides is represented by a different color). The residues that differ among the identified peptides are shown in white letters. These peptides are specific for the respective forms of hipI-SOD. Detailed analysis of the mass spectrometric data showed that among all identified peptides, two were common to all three hipI-SODs (hipI-SODC1b, hipI-SODC1s, and hipI-SODC2), and specific peptides belonging to hipI-SODC1s and hipI-SODC2 were identified. In the phloem tissue sample, in addition to common peptides, only specific peptides belonging to hipI-SODC1s were identified, whereas in xylem tissue samples, specific peptides for both hipI-SODC2 and hipI-SODC1s as well as common peptides of all isoforms were identified. Since all three hipI-SOD transcripts were found throughout the vascular tissue (Fig. 4) but their respective proteins did not exhibit a similar pattern, it might be concluded that the level of hipI-SOD proteins is restricted to a particular tissue and is independent of its mRNA level. Brockmann et al. (2007) stated that protein concentrations are determined by their corresponding mRNA concentrations in only 20% to 40% of cases. We were unable to identify any specific peptide belonging to the extended exon 6 of hipI-SODC1b from the mass spectrometry data, possibly because the expression of hipI-SODC1b is too weak and unstable and/or it is mainly involved in posttranscriptional regulation at the transcript level. Graveley (2001) discussed the fact that inappropriately spliced transcripts are either translated, in which case they usually make abnormal nonfunctional proteins, or are degraded via the mRNA surveillance system (e.g. in eukaryotes) to prevent the synthesis of potentially harmful proteins. Perhaps hipI-SOD proteins themselves affect hipI-SOD splicing in some way in order to maintain the required level within a cell (e.g. negative or positive feedback regulation). In C. elegans, the major targets of the mRNA surveillance system are not the aberrantly spliced transcripts but the deliberately spliced ones, as part of a complex autoregulatory system (Mitrovich and Anderson, 2000).
Figure 6.
hipI-SOD protein expression patterns in Populus. Western blot of partially purified protein from xylem and phloem tissues of a Populus wild-type plant. The size of the band on the blot is indicated at right.
Expression of hipI-SOD Proteins in Escherichia coli
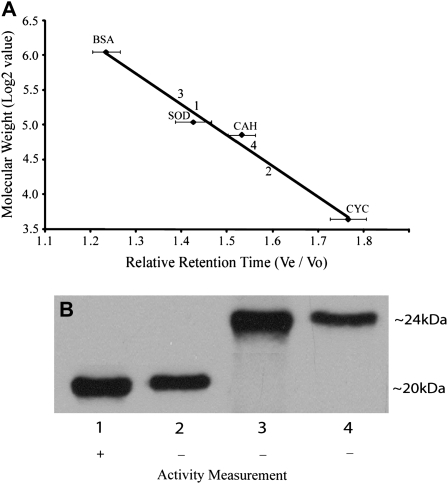
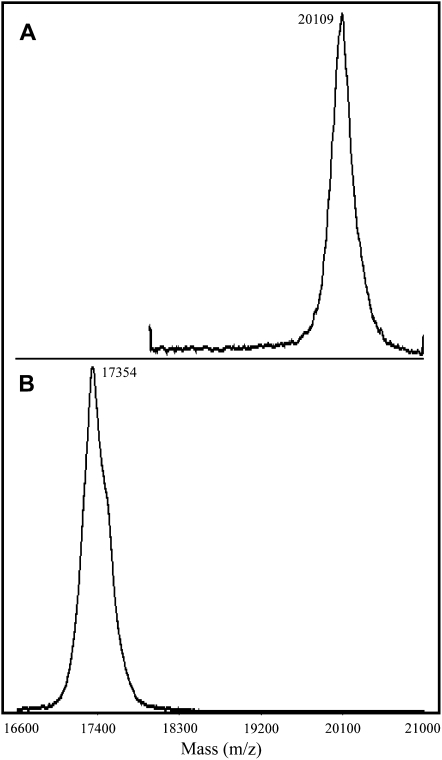
Since we had acquired evidence of the presence of hipI-SODC1b transcripts, but no specific data regarding the corresponding protein, this gene was expressed in E. coli to check the activity of its product. The hipI-SODC1s protein was also expressed in the same manner. The expression of hipI-SODC1s and hipI-SODC1b proteins was confirmed both by immunoblotting using hipI-SOD-specific antibodies and mass spectrometry (data not shown). Because both the recombinant proteins were expressed as inclusion bodies, an on-column folding of insoluble His6-tagged recombinant proteins strategy was used, as described by Zhu et al. (2005). The inclusion bodies were dissolved in urea, and the denatured hipI-SOD proteins were subjected to refolding on an immobilized metal affinity chromatography (IMAC) column. IMAC has been proved to be one of the best choices for simultaneous refolding and purification of His6-tagged recombinant proteins (Zhu et al., 2005). Several proteins expressed as inclusion bodies have been successfully renatured on-column and found to be biologically active (Zhu et al., 2005; Wang et al., 2007). After refolding, the purity of hipI-SOD proteins in the IMAC eluate was found to be high (>95%), since only the hipI-SOD protein band was visible on Coomassie Brilliant Blue-stained SDS gels (data not shown). The recovery of recombinant hipI-SODC1b after purification was lower than that for hipI-SODC1s. This may be the result of improper folding of hipI-SODC1b, which leads to an unstable structure so that precipitation may occur inside the column. Activity measurement by the pyrogallol method (Marklund, 1985) demonstrated that recombinant hipI-SODC1s was active but recombinant hipI-SODC1b was not. On the basis of these results, it appears that the presence of additional amino acids at the C terminus may interfere with proper folding and dimerization of hipI-SODC1b, thus making it nonactive. SOD has been shown to be more active as a dimer than in the monomeric form (Zhu et al., 2005). The oligomerization state of both recombinant proteins has been estimated after separation by gel filtration chromatography. All of the fractions collected were subsequently tested by western blots for the presence of hipI-SOD. After comparison with the size and relative retention time of standard proteins, the size range of fractions containing hipI-SOD protein was determined (Fig. 7A). These results indicated that fractions 1 and 3 contain dimer subunits, while fractions 2 and 4 contain monomer subunits for their respective recombinant proteins. Despite the expected molecular mass of hipI-SODC1s and hipI-SODC1b recombinant proteins (approximately 17 and 20 kD, respectively) also confirmed by matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS; Fig. 8), their migration on SDS-PAGE was found to be different, with apparent molecular masses of approximately 20 kD for hipI-SODC1s and approximately 24 kD for hipI-SODC1b (Fig. 7B). Irrespective of their exact size, hipI-SOD proteins have previously been shown to migrate differently on Tris-Gly SDS-PAGE (Srivastava et al., 2007).
Figure 7.
Gel filtration chromatography and activity test for hipI-SOD proteins. A, The system was calibrated by prerunning the following protein standards three times: bovine serum albumin (BSA; 67 kD), bovine Cu/Zn-SOD (SOD; 32.6 kD), carbonic anhydrase (CAH; 29 kD), and cytochrome c (CYC; 12.5 kD). Horizontal bars represent sds. B, Lanes 1 (size range, 41–31 kD) and 2 (size range, 24–15 kD) are fractions of hipI-SODC1s, and lanes 3 (size range, 47–36 kD) and 4 (size range, 28–16 kD) are fractions of hipI-SODC1b recombinant proteins collected after gel filtration, in which hipI-SOD proteins were found. The sizes of the bands on the blot are indicated at right. Activity in all of these fractions was measured by the pyrogallol method (Marklund, 1985). + indicates activity found, and – indicates no activity found.
Figure 8.
MALDI-MS of hipI-SOD proteins. MALDI spectra of hipI-SODC1b (A) and hipI-SODC1s (B) proteins in linear mode (see “Materials and Methods” for details).
Only fraction 1 exhibited SOD activity (Fig. 7). These results indicate that expressed hipI-SODC1s is functionally active as a dimer while its monomeric form is inactive. In spite of forming a dimer, hipI-SODC1b exhibited no activity, like its monomeric form. Similarly, one of the alternatively spliced forms of PEPc kinase in tomato has been found to encode an enzymatically inactive protein (Marsh et al., 2003).
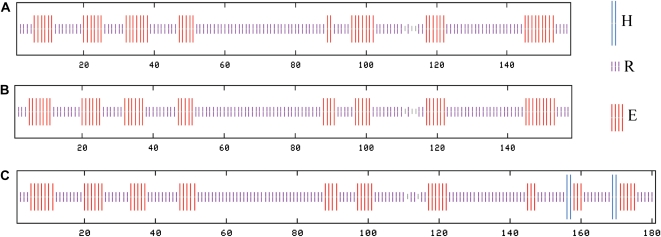
For full catalytic function, SOD requires both disulfide formation and metal coordination (Hornberg et al., 2007). From the predicted consensus secondary structures obtained for all hipI-SOD proteins (Fig. 9), we can see that all proteins are mainly composed of β-strands and some random coils, but hipI-SODC1b also contains two predicted short α-helices close to its C-terminal region. We infer that the presence of the additional β-strand and α-helices at the C terminus of hipI-SODC1b may lead to conformational changes in its catalytic active region, resulting in an inactive form. It has been stated that loops IV and VII of SOD form part of the active-site channel that connects the protein surface to the active site (Hornberg et al., 2007). Arg-143 and Cys-146 have been shown to be important amino acids for retaining these structures (Bertini et al., 1994). Both of these amino acids are in close proximity to the extra 23 amino acids in hipI-SODC1b. In mutated SOD, a slight movement of Arg-143 was accompanied by a 10-fold reduction in enzymatic turnover (Hornberg et al., 2007).
Figure 9.
Secondary structure prediction results of hipI-SOD proteins. The prediction models used for this analysis include DPM, DSC, GOR4, HNNC, Predator, SIMPA96, and SOPM. A, hipI-SODC2. B, hipI-SODC1s. C, hipI-SODC1b. H, α-Helix; R, random coils; E, β-strand. [See online article for color version of this figure.]
CONCLUSION
In plants, AS is common and generates tremendous transcriptome and proteome complexity, and many studies have shown that it is regulated by tissue-specific developmental signals and stresses. When comparing the genomes from different plant species, or even different organisms, there is also a high level of conservation (Reddy, 2007).
We have found that many genes in the Populus ROS gene network are AS and that many of the closest homologs of these genes in Arabidopsis are not reported to be AS. This type of gene regulation might thus be an important mechanism for diversification between plant species, leading to specific traits. hipI-SOD, shown to regulate ROS level and plant development (Srivastava et al., 2007), is one of the AS genes we found in the Populus ROS gene network. Transcript analysis showed that the splice variant hipI-SODC1b was expressed in cambial and xylem regions. Furthermore, the additional secondary structure in its C-terminal region suggests an interference with proper dimerization, which might lead to conformational changes in its catalytically active region, resulting in an inactive and/or instable form. Similarly, a growing body of evidence suggests that a subset of mutations located close to the dimeric interface can lead to a major destabilization of mouse Cu/Zn-SOD1 (Hough et al., 2004; Hornberg et al., 2007). Additional types of possible regulation at the posttranscriptional level (e.g. negative feedback regulation of hipI-SOD gene expression) have to be explored in future experiments.
MATERIALS AND METHODS
The Search for AS Sites in the ROS Gene Network
All high-annotation-quality EST clusters related to the ROS gene network were selected from the Populus EST database (PopulusDB; http://www.populus.db.umu.se/; Sterky et al., 2004), and their closest corresponding gene models were identified from the Populus genome database (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). All of the EST contigs belonging to each cluster were then aligned with each other and compared with the corresponding cDNA and gene models using the MultAlin software tool (Corpet, 1988) and SplicePredictor (http://deepc2.psi.iastate.edu/cgi-bin/sp.cgi). Each alignment was manually investigated three times to check the locations and types of AS. Pairs of EST contigs showing no differences were not further analyzed, and those sharing two consecutively matching regions flanking a discontinuity in the alignment were aligned with their genomic sequences. A pair was only considered to be alternatively spliced if it had at least 25-bp-long alignments, followed by an insertion or deletion of at least 5 bp, followed by a subsequent match of at least 25 bp, as described by Ner-Gaon et al. (2007). EST clusters containing only one contig were not used in this study. To avoid contamination from paralogs, two or more contigs were only included in the AS analysis if they corresponded to the same gene model, according to their best alignments with the genome. Contigs belonging to the same clusters but corresponding to different gene models were excluded from the AS investigation.
The selected genes of the ROS network were also used for sequence pattern analysis at the 5′ and 3′ splice site of 165 introns. Ten nucleotides (five from the exon and five from the intron) at each 5′ and 3′ splice site were extracted from the annotated sequence of each gene in the Populus genome database.
hipI-SOD ESTs and Gene Models
Four ESTs assembled into two contigs for hipI-SOD have been identified in PopulusDB (POPLAR.5824.C1 and POPLAR.5824.C2), as have their closest gene models in the Populus genome database: estExt_fgenesh4_pg.C_LG_XIX0416 (PthipI-SODC1) and estExt_Genewise1_v1.C_LG_XIII1983 (PthipI-SODC2), respectively (Srivastava et al., 2007). Deduced amino acid sequences of all hipI-SOD proteins from Populus (hipI-SODC1s, hipI-SODC1b, and hipI-SODC2; GenBank accession nos. ACJ13748, ACJ13749, and CAC33847, respectively) were aligned using GeneDoc (Nicholas et al., 1997; http://www.psc.edu/biomed/genedoc).
Plant Material, Isolation of RNA, Semiquantitative Reverse Transcription-PCR, and Cloning
Total RNA was extracted from xylem, phloem, and cambium tissues of actively growing (10-week-old) hybrid aspen (Populus tremula × Populus tremuloides) and aspen (Populus tremula) plants using an Aurum Total RNA Mini Kit (Bio-Rad) according to the manufacturer's protocol. First-strand cDNAs were synthesized from 1 μg of total RNA using an iScript cDNA Synthesis Kit (Bio-Rad) and then used as templates for reverse transcription (RT)-PCR amplification. As a loading control, the housekeeping ubiquitin (UBQ; Brunner et al., 2004) gene was amplified using specific primers (Table II) under the same conditions as those for the hipI-SOD amplifications. All of the PCR amplifications were repeated independently three times. 2× PCR Master Mix (Fermentas) was used for all PCRs. After electrophoresis, reaction mixtures were separated on 2% (w/v) agarose gels and then stained with ethidium bromide. Bands were excised from the gels, and PCR products were purified using a Gel Extraction Kit (Qiagen Nordic) and cloned using a TA cloning kit (Invitrogen) according to the manufacturer's instructions. For sequence confirmation, three clones from each of the inserts were sequenced.
Protein Extraction, Purification, and In-Gel Digestion
Proteins were extracted and purified from phloem (cambium attached) and xylem tissues of hybrid aspen plants as described by Srivastava et al. (2007). Approximately 20 μg of purified protein was separated on duplicate 14% SDS-polyacrylamide gels using a MiniProtean II electrophoresis assembly (Bio-Rad). One of the gels was stained with Coomassie Brilliant Blue, and the other was electroblotted onto a nitrocellulose membrane (Hybond-ECL; GE Healthcare). Western-blot analysis was performed using a specific hipI-SOD antibody as described previously (Srivastava et al., 2007). After comparison with the immunoblots, bands corresponding to hipI-SOD were excised from the Coomassie Brilliant Blue-stained gel and digested in-gel as described by Backstrom et al. (2007). Briefly, all of the selected bands were washed, reduced, alkylated, and digested in-gel with sequencing-grade modified trypsin (Promega/SDS Biosciences). The resulting peptides were extracted, dried, and redissolved in 0.1% formic acid for mass spectrometric analysis.
Mass Spectrometry and Data Analysis
Peptide analysis was done by reverse-phase liquid chromatography-electrospray ionization-tandem mass spectrometry as described by Backstrom et al. (2007) using a capillary HPLC system coupled to a quadrupole time-of-flight mass spectrometer (CapLC Q-TOF Ultima; Waters Corp.) without any further changes.
Raw data were processed by ProteinLynx Global Server (version 2.2.5) software for database searches. Proteins were identified by an in-house Mascot server and the Mascot Daemon application (version 2.1.6; Matrix Science; www.matrixscience.com) using the Populus EST database (Sterky et al., 2004). The following settings were used for the database search: trypsin-specific digestion with one missed cleavage allowed, carbamidomethylated Cys set as fixed modification, oxidized Met and deamidation in variable mode, peptide tolerance of 100 ppm, and fragment tolerance of 0.1 D. Peptides with Mascot ion scores exceeding the threshold for statistical significance (P < 0.05) were selected. For reconfirmation of the significant peptides, those that were unique for their respective form of hipI-SOD proteins were also manually reprocessed, checked, and found to match the criteria of Mascot ion score with at least four consecutive y- or b-ions with a significant signal-to-background ratio.
Prior to analyzing molecular sizes of recombinant hipI-SOD proteins by MALDI-MS, samples were purified with 3M Empore C8 extraction discs (3M Bioanalytical Technologies). A microcolumn was prepared by stamping out a small plug of C8 disc and placing it in a GELoader tip (Eppendorf) as described by Thingholm et al. (2006). Small portions from eluates obtained after IMAC purification of recombinant proteins were acidified by trifluoroacetic acid (TFA) to its final concentration of 1%. The acidified protein was loaded onto the column after equilibrating it with 0.1% TFA. After washing the column with 0.1% TFA, proteins were eluted with 80% acetonitrile containing 0.1% TFA directly on the MALDI plate. The matrix used was a ready-made solution of sinapic acid (G2038A; Agilent Technologies). MALDI-TOF analysis was performed using a Voyager-DE STR mass spectrometer operated in positive linear mode. Data from 300 laser shots (337-nm nitrogen laser) were collected, signal averaged, and processed with the instrument manufacturer's Data Explorer software. External mass calibration was carried out using [M+H]+ ions of horse myoglobin.
Light Microscopy, Transmission Electron Microscopy, and Immunolocalization
Samples from the 10th internode of hybrid aspen plants were used for fixation, substitution, resin embedding, and immunogold labeling as described previously (Karpinska et al., 2001). For light microscopy, semithin sections (approximately 3 μm thick) were cut from the embedded samples, mounted on slides, and stained for 2 min with 1% (w/v) methylene blue/1% (w/v) Azur II in 1% (w/v) aqueous borax at 60°C prior to light microscopic examination with a Zeiss Axiovert 135 microscope.
Cloning, Expression, and Purification of Recombinant hipI-SOD Proteins
First template plasmids were transformed and amplified in Escherichia coli strain DH5α, and NdeI and SmaI sequences were introduced into forward and reverse primers, respectively. hipI-SODC1s and hipI-SODC1b cDNAs were amplified by PCR from their EST clones (FJ393058 and FJ393059, respectively) with the help of Pwo DNA polymerase (Roche Diagnostics). Both the amplified hipI-SOD coding sequences were subcloned into the NdeI and SmaI sites of linearized pIVEX2.3d plasmid (Roche) to construct pIVEX2.3d-hipI-SODC1s and pIVEX2.3d-hipI-SODC1b expression vectors, respectively. In addition to sequencing, the expression vectors were also checked by digestion using restriction endonucleases purchased from Fermentas. Each plasmid construct was then transformed into the BL21 (DE3) E. coli strain to overexpress His6-tagged hipI-SOD recombinant proteins. Briefly, bacterial cells were cultured overnight at 37°C in Luria-Bertani medium containing 100 μg mL−1 ampicillin, 500 μm Cu2+, and 100 μm Zn2+. Bacterial cells containing only the pIVEX2.3d plasmid, without any insert, were also cultured, in parallel, as a negative control. Cell pellets from both the negative control culture and the culture with hipI-SOD inserts were collected and analyzed by 14% SDS-PAGE; it was found after cell lysis that hipI-SOD proteins were expressed as inclusion bodies.
A cell pellet from 500 mL of culture was resuspended in 25 mL of lysis buffer (25 mm Tris-HCl, pH 8.0, and 500 mm NaCl) and disrupted by passing it twice through a French press at 6.9 MPa (1,000 p.s.i.). The lysates were centrifuged at 18,500g for 30 min at 4°C. The pellet, containing inclusion bodies, was washed three times with lysis buffer. Purified inclusion bodies were solubilized in 25 mm Tris-HCl (pH 8.0) containing 8 m urea and 2 mm β-mercaptoethanol for 2 h at room temperature. His6-tagged hipI-SOD proteins were purified under denaturing conditions using a HiTrap chelating column (GE Healthcare) and then refolded on the column as described by Zhu et al. (2005). The IMAC eluates were concentrated and applied onto an FPLC column (Superdex 75; Pharmacia) preequilibrated with equilibration buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1.5 m urea, 1 mm reduced glutathione, and 0.1 mm oxidized glutathione) at a flow rate of 0.75 mL min−1. Different fractions were collected over a broad range, and those in which hipI-SOD proteins were detected by western blot were checked for SOD activity by the pyrogallol method (Marklund, 1985).
Prediction of Secondary Structure
Several secondary structure prediction models (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html; Combet et al., 2000), including DPM, DSC, GOR4, HNNC, Predator, SIMPA96, and SOPM, were used in combination to predict the consensus secondary structures of the deduced hipI-SOD proteins.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FJ393058, FJ393059, AJ278671, ACJ13748, ACJ13749, and CAC33847.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. cDNA sequences of hipI-SODC1s (FJ393058), hipI-SODC1b (FJ393059), and hipI-SODC2 (AJ278671; A) and genomic sequences of PthipI-SODC1 (estExt_fgenesh4_pg.C_LG_XIX0416) and PthipI-SODC2 (estExt_Genewise1_v1.C_LG_XIII1983; B). Red, Exons; black, introns; blue, UTRs.
Supplemental Table S1. List of ROS-related EST clusters selected for the study of AS in Populus.
Supplemental Table S2. Description of positions and types of various AS found within genes shown in Table I.
Supplementary Material
Acknowledgments
We thank Tatiana Shutova and Göran Samuelsson for kindly providing and helping with the protein purification system. We are also grateful to Laszlo Bako and Lena Tibell for their excellent technical assistance.
This work was supported by grants to the Swedish University of Agricultural Sciences from the Swedish Council for FORMAS/SIDA, the Swedish Research Council, and the Swedish Foundation for Strategic Research, by the Swedish Foundation for National Cooperation in Research and Higher Education, and by the Kempe Foundation. The instruments and bioinformatics infrastructure of Umeå Protein Analysis Facility were supported by the Wallenberg Foundation and the Kempe Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gunnar Wingsle (gunnar.wingsle@genfys.slu.se).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5 773–782 [DOI] [PubMed] [Google Scholar]
- Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26 717–729 [DOI] [PubMed] [Google Scholar]
- Bertini I, Piccioli M, Viezzoli MS, Chiu CY, Mullenbach GT (1994) A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase. Eur Biophys J 23 167–176 [DOI] [PubMed] [Google Scholar]
- Brendel V, Kleffe J, Carle-Urioste JC, Walbot V (1998) Prediction of splice sites in plant pre-mRNA from sequence properties. J Mol Biol 276 85–104 [DOI] [PubMed] [Google Scholar]
- Brockmann R, Beyer A, Heinisch JJ, Wilhelm T (2007) Posttranscriptional expression regulation: what determines translation rates? PLoS Comput Biol 3 0531–0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deléage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25 147–150 [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Vossem JH, Vermeer JEM, Vroomen MJ, Gadella TWJ, Haring MA, Cornelissen BJC (2003) The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol 133 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Hongbin W, Bing L, Jinfa W (2006) Cloning and characterization of a novel splicing isoform of the iron-superoxide dismutase gene in rice (Oryza sativa L.). Plant Cell Rep 24 734–742 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Fujii M, Ishii N, Joguchi A, Yasuda K, Ayusawa D (1998) A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res 5 25–30 [DOI] [PubMed] [Google Scholar]
- Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17 100–107 [DOI] [PubMed] [Google Scholar]
- Hornberg A, Logan DT, Marklund SL, Oliveberg M (2007) The coupling between disulphide status, metallation and dimer interface strength in Cu/Zn-superoxide dismutase. J Mol Biol 365 333–342 [DOI] [PubMed] [Google Scholar]
- Hough MA, Grossmann JG, Antonyuk SV, Strange RW, Doucette PA, Rodriguez JA, Whitson LJ, Hart PJ, Hayward LJ, Valentine JS, et al (2004) Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc Natl Acad Sci USA 101 5976–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Hirochika H, Kurata N (2002) Organ-specific alternative transcripts of KNOX family class 2 homeobox genes of rice. Gene 288 41–47 [DOI] [PubMed] [Google Scholar]
- Jansson S, Douglas CJ (2007) Populus: a model system for plant biology. Annu Rev Plant Biol 58 435–458 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Melzer M, Prokhorenko I, Johansson T, Wingsle G (2005) Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J Exp Bot 56 2085–2093 [DOI] [PubMed] [Google Scholar]
- Karpinska B, Karlsson M, Schinkel H, Streller S, Suss KH, Melzer M, Wingsle G (2001) A novel superoxide dismutase with a high isoelectric point in higher plants: expression, regulation and protein localization. Plant Physiol 126 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284 654–657 [DOI] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Green RE, Bhatnagar RS, Brenner SE (2004) The evolving roles of alternative splicing. Curr Opin Struct Biol 14 273–282 [DOI] [PubMed] [Google Scholar]
- Liochev S, Fridovich I (1994) The role of superoxide anion radicals in the production of hydroxyl radicals: in vitro and in vivo. Free Radic Biol Med 16 29–33 [DOI] [PubMed] [Google Scholar]
- Marklund SL (1985) Pyrogallol autooxidation. In RA Greenwald, ed, Handbook of Methods for Oxygen Radical Research. CRC Press, Boca Raton, FL, pp 243–247
- Marsh JT, Sullivan S, Hartwell J, Nimmo HG (2003) Structure and expression of phosphoenolpyruvate carboxylase kinase genes in Solanaceae: a novel gene exhibits alternative splicing. Plant Physiol 133 2021–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H (2003) Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J Biol Chem 278 18945–18952 [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, Anderson P (2000) Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev 14 2173–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Breusegem FV (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30 13–19 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39 877–885 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Leviatan N, Rubin E, Fluhr R (2007) Comparative cross-species alternative splicing in plants. Plant Physiol 144 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DWK, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEWNEWS 4 14 [Google Scholar]
- Overmyer K, Brosche M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8 335–342 [DOI] [PubMed] [Google Scholar]
- Pajares M, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM (2007) Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol 8 349–357 [DOI] [PubMed] [Google Scholar]
- Reddy ASN (2007) Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58 267–294 [DOI] [PubMed] [Google Scholar]
- Scandalios JG (2005) Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz J Med Biol Res 38 995–1014 [DOI] [PubMed] [Google Scholar]
- Schinkel H, Hertzberg M, Wingsle G (2001) A small family of novel Cu/Zn-superoxide dismutases with high isoelectric points in hybrid aspen. Planta 213 272–279 [DOI] [PubMed] [Google Scholar]
- Schinkel H, Streller S, Wingsle G (1998) Multiple forms of extracellular superoxide dismutase in needles, stem tissues and seedlings of Scots pine. J Exp Bot 49 931–936 [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G (2004) A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53 1305–1319 [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, Pozo OD, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99 11640–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Schinkel H, Witzell J, Hertzberg M, Torp M, Srivastava MK, Karpinska B, Melzer M, Wingsle G (2007) Downregulation of high-isoelectric-point extracellular superoxide dismutase mediates alterations in the metabolism of reactive oxygen species and developmental disturbances in hybrid aspen. Plant J 49 135–148 [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H (2005) Functions of alternative splicing. Gene 344 1–20 [DOI] [PubMed] [Google Scholar]
- Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Campaa L, Jonsson Lindvall J, Tandre K, Strauss SH, et al (2004) A Populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101 13951–13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Wang J, Yu J, Huang X, Gu X (2006) Evolution of alternative splicing after gene duplication. Genome Res 16 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki M, Tsugawa H, Minami E, Kayano T, Yamamoto N, Kano-Murakami Y, Matsuoka M (1995) Alternative RNA products from a rice homeobox gene. Plant J 7 927–938 [DOI] [PubMed] [Google Scholar]
- Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protocols 1 1929–1935 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Valdivia HH (2007) One gene, many proteins: alternative splicing of the ryanodine receptor gene adds novel functions to an already complex channel protein. Circ Res 100 761–763 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genome wide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang L, Geng X (2007) Renaturation with simultaneous purification of rhG-CSF from Escherichia coli by ion exchange chromatography. Biomed Chromatogr 21 1291–1296 [DOI] [PubMed] [Google Scholar]
- Wiebauer K, Herrero JJ, Filipowicz W (1988) Nuclear pre-mRNA processing in plants: distinct modes of 3′-splice-site selection in plants and animals. Mol Cell Biol 8 2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Weber DJ, Carrier F (2006) Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res 34 1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XQ, Li SX, He HJ, Yuan QS (2005) On-column refolding of an insoluble His6-tagged recombinant EC-SOD overexpressed in Escherichia coli. Acta Biochim Biophys Sin (Shanghai) 37 265–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.