Abstract
Jasmonic acid (JA) is involved in plant development and the defense response. Transgenic overexpression of the Arabidopsis (Arabidopsis thaliana) jasmonic acid carboxyl methyltransferase gene (AtJMT) linked to the Ubi1 promoter increased levels of methyl jasmonate (MeJA) by 6-fold in young panicles. Grain yield was greatly reduced in Ubi1:AtJMT plants due to a lower numbers of spikelets and lower filling rates than were observed for nontransgenic (NT) controls. Ubi1:AtJMT plants had altered numbers of spikelet organs, including the lemma/palea, lodicule, anther, and pistil. The loss of grain yield and alteration in spikelet organ numbers were reproduced by treating NT plants with exogenous MeJA, indicating that increased levels of MeJA in Ubi1:AtJMT panicles inhibited spikelet development. Interestingly, MeJA levels were increased by 19-fold in young NT panicles upon exposure to drought conditions, resulting in a loss of grain yield that was similar to that observed in Ubi1:AtJMT plants. Levels of abscisic acid (ABA) were increased by 1.9- and 1.4-fold in Ubi1:AtJMT and drought-treated NT panicles, respectively. The ABA increase in Ubi1:AtJMT panicles grown in nondrought conditions suggests that MeJA, rather than drought stress, induces ABA biosynthesis under drought conditions. Using microarray and quantitative polymerase chain reaction analyses, we identified seven genes that were regulated in both Ubi1:AtJMT and drought-treated NT panicles. Two genes, OsJMT1 and OsSDR (for short-chain alcohol dehydrogenase), are involved in MeJA and ABA biosynthesis, respectively, in rice (Oryza sativa). Overall, our results suggest that plants produce MeJA during drought stress, which in turn stimulates the production of ABA, together leading to a loss of grain yield.
Rice (Oryza sativa), the model system for the study of monocotyledonous plants, is a cereal crop consumed by more than half of the world's population. As such, improvements in grain yield are an important focus of research. Since rice plants grow in a paddy field, they are susceptible to water stress and in particular to drought (Yang et al., 2004, 2007). Approximately 20% of the total worldwide rice growing area is prone to drought (Pandey et al., 2007), and drought is one of the major constraints to rice production worldwide. Although drought conditions can alter the growth and development of rice at any time during its life cycle, drought stress during reproductive growth, but not during vegetative growth, results in a loss of grain yield.
Immediately following the transition of rice plants to the reproductive phase, the vegetative meristem is converted into the panicle meristem. The panicle meristem subsequently differentiates in an orderly fashion into primary branches, secondary branches, and spikelet meristems (Ikeda et al., 2004). Rice grain yield is determined by four parameters: number of panicles per plant, number of spikelets per panicle, filling rate, and total seed weight. The number of panicles and spikelets is determined soon after formation of the panicle and spikelet meristems (Sakamoto and Matsuoka, 2008). Drought exposure during the earlier stages of this transition affects the first two parameters more than the other parameters, while drought exposure at later stages of reproductive development affects the filling rate more than the other parameters. The total seed weight is the combined result of the other three parameters. The development of the panicle and/or spikelet meristem is repressed under the drought conditions, resulting in a reduction in the number of panicles per plant and/or the number of spikelets per panicle (O'Toole and Namuco, 1983; Boonjung and Fukai, 1996; Wopereis et al., 1996; Asch et al., 2005).
Abscisic acid (ABA) has been implicated in a reduction of grain yield following water stress during reproductive plant development. ABA levels were increased upon exposure of plants to drought conditions, which reduces the filling rate by increasing sterility in cereal plants (Morgan, 1980; Ober et al., 1991; Beltrano et al., 1999; Yang et al., 2004, 2007). These studies focused on plant hormone levels during the late stages of reproductive development, including meiosis and/or grain-filling periods following exposure to drought conditions. However, studies on plant hormone levels during the early stages of reproductive development, which determine the number of panicles and spikelets, are currently lacking.
Methyl jasmonate (MeJA) and jasmonic acid (JA) are important cellular regulators involved in diverse plant developmental processes, including seed germination (Nojavan-Asghari and Ishizawa, 1998), callus growth (Ueda and Kato, 1982), primary root growth (Staswick et al., 1992), flowering (Albrechtová and Ullmann, 1994), formation of gum and bulb (Saniewski et al., 1998), and senescence (Ueda and Kato, 1980). They are also involved in plant defense responses to insect wounding, attack by various pathogens, and water deficit (Creelman and Mullet, 1995; Wasternack and Parthier, 1997; Seo et al., 2001; Rakwal et al., 2002). The levels of endogenous jasmonates were reported to be increased following pathogen exposure (Reymond and Farmer, 1998; Thomma et al., 1998). Likewise, exogenous application of jasmonates to plants induced stress-related or pathogenesis-related (PR) genes (Moons et al., 1997; Mei et al., 2006). Thus, the role(s) of jasmonate in response to biotic stresses has been well documented; however, relatively little is known about its involvement in response to abiotic stress. Jasmonate levels were increased in soybean (Glycine max; Creelman and Mullet, 1995) and Pinus pinaster (Pedranzani et al., 2007) upon plant exposure to drought and in tomato (Solanum lycopersicum; Pedranzani et al., 2003) and Iris hexagona (Wang et al., 2001) upon exposure to high salinity. In rice, both drought and high salinity increased jasmonate levels in the leaves and roots, resulting in the induction of stress-related PR and JA biosynthetic genes (Moons et al., 1997; Kiribuchi et al., 2005; Tani et al., 2008). These abiotic stress-induced increases in jasmonate levels were observed only in vegetative tissues. Whether drought conditions increase jasmonate levels in reproductive organs remains to be determined.
The jasmonic acid carboxyl methyltransferase (JMT) enzyme converts JA to a volatile component, MeJA. Expression of endogenous JMT was not detected in young seedlings, but its expression was initiated in conjunction with the nectar in the developing flowers of Chinese cabbage (Brassica campestris pekinensis; Song et al., 2000). Transgenic overexpression of the Arabidopsis (Arabidopsis thaliana) JMT gene (AtJMT) in Arabidopsis plants using the 35S promoter increased leaf MeJA levels by 3-fold (Seo et al., 2001). This overexpression also caused a significant decline in transgenic Arabidopsis seed production (Cipollini, 2007). In this study, we attempted to show that MeJA plays an important role in stress-induced loss of grain yield. Transgenic overexpression of AtJMT in rice resulted in a large reduction in grain yield through increased MeJA and ABA levels in young panicles. Exposure of nontransgenic plants to drought conditions also increased MeJA and ABA levels in young panicles and significantly reduced grain yield, in a similar manner to that observed in AtJMT plants. These results suggest a role for MeJA in the plant response to drought stress.
RESULTS
Ubi1:AtJMT Transgenic Rice That Produce High Levels of MeJA in Their Panicles
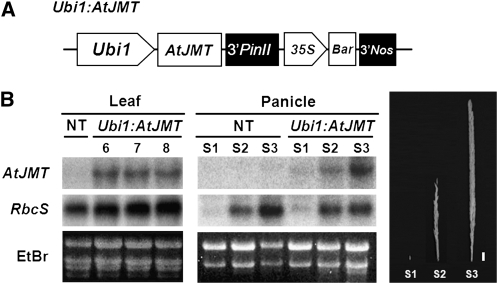
To study the role of MeJA in reproductive development of rice panicles, rice were transformed with the construct Ubi1:AtJMT (Fig. 1A), in which AtJMT was under the control of the maize (Zea mays) ubiquitin1 promoter, including its first intron (Ubi1; Christensen and Quail, 1996). Twelve independent transgenic lines were produced using the Agrobacterium tumefaciens-mediated transformation method (Hiei et al., 1994). Of the T1-6 seeds collected, T4-6 homozygous seeds were used for further analysis. Since phenotype was similarly observed in more than three independent transgenic lines, we chose one for extensive study. RNA gel-blot analysis showed that AtJMT was expressed in the leaves and developing panicles of Ubi1:AtJMT plants (Fig. 1B). In the panicles, AtJMT expression increased with development from the 1.5-cm-long young panicle (S1) to the fully developed panicle just before the emergence stage (S3; Fig. 1B). Such an increase in expression levels of AtJMT appears to be due to different activities of the Ubi1 promoter in developing panicles (Cornejo et al., 1993). To our knowledge, there is no previous report that directly describes the activity of the Ubi1 promoter in panicles. MeJA levels were assessed in S1 panicles from transgenic plants and nontransgenic (NT) controls (Table I). Ubi1:AtJMT panicles had MeJA levels that were 6-fold higher than those of NT panicles. Grain yield, number of spikelets per panicle, and filling rate were also largely reduced in Ubi1:AtJMT plants as compared with NT controls, a typical phenomenon observed in plants exposed to drought stress. These observations led us to investigate whether MeJA levels were increased by drought stress during reproductive development. Since similar phenotypes were observed in more than three independent transgenic lines, we chose one for further study. NT plants were treated with drought stress at the panicle initiation stage (approximately 30–35 d before heading), and levels of MeJA were measured in S1 panicles. MeJA was 19-fold higher in drought-treated NT panicles than in untreated NT panicles. Levels of ABA, another plant stress hormone, were increased by 1.4-fold in the drought-treated NT panicles as compared with untreated NT panicles. ABA levels were 1.9-fold higher in S1 panicles from Ubi1:AtJMT plants than in S1 panicles from untreated NT control plants under nondrought conditions, suggesting that ABA levels were increased by MeJA rather than by drought stress.
Figure 1.
Production of Ubi1:AtJMT transgenic rice plants. A, Ubi1:AtJMT consists of the maize ubiquitin1 promoter (Ubi1) linked to the AtJMT coding region from Arabidopsis (AY008434; Seo et al., 2001), the 3′ region of the potato (Solanum tuberosum) proteinase inhibitor II gene (3′PinII), and a bar expression cassette containing the 35S promoter of the Cauliflower mosaic virus, the bar coding region, and the 3′ region of the nopaline synthase gene (3′Nos). B, RNA gel-blot analysis was performed using total RNAs from the leaves of 4-week-old plants from three independent transgenic lines (lanes 6, 7, and 8) and panicles from Ubi1:AtJMT and NT plants at three developmental stages. The blots were hybridized with the AtJMT probe and reprobed with the rice RbcS gene (Kyozuka et al., 1993). RbcS transcript levels in the transgenic leaves and panicles were similar to levels in the NT controls. Ethidium bromide (EtBr) staining of total RNA was performed for verification of equal RNA loading. S1 panicles are less than 1.5 cm in length. S2 panicles are not fully elongated (3–10 cm) and are pale white/yellow. S3 panicles are fully elongated and enclosed within the flag leaf sheath during the booting stage. Bar = 1 cm.
Table I.
Levels of MeJA and ABA in S1 panicles of NT, Ubi1:AtJMT, and drought-treated NT plants
The values (ng g−1 fresh weight) represent means ± sd of three independent experiments.
| Compound | NT | Ubi1:AtJMT | Drought-Treated NT |
|---|---|---|---|
| MeJA | 222.0 ± 67.5 | 1,401.8 ± 11.2 | 4,176.8 ± 223.8 |
| ABA | 24.8 ± 2.6 | 46.3 ± 3.9 | 34.5 ± 2.2 |
High Levels of MeJA Reduce Grain Yield in Both Ubi1:AtJMT and Drought-Treated NT Plants
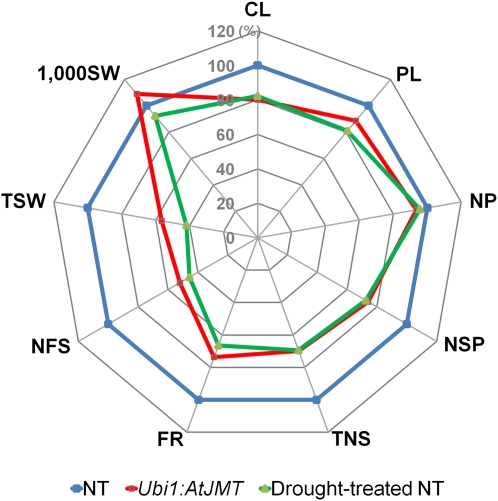
Phenotypic evaluation of Ubi1:AtJMT and NT plants revealed no major differences in the vegetative growth of the entire plants and the time to flowering, although the transgenic plants were slightly smaller than the NT controls. However, grain yield was significantly reduced in the Ubi1:AtJMT plants (Fig. 2). A similar reduction in plant height and grain yield was also observed in NT plants following drought stress or treatment with exogenous MeJA at the panicle initiation stage (Fig. 2). These observations prompted us to investigate yield components of the Ubi1:AtJMT and NT plants treated either with drought stress or with exogenous MeJA (Fig. 3; Table II). When compared with untreated NT controls, decreases in the yield parameters of Ubi1:AtJMT plants were strikingly similar to those observed in drought-treated NT plants (Fig. 3). These experiments were performed twice in the summer and again in the winter of 2007 in a greenhouse, as shown in Table II, obtaining comparable results. Similar reductions in yield parameters were observed in NT plants that were treated with exogenous MeJA, suggesting that MeJA is involved in the loss of grain yield. Specifically, in Ubi1:AtJMT, MeJA-treated NT, and drought-treated NT plants, the number of spikelets per panicle was reduced by 40.9%, 59.4%, and 25.6%, respectively, and filling rates were reduced by 74.1%, 75.2%, and 38.5%, respectively, as compared with untreated NT control plants. In contrast, 1,000 seed weight of experimental plants remained unchanged. Together, these results suggested that MeJA reduces grain yield by affecting the development of spikelets.
Figure 2.
Phenotypes of NT, drought-treated NT, MeJA-treated NT, and Ubi1:AtJMT plants. A, Plant phenotypes at the grain-filling stage. B, Panicles taken from plants that had reached maturity with ripened grains. C, Panicle architecture.
Figure 3.
Comparison of agronomic traits between Ubi1:AtJMT and drought-treated NT plants. A spider plot of Ubi1:AtJMT plant (red) and drought-treated NT plant (green) agronomic traits as compared with untreated NT plants (blue). Each data point represents a percentage of the mean values (n = 15) listed in Table II (summer). Mean values from untreated NT plants were set at 100% as a reference. CL, Culm length; PL, panicle length; NP, number of panicles per plant; NSP, number of spikelets per panicle; TNS, total number of grains; FR, filling rate; NFS, number of filled seeds; TSW, total seed weight; 1,000SW, 1,000 seed weight.
Table II.
Analysis of seed production parameters in NT, Ubi1:AtJMT, and drought-treated and MeJA-treated NT plants
Each parameter represents the mean ± sd (n = 15 or 30) for NT, Ubi1:AtJMT, MeJA-treated NT, and drought-treated NT plants. The percentage differences (%Δ) between the values for NT and the values for Ubi1:AtJMT, MeJA-treated NT, and drought-treated NT was calculated. P values were determined by Student's t test.
| Plants | No. of Panicles | Spikelets per Panicle | Filling Rate | Total Seed Weight | 1,000 Seed Weight |
|---|---|---|---|---|---|
| count | count | % | g | g | |
| 2007 (summer; n = 15) | |||||
| NT | 23.4 ± 5.2 | 86.1 ± 6.8 | 91.2 ± 2.9 | 42.3 ± 5.4 | 23.4 ± 0.3 |
| Ubi1:AtJMT | 21.9 ± 4.2 | 63.9 ± 7.4 | 67.2 ± 10.1 | 23.9 ± 7.5 | 25.5 ± 3.2 |
| %Δ | −6.6 | −25.8 | −26.3 | −43.5 | 8.9 |
| P | 0.5837 | 0.0004 | 0.0005 | 0.0009 | 0.1896 |
| Drought-treated NT | 22.4 ± 2.9 | 62.5 ± 8.0 | 60.5 ± 18.0 | 17.7 ± 1.8 | 21.7 ± 1.0 |
| %Δ | −4.3 | −27.4 | −33.7 | −58.1 | −7.3 |
| P | 0.6997 | 0.0009 | 0.0904 | 0.0023 | 0.0046 |
| 2007 (winter; n = 30) | |||||
| NT | 7.0 ± 1.7 | 76.4 ± 13.5 | 45.6 ± 13.9 | 5.2 ± 2.2 | 21.4 ± 1.7 |
| Ubi1:AtJMT | 5.4 ± 2.2 | 45.2 ± 10.8 | 11.8 ± 7.6 | 0.7 ± 0.7 | 22.3 ± 2.5 |
| %Δ | −22.28 | −40.9 | −74.1 | −86.0 | 4.2 |
| P | 0.0098 | 0.0000 | 0.0335 | 0.0000 | 0.1668 |
| MeJA-treated NT | 7.3 ± 2.1 | 31.0 ± 6.8 | 11.3 ± 13.5 | 0.6 ± 0.7 | 22.7 ± 4.9 |
| %Δ | 5.0 | −59.4 | −75.2 | −89.3 | 6.1 |
| P | 0.8743 | 0.0000 | 0.0000 | 0.0000 | 0.1825 |
| Drought-treated NT | 6.9 ± 1.5 | 56.9 ± 11.2 | 28.3 ± 20.3 | 2.6 ± 2.3 | 21.7 ± 2.2 |
| %Δ | −1.0 | −25.6 | −38.5 | −48.8 | 1.3 |
| P | 0.5112 | 0.00000 | 0.0003 | 0.0001 | 0.5782 |
High MeJA Levels Alter Spikelet Organ Numbers in Both Ubi1:AtJMT and Drought-Treated NT Plants
Several alterations in spikelet organ numbers were noted in the developing spikelets of Ubi1:AtJMT plants (Fig. 4). NT spikelets are normally composed of a pair of glumes at the base and four whorls of spikelet organs, a lemma/palea, two lodicules, six stamens, and a pistil extending from the periphery to the center (Fig. 4, C1–C4). With the exception of the glumes, numbers of four spikelet organ types were altered in the Ubi1:AtJMT spikelets (Fig. 4, D1–D7). For example, the numbers of lemma/palea and lodicules were increased, and these extra organs were often elongated. The number of stamens varied from five to 10 stamens. The number of pistils was increased, and a compound ovary with three stigma branches was observed. To investigate the developmental changes of Ubi1:AtJMT spikelets in detail, spikelets at the early stage of development were fixed and examined with a scanning electron microscope. Compared with the NT controls, the spikelet meristem of Ubi1:AtJMT plants was enlarged, the number of spikelet organ primordia was altered, and the extra organ structures were modified in appearance (Fig. 4, B1–B4).
Figure 4.
Spikelet morphology of NT, Ubi1:AtJMT, MeJA-treated NT, and drought-treated NT plants. Scanning electron micrographs of developing NT (A1–A4) and Ubi1:AtJMT (B1–B4) spikelets, a larger spikelet primordium (B1) than NT (A1), a spikelet with three extra stamen primordia (B2), a spikelet with extra stamens and extended lodicules (B3), and a spikelet with four stigma branches (B4). Light microscope images of NT (C1–C4), Ubi1:AtJMT (D1–D7), MeJA-treated NT (E1–E7), and drought-treated NT (F1–F4) spikelets. Ubi1:AtJMT spikelets with an extra whorl 1(D1), with lemma and palea removed showing two extra whorl 1 (D2), with normal lodicules and three extra, elongated lodicules (D3), with a stamen attached with two anthers (D4), with two extra stamens (D5), with a pistil with an extra stigma branch (D6), and with a gynoecium with an extra pistil (D7). MeJA-treated NT spikelets with an extra whorl 1 (E1), with two extra whorl 1 (E2), with extended lodicules (E3), with a stamen attached with two anthers (E4), with an extra stamen (E5), with a pistil with an extra stigma branch (E6), and with a gynoecium with an extra pistil (E7). Drought-treated NT spikelets with an extra whorl 1 (F1), with two elongated extra lodicules (F2), with an androecium with five stamens (F3), and with a pistil with an extra stigma branch (F4). an, Anther; f, filament; g, glume; l, lemma; lo, lodicule; ov, ovary; p, palea; pi, pistil; s, stamen; sp, spikelet primordium; st, stigma; sty, style. Bars = 50 μm (A1) and 1 mm (C1).
The proportion of altered spikelets in Ubi1:AtJMT plants ranged from 34.8% to 60% (Table III). In NT plants that were treated either with exogenous MeJA or with drought, the proportion of altered spikelets was 11.1% or 10.0%, respectively. These proportions of altered NT spikelets were lower than those observed for Ubi1:AtJMT spikelets, possibly because MeJA levels were maintained at a high level throughout all stages of panicle development in Ubi1:AtJMT plants due to the constitutive expression of AtJMT, as opposed to transient increases of MeJA in NT plants. Thus, our results demonstrate that increased levels of MeJA in Ubi1:AtJMT plants are responsible for alteration in spikelet organ numbers.
Table III.
Alteration in organ numbers of NT, Ubi1:AtJMT, and MeJA-treated and drought-treated NT spikelets
| Plants | Totala | Alteredb | Percentage of Altered/Total |
|---|---|---|---|
| NT | 640 | 3 | 0.5 |
| Ubi1:AtJMT | |||
| 1-3-2 | 648 | 351 | 54.2 |
| 6-7-1 | 590 | 354 | 60.0 |
| 8-4-4 | 690 | 240 | 34.8 |
| MeJA-treated NT | 596 | 66 | 11.1 |
| Drought-treated NT | 856 | 86 | 10.0 |
Number of total spikelets used in each assay.
Number of altered spikelets.
Identification of Genes Regulated by MeJA and Drought in Young Panicles
To identify genes that are regulated by MeJA and drought, global expression profiling was performed on panicles from Ubi1:AtJMT, drought-treated NT, and untreated NT plants. The underlying assumption of this approach was that high levels of MeJA produced either by overexpression of AtJMT in the transgenic panicles or by drought treatment in the NT panicles regulate genes that are involved in spikelet and/or panicle development. Profiling was conducted using the Rice 3′-Tiling Microarray (GreenGene Biotech). RNA samples from S1 panicles of Ubi1:AtJMT, drought-treated NT, and untreated NT plants were used to generate cyanine-3 (Cy3)-labeled cDNA probes, which were then hybridized to the microarray. Each data set was obtained from three biological repeats. When three replicates were averaged and compared with untreated NT panicles, 157 and 372 genes were up-regulated and 127 and 700 genes were down-regulated in Ubi1:JMT and drought-treated NT panicles, respectively.
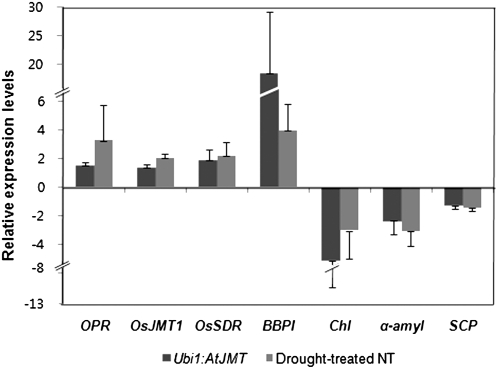
From this list, we further selected genes that were regulated in both Ubi1:JMT and drought-treated NT panicles in all three replicates. The resulting collection of 10 up-regulated and 17 down-regulated genes is presented in Table IV. The regulated patterns of gene expression were subsequently confirmed by quantitative real-time PCR using the same RNAs that were used for the microarrays (Fig. 5). Some of the up-regulated genes have been reported to be associated with JA (Cheong and Choi, 2003) and ABA (González-Guzmán et al., 2002) biosynthesis, including 12-oxo-phytodienoate reductase (OPR), OsJMT1, and short-chain alcohol dehydrogenase (OsSDR). ABA levels are dramatically reduced in the aba2 mutant of Arabidopsis, in which an ortholog of OsSDR has been knocked out (González-Guzmán et al., 2002), suggesting that OsSDR is a key enzyme for ABA biosynthesis. Senescence-related genes were also up-regulated in Ubi1:AtJMT panicles as compared with untreated NT panicles. These include ERD1, which mediates chloroplast protein degradation upon senescence in plants (Zheng et al., 2006; Jung et al., 2007), and the blue copper-binding protein gene that is activated during ozone exposure in Arabidopsis (Miller et al., 1999).
Table IV.
List of genes that are up- or down-regulated in Ubi1:AtJMT and drought-treated NT panicles as compared with untreated NT panicles
Numbers appearing in boldface are those that are up- or down-regulated by more than 1.5-fold or more in Ubi1:AtJMT or drought-treated NT plants.
| Gene Name | Accession No.a | Ubi1:AtJMTb | Drought-Treated NTb |
|---|---|---|---|
| Flavin-containing monooxygenase | AK071801 | 3.2 | 3.4 |
| Ser/Thr kinase | AK069697 | 7.3 | 3.5 |
| Hypothetical protein | AK059506 | 5.1 | 4.3 |
| Ser carboxypeptidase | NM_001069939 | 2.0 | 1.9 |
| Flavonol reductase | AK106089 | 4.8 | 2.3 |
| Cyclin-like F-box domain-containing protein | AK068342 | 2.2 | 3.7 |
| Hypothetical protein | AK063829 | 4.5 | 2.8 |
| Flagelliform silk protein | AK062794 | 5.0 | 1.5 |
| Floral nectary-specific protein (OsJMT1) | AK067321 | 1.9 | 1.8 |
| OsSDR | AK064532 | 1.9 | 2.4 |
| RING domain-containing protein | AK067013 | 1.8 | 3.5 |
| Disease resistance protein | AK121159 | 3.3 | −0.9 |
| Galactose oxidase | AK060256 | 6.8 | −0.7 |
| Iron/ascorbate-dependent oxidoreductase | NM_001063498 | 2.2 | −0.7 |
| F-box protein interaction domain-containing protein | AK100978 | 12.7 | −0.3 |
| TPR-like domain-containing protein | AK069320 | 1.7 | −0.1 |
| Iron/ascorbate-dependent oxidoreductase | NM_001063495 | 2.4 | 0.4 |
| Ser/Thr protein kinase | AK073168 | 5.1 | 0.6 |
| MYB transcription factor (MYBS2) | AK071611 | 2.0 | 0.7 |
| Leu-rich repeat-containing protein | NM_001052513 | 2.4 | 0.8 |
| Pectinesterase family protein | AK106909 | 5.8 | 1.2 |
| Cyclin-like F-box domain-containing protein | AK101804 | 3.0 | 1.4 |
| Oxophytodienoic acid reductase (OPR) | AK072596 | 2.6 | −1.0 |
| Bowman-Birk proteinase inhibitor (BBPI) | AK102138 | 4.8 | 0.4 |
| Disease resistance protein | NM_001060449 | 2.3 | −10.3 |
| Chitinase (OsChib3H-a) | AB027417 | 4.4 | −1.6 |
| WRKY61 | NM_001075006 | 2.0 | −1.8 |
| Von Willebrand factor type A | AK070289 | 2.1 | −6.5 |
| Allyl alcohol dehydrogenase | AK060453 | −2.4 | −2.3 |
| Met adenosyltransferase 1 | NM_001049329 | −2.4 | −2.1 |
| Acetyl-CoA synthetase | NM_001059240 | −2.0 | −2.1 |
| RbcS | NM_001073137 | −1.8 | −3.3 |
| WRKY45 | AK066255 | −2.6 | −2.2 |
| Cereal-type α-amylase inhibitor (α-amyl) | AK062381 | −2.0 | −6.0 |
| PSII protein (PsbW) | AF366557 | −2.1 | −2.6 |
| Isopenicillin nitrogen synthase | NM_001068348 | −1.6 | −2.0 |
| Ser carboxypeptidase (SCP) | AK064814 | −2.4 | −2.3 |
| RbcS | AK099574 | −2.3 | −1.8 |
| Wound-induced protein | NM_001060540 | −2.1 | −2.0 |
| Hsp70 | X75616 | −3.3 | −1.7 |
| Chlorophyll a/b-binding protein | AK062725 | −2.3 | −1.5 |
| Glycoside hydrolase | AK061093 | −2.5 | −1.4 |
| Small heat stress protein | AK119261 | −5.5 | −1.2 |
| Naringenin-chalcone synthase family protein | AK119922 | −2.0 | −0.8 |
| RbcS | AK068266 | −2.3 | −0.8 |
| Chloroplast precursor (Chl) | AK120809 | −4.3 | −0.5 |
| bZIP transcription factor (CAT103) | AK059435 | −1.9 | −0.5 |
| Ethylene-responsive transcriptional coactivator | AK119729 | −2.7 | 0.5 |
| Terpene synthase family protein | AK067451 | −9.0 | 0.5 |
| Cyclin-like F-box domain-containing protein | NM_001074567 | −5.4 | 1.4 |
GenBank accession numbers for full-length cDNA sequences of corresponding genes.
The microarray data sets can be found at http://www.ncbi.nlm.nih.gov/geo/ (Gene Expression Omnibus). The Gene Expression Omnibus accession number of microarray data sets is GSE14508. Numbers represent means of relative expression levels of three independent biological replicates.
Figure 5.
Relative transcript levels of regulated genes in Ubi1:AtJMT and drought-treated NT panicles relative to levels in untreated NT controls. Transcript levels of the regulated genes OPR (AK072596), OsJMT1 (AK067321), OsSDR (AK064532), BBPI (AK102138), Chl (AK120809), α-amyl (AK062381), and SCP (AK064814) in the S1 panicles of Ubi1:AtJMT, drought-treated NT, and untreated NT plants were measured by quantitative real-time PCR. For each experiment, results were normalized using rice tubulin (AK102560) transcript levels. Values are means ± sd of three independent experiments.
DISCUSSION
In this study, we show that transgenic overexpression of the AtJMT gene in rice (Ubi1:AtJMT) increases MeJA levels in young panicles. Surprisingly, the increased MeJA levels caused dramatic changes in Ubi1:AtJMT plant reproductive development. These changes included a reduced number of spikelets per panicle, low filling rate, and alterations in floral organ numbers, collectively resulting in a large loss of grain yield. Similar phenotypes were also observed in NT plants that were treated with either exogenous MeJA or with drought at the panicle initiation stage, suggesting that MeJA is the key component for the observed alterations in reproductive development.
The effects of MeJA on plant development have also been examined in other model plant species. Unlike our Ubi1:AtJMT rice, Arabidopsis plants transformed with 35S:AtJMT had increased levels of MeJA, but their flowers were visually indistinguishable from those of NT plants (Seo et al., 2001). This is likely because the anatomy and development of rice spikelets differ from those of Arabidopsis flowers (Ikeda et al., 2004; Dreni et al., 2007). Interestingly, Cipollini (2007) reported that Arabidopsis plants transformed with 35S:AtJMT produced 40% less total seed weight than NT controls. In this case, the reduction in total seed weight was due to a reduced number of flowers. Similarly, application of exogenous MeJA on Pharbitis nil (Maciejewska and Kopcewicz, 2002) and Nicotiana sylvestris (Baldwin and Hamilton, 2000) led to a dramatic reduction in the number of flowers. Collectively, these reports from different plant species support our observations that the increased levels of MeJA in both Ubi1:AtJMT and drought-treated NT plants are responsible for the reduced number of spikelets per panicle.
Rice plants containing high levels of MeJA in their young panicles had not only a reduced number of spikelets but also a reduced filling rate. For example, in drought-treated NT plants, total seed weight was reduced by 48.8%. This reduction was the result of a 25% decrease in the number of spikelets per panicle and a 38.5% decrease in the filling rate as compared with results from untreated NT plants (Table II). The reduction in filling rate may have been due to altered levels of JA rather than MeJA. This possibility is supported by reports of decreased levels of JA, impaired floral development such as flower opening, and impaired development and release of pollen in the Arabidopsis mutants fad3-1/fad7-2/fad8, dde1, and dad1 (McConn and Browse, 1996; Sanders et al., 2000; Ishiguro et al., 2001). Lack of JA sensitivity in the Arabidopsis coi1 mutant also led to male sterility (Xie et al., 1998). This does not appear to be the case in our Ubi1:AtJMT rice, in which levels of JA in young panicles remained unchanged. Similarly, JA levels were not altered in transgenic Arabidopsis and soybean overexpressing AtJMT and NTR1 (a Chinese cabbage ortholog of AtJMT), respectively (Seo et al., 2001; Xue and Zhang, 2007). Thus, our results demonstrated that increased levels of MeJA altered the number of spikelet organs in Ubi1:AtJMT as well as in drought-treated NT plants. The alteration in spikelet organ numbers was correlated with a reduction in the filling rate in these plants, which subsequently led to a loss of grain yield. This possibility is supported by the reported observation that exogenous application of MeJA during the anther dehiscence stage of rice development resulted in increased sterility (Zhu et al., 2004). It is possible that the reductions in seed production of Ubi1:AtJMT plants were due partly to energetic costs associated with constitutive overexpression of AtJMT, overproduction of MeJA, and activation of defense and fitness responses to nutrients, as observed previously in Arabidopsis (Cipollini, 2007, 2009). Since MeJA stimulates the production of ABA in Ubi1:AtJMT plants, the reduction in grain yield was also due partly to ABA-mediated pollen abortion that was reported to occur under drought and cold stresses in rice and maize (Morgan, 1980; Oliver et al., 2007).
Application of exogenous MeJA was reported to inhibit P. nil shoot growth in a dose-dependent manner (Maciejewska and Kopcewicz, 2002). MeJA is also reported to inhibit seed germination, callus growth, and primary root growth (Ueda and Kato, 1982; Staswick et al., 1992; Berger et al., 1996; Nojavan-Asghari and Ishizawa, 1998). More recently, Pauwels et al. (2008) reported that high levels of MeJA repressed cell cycle progression by arresting cells in the G2 phase. It is likely that increased levels of MeJA reduce the number of spikelets in our Ubi1:AtJMT panicles by repressing cell cycle progression.
ABA is generally thought to play a role in plant response to drought stress. ABA was reported to increase by 1.8-fold in rice panicles in response to drought treatment at the mature panicle stage (Yang et al., 2001, 2004, 2007). In this study, we exposed NT plants to drought conditions for 2 weeks during the panicle initiation stage. In young panicles from drought-treated NT plants, the fold increase in levels of MeJA (19-fold) was much higher than that of ABA (1.4-fold) as compared with levels in untreated NT controls. In addition, in young panicles of Ubi1:AtJMT plants grown in nondrought conditions, levels of ABA were 1.9-fold higher than those of untreated NT controls, suggesting that ABA levels were increased due to MeJA exposure rather than by drought stress. These results led us to postulate that plants start to produce MeJA upon exposure to drought stress, which in turn stimulates the production of ABA. This hypothesis is supported by the fact that MeJA treatment increased ABA concentrations at an early stage of fruit development in sweet cherry (Prunus avium; Kondo et al., 2002). The OsSDR gene, an Arabidopsis ortholog of which is essential for ABA biosynthesis (González-Guzmán et al., 2002), was up-regulated in both Ubi1:AtJMT panicles and drought-treated NT panicles, suggesting its involvement in MeJA-dependent increases in ABA levels. The activity of the Arabidopsis SDR gene was reported to be required for JA biosynthesis (Adie et al., 2007), suggesting that ABA either precedes or cooperates with JA in signaling pathways. It is possible, therefore, that a positive feedback exists between MeJA and ABA under drought conditions. Determination of MeJA and ABA levels after exogenous application of ABA and MeJA or in ABA- and JA-defective mutants will clarify the feedback relationship further. Expression of OsJMT1, a rice ortholog of AtJMT, was increased in Ubi1:AtJMT and drought-treated NT panicles, indicating that OsJMT1 may be responsible for the biosynthesis of MeJA upon exposure to drought conditions. OsJMT1 shares 36% homology with AtJMT in nucleotide sequence without any stretch of 21 to 24 nucleotides that is required for a small RNA to potentially cross-react.
Overall, our results suggest that MeJA plays a role in a stress-induced loss of grain yield in rice.
MATERIALS AND METHODS
Plasmid Construction and Transformation of Rice
The expression plasmid Ubi1:AtJMT contained the bar gene under the control of the cauliflower mosaic virus 35S promoter for use with herbicide-based plant selection. The ubiquitin1 promoter, together with its intron (Ubi1), was used to drive constitutive plasmid gene expression (Christensen and Quail, 1996). The AtJMT cDNA clone was kindly provided by Dr. Y.D. Choi (Seo et al., 2001). Vectors were introduced into Agrobacterium tumefaciens LBA4404 by triparental mating. Embryonic callus initiated from the embryo of dehulled rice (Oryza sativa ‘Nakdong’) grains was transformed by cocultivation (Hiei et al., 1994), selected with 7 mg L−1 phosphinothricin, and used to regenerate transgenic plants according to the method of Jang et al. (2002).
Drought and MeJA Treatments
Transgenic and NT rice seeds were germinated in Murashige and Skoog solid medium in a growth chamber in the dark at 28°C for 3 d, transplanted to soil pots, and grown in a greenhouse (16-h-light/8-h-dark cycle) at 28°C to 30°C. Each pot (10 × 10 × 10 cm) was filled with rice nursery soil (Bio-media) according to the method of Oh et al. (2007, 2008). Twelve weeks after transplanting, rice plants (before panicle initiation) were subjected to either 2 weeks of drought or 60 μm MeJA (95%; Sigma-Aldrich) solution. When the plants had reached maturity and grains had ripened, the plants were harvested and threshed (separation of seeds from the vegetative parts). The unfilled and filled grains were taken apart, independently counted, and weighed. The following agronomic traits were scored: number of panicles, spikelets per panicle, filling rate (%), total seed weight (g), and 1,000 seed weight (g). Resultant data were separately analyzed by Student's t test for Table II. A spider plot of Ubi1:AtJMT and drought-treated NT plant agronomic traits as compared with untreated NT plants was drawn using Microsoft Excel software (Fig. 3).
Measurements of MeJA and ABA
Levels of MeJA were measured following the method of Engelberth et al. (2003). Approximately 0.5 g of S1 panicles was collected and extracted in methanol with an internal standard, [9,10-2H2]JA. Each sample was separated using a solid-phase extraction cartridge (reverse-phase C18, 12 mL; Mallinckrodt Baker), and sample pH was adjusted to pH 3.5 with 10% (v/v) formic acid. The oxylipin fraction was eluted with diethyl ether, and the eluate was completely dried under N2 gas. Methanolysis was performed by incubation with a 1:2 (v/v) HCl:methanol mixture for 12 h at 30°C. The HCl:methanol was then completely removed under a stream of N2 gas, and each sample was eluted with dichloromethane. Samples were analyzed by gas chromatography-mass spectrometry (Agilent Technologies).
Levels of ABA were measured following the method of Kang et al. (2005). Approximately 0.1 g of S1 panicles was collected and extracted with an extraction solution containing 95% isopropanol, 5% glacial acetic acid, and an internal standard, [(±)-3,5,5,7,7,7-d6]ABA. The aqueous phase was extracted with ethyl acetate and separated using a silica cartridge (Sep-Pak; Waters Associates). The extracts were dried and methylated by adding diazomethane prior to gas chromatography-mass spectrometry (Agilent Technologies) analysis.
Scanning Electron Micrographs of Spikelets
Samples of panicles (<1 cm long) were prepared for scanning electron micrography analysis by prefixation in 0.1 m phosphate buffer (pH 7.4) containing 2.5% (v/v) glutaraldehyde and 4% (v/v) paraformaldehyde. Air was removed from the samples, and they were rinsed with phosphate buffer. Postfixation was carried out using OsO4, and samples were dehydrated with 60% to 100% ethanol. The samples were treated with isoamyl acetate, dried, and ion coated. The mounted specimens were observed using an S-4300 scanning electron microscope (Hitachi).
Rice 3′-Tiling Microarray Analysis
Expression profiling was conducted using a Rice 3′-Tiling Microarray. Information on the microarray can be found at http://www.ggbio.com (GreenGene Biotech). The Rice 3′-Tiling Microarray was designed from 27,448 genes deposited at the International Rice Genome Sequencing Project RAP1 database (http://rapdb.lab.nig.ac.jp). Among these, 20,507 genes were from representative RAP1 sequences with cDNA/EST supports and 6,941 genes were predicted without cDNA/EST supports. Ten 60-nucleotide-long probes were designed from each gene, starting 60 bp ahead of the end of the stop codon with 10-bp shifts in position, so that 10 probes covered 150 bp in the 3′ region of the gene. In total, 270,000 probes were designed (average size, 60 nucleotides) to have Tm values of 75°C to 85°C. The microarray was manufactured by NimbleGen (http://www.nimblegen.com/). Random gas chromatography probes (38,000) were used to monitor the hybridization efficiency, and fiducial markers at the four corners (225) were included to assist with overlaying the grid on the image.
The microarray was used to profile gene expression in Ubi1:AtJMT, drought-treated NT, and untreated NT plants. Cy3-labeled target cDNA fragments were synthesized from S1 panicles using a Cy3-9mer primer. For normalization, data were processed with cubic alpine normalization using quartiles to adjust signal variation between chips and with Robust Multi-Chip Analysis using a median polish algorithm implemented in NimbleScan (Workman et al., 2002; Irizarry et al., 2003). To assess the reproducibility of the microarray analysis, we repeated the experiment three times with independently prepared total RNAs.
Real-Time PCR
Total RNA was prepared as reported previously (Oh et al., 2007). PCR products were amplified using the primers designed with Primer Designer 4 software (Sci-ed Software) listed in Supplemental Table S1. For quantitative real-time PCR experiments, the SuperScript III Platinum One-Step Quantitative RT-PCR system (Invitrogen) was used. For PCR, a master mix of reaction components was prepared according to the manufacturer's protocol for Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen). Thermocycling and fluorescence detection were performed using a Stratagene Mx3000p Real-Time PCR machine (Stratagene). PCR was performed at 95°C for 10 min, followed by 20 to 25 cycles of 94°C for 30 s, 57°C for 30 s, and 68°C for 1 min. To validate our quantitative PCR results, we repeated each experiment three times.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used for real-time PCR.
Supplementary Material
Acknowledgments
We are grateful to Dr. Sang-Ik Song at Myongji University for making critical comments on the experimental results.
This work was supported by the Ministry of Education, Science, and Technology, Korea, through the Crop Functional Genomics Center (grant no. CG2111 to J.-K.K.), by the Biogreen21 Program (grant to J.-K.K.), and by the Korea Science and Engineering Foundation through the Plant Metabolism Research Center at Kyung-Hee University (grant to J.-K.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ju-Kon Kim (jukon306@gmail.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtová JPT, Ullmann J (1994) Methyl jasmonate inhibits growth and flowering in Chenopodium rubrum. Biol Plant 36 317–319 [Google Scholar]
- Asch F, Dingkuhn M, Sow A, Audebert A (2005) Drought-induced changes in rooting patterns and assimilate partitioning between root and shoot in upland rice. Field Crops Res 93 223–236 [Google Scholar]
- Baldwin IT, Hamilton W (2000) Jasmonate-induced responses of Nicotiana sylvestris result in fitness costs due to impaired competitive ability for nitrogen. J Chem Ecol 26 915–952 [Google Scholar]
- Beltrano J, Ronco MG, Montaldi ER (1999) Drought stress syndrome in wheat is provoked by ethylene evolution imbalance and reversed by rewatering, aminoethoxyvinylglycine, or sodium benzoate. J Plant Growth Regul 18 59–64 [DOI] [PubMed] [Google Scholar]
- Berger S, Bell E, Mullet JE (1996) Two methyl jasmonate-insensitive mutants show altered expression of AtVSP in response to methyl jasmonate and wounding. Plant Physiol 111 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonjung H, Fukai S (1996) Effects of soil water deficit at different growth stages on rice growth and yield under upland conditions. 2. Phenology, biomass production and yield. Field Crops Res 48 47–55 [Google Scholar]
- Cheong JJ, Choi YD (2003) Methyl jasmonate as a vital substance in plants. Trends Genet 19 409–413 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5 213–218 [DOI] [PubMed] [Google Scholar]
- Cipollini D (2007) Consequences of the overproduction of methyl jasmonate on seed production, tolerance to defoliation and competitive effect and response of Arabidopsis thaliana. New Phytol 173 146–153 [DOI] [PubMed] [Google Scholar]
- Cipollini D (2009) Constitutive expression of methyl jasmonate-inducible responses delays reproduction and constrains fitness responses to nutrients in Arabidopsis thaliana. Evol Ecol (in press)
- Cornejo M, Luth D, Blankenship KM, Anderson OD, Blechl AE (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol Biol 23 567–581 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1995) Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA 92 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PBF, An G, Colombo L, Kater MM (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52 690–699 [DOI] [PubMed] [Google Scholar]
- Engelberth J, Schmelz EA, Alborn HT, Cardoza YJ, Huang J, Tumlinson JH (2003) Simultaneous quantification of jasmonic acid and salicylic acid in plants by vapor-phase extraction and gas chromatography-chemical ionization mass spectrometry. Anal Biochem 312 242–250 [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54 147–156 [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Choi WB, Lee KH, Song SI, Nahm BH, Kim JK (2002) High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol 129 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Lyou SH, Yeu SY, Kim MA, Rhee S, Kim M, Lee JS, Choi YD, Cheong JJ (2007) Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep 26 1053–1063 [DOI] [PubMed] [Google Scholar]
- Kang DJ, Seo YJ, Lee JD, Ishii R, Kim KU, Shin DH, Park SK, Jang SW, Lee IJ (2005) Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J Agron Crop Sci 191 273–282 [Google Scholar]
- Kiribuchi K, Jikumaru Y, Kaku H, Minami E, Hasegawa M, Kodama O, Seto H, Okada K, Nojiri H, Yamane H (2005) Involvement of the basic helix-loop-helix transcription factor RERJ1 in wounding and drought stress responses in rice plants. Biosci Biotechnol Biochem 69 1042–1044 [DOI] [PubMed] [Google Scholar]
- Kondo S, Motoyama M, Michiyama H, Kim M (2002) Roles of jasmonic acid in the development of sweet cherries as measured from fruit or disc samples. Plant Growth Regul 37 37–44 [Google Scholar]
- Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K (1993) Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion genes in transgenic rice. Plant Physiol 102 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska B, Kopcewicz J (2002) Inhibitory effect of methyl jasmonate on flowering and elongation growth on Pharbitis nil. J Plant Growth Regul 21 216–223 [Google Scholar]
- McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19 1127–1137 [DOI] [PubMed] [Google Scholar]
- Miller JD, Arteca RN, Pell EJ (1999) Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol 120 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A, Prinsen E, Bauw G, Montagu MV (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9 2243–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JM (1980) Possible role of abscisic acid in reducing seed set in water-stressed wheat plants. Nature 285 655–657 [Google Scholar]
- Nojavan-Asghari M, Ishizawa K (1998) Inhibitory effects of methyl jasmonate on the germination and ethylene production in cocklebur seeds. J Plant Growth Regul 17 13–18 [Google Scholar]
- Ober ES, Setter TL, Madison JT, Thompson JF, Shapiro PS (1991) Influence of water deficit on maize endosperm development: enzyme activities and RNA transcripts of starch and zein synthesis, abscisic acid, and cell division. Plant Physiol 97 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Kim SJ, Kim YS, Park SH, Ha SH, Kim JK (2008) Arabidopsis cyclin D2 expressed in rice forms a functional cyclin-dependent kinase complex that enhances seedling growth. Plant Biotechnol Rep 2 227–231 [Google Scholar]
- Oh SJ, Kwon CW, Choi DW, Song SI, Kim JK (2007) Expression of barley hvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol J 5 646–656 [DOI] [PubMed] [Google Scholar]
- Oliver SN, Dennis ES, Dolferus R (2007) ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol 48 1319–1330 [DOI] [PubMed] [Google Scholar]
- O'Toole JC, Namuco OS (1983) Role of panicle exertion in water-stress induced sterility. Crop Sci 23 1093–1097 [Google Scholar]
- Pandey S, Bhandari H, Hardy B (2007) Economic Costs of Drought and Rice Farmers' Coping Mechanisms: A Cross-Country Comparative Analysis. International Rice Research Institute, Los Banos, Philippines, pp 1–9
- Pauwels L, Morreel K, Witte ED, Lammertyn F, Montagu MV, Boerjan W, Inzé D, Goossens A (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 29 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedranzani H, Racagni G, Alemano S, Miersch O, Ramírez I, Peña-Cortés H, Taleisnik E, Machado-Domenech E, Abdala G (2003) Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul 41 149–158 [Google Scholar]
- Pedranzani H, Sierra-de-Grado R, Vigliocco A, Miersch O, Abdala G (2007) Cold and water stresses produce changes in endogenous jasmonates in two populations of Pinus pinaster Ait. Plant Growth Regul 52 111–116 [Google Scholar]
- Rakwal R, Tamogami S, Agrawal GK, Iwahashi H (2002) Octadecanoid signaling component “burst” in rice (Oryza sativa L.) seedling leaves upon wounding by cut and treatment with fungal elicitor chitosan. Biochem Biophys Res Commun 295 1041–1045 [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1 404–411 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M (2008) Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol 11 209–214 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saniewski M, Miyamoto K, Ueda J (1998) Methyl jasmonate induces gums and stimulates anthocyanin accumulation in peach shoots. J Plant Growth Regul 17 121–124 [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant response. Proc Natl Acad Sci USA 98 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Seo HS, Song SI, Lee JS, Choi YD (2000) NTR1 encodes a floral nectar-specific gene in Brassica campestris L. ssp. pekinensis. Plant Mol Biol 42 647–655 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Sobajima H, Okada K, Chujo T, Arimura S, Tsutsumi N, Nishimura M, Seto H, Nojiri H, Yamane H (2008) Identification of the OsOPR7 gene encoding 12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta 227 517–526 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckk IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Kato J (1980) Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol 66 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Kato J (1982) Inhibition of cytokinin-induced plant growth by jasmonic acid and its methyl ester. Physiol Plant 54 249–252 [Google Scholar]
- Wang Y, Mopper S, Hasenstein KH (2001) Effects of salinity on endogenous ABA, IAA, JA, and SA in Iris hexagona. J Chem Ecol 27 327–342 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Parthier B (1997) Jasmonate-signalled plant gene expression. Trends Plant Sci 2 302–307 [Google Scholar]
- Wopereis MCS, Kropff MJ, Maligaya AR, Tuong TP (1996) Drought-stress responses of two lowland rice cultivars to soil water status. Field Crops Res 46 21–39 [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Saxild HH, Nielsen C, Brunak S, Knudsen S (2002) A new non-linear normalization method to reduce variability in DNA microarray experiments. Genome Biol 3 research0048.1–research0048.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xue R, Zhang B (2007) Increased endogenous methyl jasmonate altered leaf and root development in transgenic soybean plants. J Genet Genomics 34 339–346 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Liu K, Wang Z, Liu L (2007) Abscisic acid and ethylene interact in rice spikelets in response to water stress during meiosis. J Plant Growth Regul 26 318–328 [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JC, Zhang JH, Ye YX, Wang ZQ, Zhu QS, Liu LJ (2004) Involvement of abscisic acid and ethylene in the responses of rice grains to water stress during filling. Plant Cell Environ 27 1055–1064 [Google Scholar]
- Zheng B, MacDonald TM, Sutinen S, Hurry V, Clark AK (2006) A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224 1103–1115 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM (2004) The ANTHER INDIHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.