Abstract
Many G2/M phase-specific genes in plants contain mitosis-specific activator (MSA) elements, which act as G2/M phase-specific enhancers and bind with R1R2R3-Myb transcription factors. Here, we examined the genome-wide effects of NtmybA2 overexpression, one of the R1R2R3-Myb transcription factors in tobacco (Nicotiana tabacum). We used a custom-made 16-K cDNA microarray for comparative transcriptome analysis of transgenic tobacco BY-2 cell lines that overexpress NtmybA2 or its truncated hyperactive form. The microarray was also used to determine the transcript profile during the cell cycle in synchronized cultures of BY-2 cells. Combined microarray data from transgenic lines and synchronized cells revealed that overexpression of the truncated hyperactive form of NtmybA2, but not its full-length form, preferentially up-regulated many G2/M phase-specific genes in BY-2 cells. We determined promoter sequences of several such up-regulated genes and showed that all contain MSA-like motifs in the proximal regions of their promoters. One of the up-regulated genes, NtE2C, encoding for cyclin-specific ubiquitin carrier proteins, contained a single functional MSA-like motif, which specifically controlled the expression of a reporter gene in the G2/M phase in BY-2 cells. Furthermore, a genomic footprint experiment showed that the MSA element in the NtE2C promoter interacted with nuclear proteins in vivo. Therefore, we propose that the transcription of many G2/M phase-specific genes in tobacco is positively regulated by NtmybA2, in most cases through direct binding to the MSA elements.
Periodic gene expression is fundamental for the ordered occurrence of events through the eukaryotic cell cycle (Cho et al., 2001; Breeden, 2003; De Veylder et al., 2007). Many studies have shown that E2F transcription factors are critical regulators of the G1/S transition in animals and plants (De Veylder et al., 2003). In Arabidopsis (Arabidopsis thaliana), E2Fa and E2Fb function as transcriptional activators and positive regulators of the cell cycle (Sozzani et al., 2006). Transcript levels of many genes involved in DNA replication are strongly up-regulated in transgenic Arabidopsis plants, which co-overexpress E2Fa and its dimerization partner DPa (De Veylder et al., 2002). Microarray analysis of the transgenic plants revealed 181 putative E2F target genes, which contain the E2F-binding motif TTTCCCGCC or its variants in their promoters (Vandepoele et al., 2005).
In contrast to the E2F-dependent regulation of G1/S phase-specific genes, less is known about the transcriptional regulation of genes expressed at the G2/M phase. In plants, many G2/M phase-specific genes contain a common cis-acting element, the so-called mitosis-specific activator (MSA) elements (Ito, 2000, 2005). In tobacco (Nicotiana tabacum), three R1R2R3-Myb proteins, NtmybA1, NtmybA2, and NtmybB, bind to the MSA element in vitro. NtmybA1 and NtmybA2 are structurally closely related and, when transiently overexpressed, activate promoters from NACK1 and CYCB1;3 genes in an MSA-dependent manner. In contrast, NtmybB is able to repress these promoters antagonistically to NtmybA1 and NtmybA2 (Ito et al., 2001). We recently showed that two R1R2R3-Myb proteins in Arabidopsis, MYB3R1 and MYB3R4, are highly homologous to NtmybA1 and NtmybA2 and act as transcriptional activators on multiple G2/M phase-specific genes (Haga et al., 2007). The myb3r1 myb3r4 double mutant showed decreased expression of G2/M phase-specific genes, including CYCB2;1, CDC20.1, and KNOLLE (KN). Incomplete cytokinesis was frequently observed in the double mutant during embryogenesis and postembryonic development, and it caused various developmental defects. This incomplete cytokinesis in the double mutant was mainly due to reduced expression of the KN gene, which encodes for plant-specific syntaxin required for membrane fusion during cell plate formation. Similar functions at the G2/M phase were recently reported in Drosophila Myb (Georlette et al., 2007) and human B-Myb (Osterloh et al., 2007). Myb proteins in these organisms are present in conserved multiprotein complexes called dREAM or Myb-MuvB, which are involved in the activation of various target genes, including many G2/M phase-specific genes.
In Arabidopsis, whose entire genome sequence has been determined, 65 of 82 G2/M phase-specific genes contain at least one MSA motif in their promoter regions (Menges et al., 2005). This finding, together with our previous observations in tobacco cells (Ito et al., 1998; Ito, 2000), led us to hypothesize that MSA elements may be present in a large number of G2/M phase-specific genes in tobacco and that such genes are transcriptionally regulated by NtmybA1 and/or NtmybA2 through the MSA elements. To test this idea, we examined the effects of NtmybA2 overexpression using tobacco BY-2 cells, which can be highly synchronized and easily transformed by the Agrobacterium tumefaciens-mediated method (Nagata et al., 1992). We used a custom-made 16-K cDNA microarray for comparative transcriptome analysis of the transgenic BY-2 cell lines overexpressing NtmybA2 or its truncated form lacking the C-terminal region, which negatively regulates its own activity for transcriptional activation (Araki et al., 2004). The microarray was also used to determine the transcript profile during the cell cycle in synchronized cultures of BY-2 cells. The transcriptome data from the transgenic lines, in combination with those from synchronized cultures, revealed that many G2/M phase-specific transcripts were selectively and significantly up-regulated by stable overexpression of the truncated hyperactive form of NtmybA2 but not of wild-type NtmybA2. The up-regulation was preferentially observed in quiescent cells during the stationary phase. Corroborating our initial hypothesis, we found that up-regulated genes consistently contained motifs that closely matched the consensus MSA sequence, TCYAACGGYYA (Ito, 2000). Our data strongly suggest that NtmybA2 functions as a general positive regulator acting directly on G2/M phase-specific genes.
RESULTS
Overexpression of C-Terminally Truncated NtmybA2 Enhances the Expression of G2/M Phase-Specific Genes
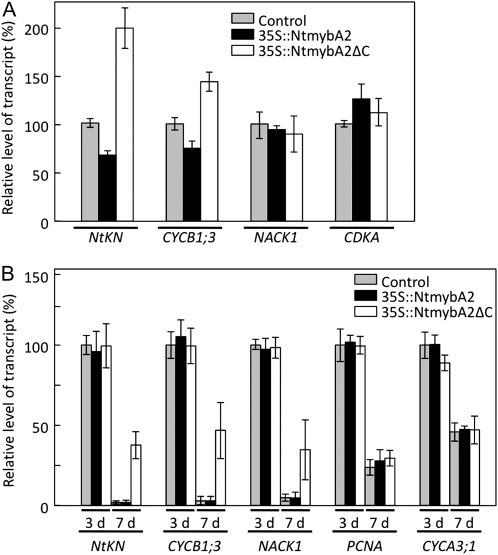
We have previously shown that transcription of CYCB1;3 and NACK1 is positively regulated by NtmybA1 and NtmybA2 through direct binding to the MSA elements (Ito et al., 2001). To test if constitutive overexpression of NtmybA2 can activate the G2/M phase-specific genes, we stably transformed BY-2 cells with a fusion between the cauliflower mosaic virus 35S promoter and NtmybA2 cDNA (35S∷NtmybA2). However, the resulting transgenic calli that expressed excess NtmybA2 mRNA did not show increased levels of the mRNAs for CYCB1;3, NACK1, or a tobacco gene homologous to KN (hereafter designated NtKN; Fig. 1A). Our previous study suggested that NtmybA2 contains a negative regulation domain in its C terminus, which inhibits its own activity for transactivation (Araki et al., 2004). To test the effects of the C-terminally truncated version of NtmybA2 (NtmybA2ΔC), we prepared transgenic BY-2 cells carrying 35S∷NtmybA2ΔC. In the 35S∷NtmybA2ΔC calli, the CYCB1;3 and NtKN genes were significantly up-regulated compared with the control calli that were transformed with a vector alone (Fig. 1A). We then established liquid cultures from the transgenic calli in order to examine the mRNA levels of G2/M phase-specific genes in cell suspensions. Our real-time reverse transcription (RT)-PCR analysis showed that the levels of G2/M phase-specific transcripts in the control cells were correlated with cell division activity, being high on day 3 during the logarithmic phase of growth but decreasing dramatically on day 7, when the cells entered the stationary phase (Fig. 1B). A similar pattern of expression was observed in 35S∷NtmybA2 cells. However, we found significant effects of NtmybA2ΔC overexpression on the expression of G2/M phase-specific genes, which were strongly dependent on the growth stage of the suspension cultures. On day 7 after subculture, the 35S∷NtmybA2ΔC cells accumulated significantly higher levels of G2/M phase-specific transcripts compared with the control cells transformed with the vector alone. However, in 3-d-old cells, NtmybA2ΔC overexpression could not further increase the already elevated levels of the G2/M phase-specific transcripts (Fig. 1B).
Figure 1.
G2/M phase-specific genes are up-regulated by the overexpression of the truncated version of NtmybA2. A, Up-regulation of G2/M phase-specific genes in transgenic calli carrying 35S∷NtmybA2ΔC. Tobacco BY-2 cells were transformed with the vector alone (Control), 35S∷NtmybA2, and 35S∷NtmybA2ΔC by the Agrobacterium-mediated method. Total RNA was extracted from the transformed calli that were generated on the selection agar medium. Real-time RT-PCR analysis was performed to quantify the transcript abundance of the G2/M phase-specific genes (NtKN, CYCB1;3, and NACK1). In addition, the CDKA gene, which is constitutively expressed during the cell cycle, was analyzed in the same way as the control. B, Up-regulation of G2/M phase-specific genes in 35S∷NtmybA2ΔC cells during the stationary phase in cell suspension cultures. Cell suspension cultures were established from transformed BY-2 cells carrying the vector alone (Control), 35S∷NtmybA2, and 35S∷NtmybA2ΔC. For each construct, total RNA was extracted from cells on day 3 (logarithmic phase) and day 7 (stationary phase) after subculture. Transcript abundance of the G2/M phase-specific genes was determined by real-time RT-PCR analysis. Two S phase-specific genes, PCNA and CYCA3;1, were analyzed in the same way for comparison. In both A and B, the results of the real-time PCR were normalized to the expression of EF1α mRNAs. The relative transcript levels were averaged over the three biological replicates and are shown with the sds (error bars).
The S phase-specific genes PCNA and CYCA3;1 also showed preferential expression during the logarithmic phase, but their expression during the stationary phase was not affected by NtmybA2ΔC overexpression (Fig. 1B), thus suggesting that NtmybA2ΔC overexpression specifically affects the expression of G2/M phase-specific genes. Flow cytometric analysis showed that there was no significant difference between DNA distribution patterns of 35S∷NtmybA2ΔC, 35S∷NtmybA2, and control cells, although the 35S∷NtmybA2ΔC cells showed a slightly decreased growth rate compared with the 35S∷NtmybA2 and control cells (data not shown). These results eliminated the possibility that NtmybA2ΔC overexpression primarily affects cell division activity, which secondarily affects the expression of G2/M phase-specific genes. Instead, the increased expression of NtmybA2ΔC may directly activate the transcription of NtmybA2 target genes, which are normally expressed in the G2/M phase.
In summary, overexpression of the full-length NtmybA2 had little effect on the transcription of G2/M phase-specific genes. The C-terminal truncation, which eliminated the negative regulation of NtmybA2, elicited its ability to activate their transcription. Such activation could be observed only when the cells were at a stationary phase in which the G2/M phase-specific genes were normally repressed (Fig. 1B).
Microarray Analysis of 35S∷NtmybA2ΔC
The observed effects of NtmybA2ΔC led us to hypothesize that many other G2/M phase-specific genes might be regulated by NtmybA2 and that such genes might also be up-regulated by NtmybA2ΔC overexpression. To test this possibility, we analyzed the effects of overexpression of the full-length and truncated versions of NtmybA2 in BY-2 cells using a custom-made 16-K cDNA microarray prepared from tobacco BY-2 cells (Matsuoka et al., 2004; Gális et al., 2006). cDNA probes were prepared by reverse transcription of RNA extracted from cells in the stationary phase (7–8 d after subculture) and were hybridized to the microarrays (Gális et al., 2006). Transformation of the BY-2 cells was repeated three times independently, and the transcript profiles from each transgenic line were analyzed statistically (Supplemental Table S1). The transcript profile of the 35S∷NtmybA2 cells was not very different from that of the control cells transformed with an empty vector. There were only eight transcripts whose levels increased or decreased more than 1.5-fold by NtmybA2 overexpression (data not shown). In contrast, NtmybA2ΔC overexpression up- and down-regulated 227 and 85 transcripts more than 1.5-fold, respectively (Supplemental Table S2). The up-regulated group contained NtKN, NACK1, and CYCB1 transcripts, corroborating the results of the RT-PCR analysis (Fig. 1).
Microarray Analysis of the Cell Cycle-Regulated Transcripts
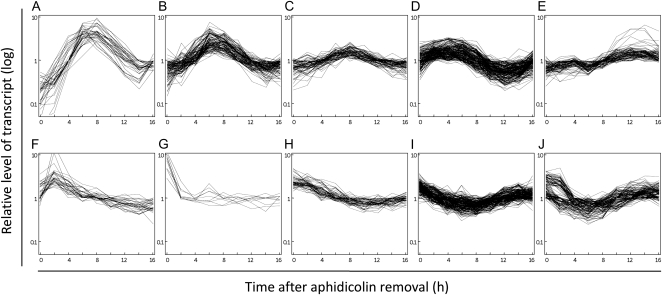
To examine whether the genes up-regulated by NtmybA2ΔC were actually G2/M phase specific, we performed a microarray analysis on synchronized cultures of BY-2 cells, which were induced by the aphidicolin method (Nagata et al., 1992). After release from the aphidicolin block, cells were sampled every 2 h from 0 to 16 h. The synchrony of the cell cycle progression was confirmed by measuring the mitotic index, conducting flow cytometry, and RNA gel blot analysis of the phase-specific transcripts such as CYCB1;3 and PCNA (Supplemental Fig. S1). cDNA probes prepared from these samples were hybridized to the 16-K microarrays. Synchronization of BY-2 cells was repeated twice independently for microarray analysis to identify transcripts that reproducibly showed similar cell cycle-regulated expression (Supplemental Table S3). A total of 883 putative cell cycle-regulated transcripts were selected from 16,896 duplicated spots on the microarrays (Supplemental Fig. S2). These transcripts were then categorized into 10 classes (A–J) on the basis of transcript expression patterns in synchronous cultures using a GeneSpring-implemented self-organizing map clustering method followed by a hand selection of clusters (Fig. 2; Supplemental Table S4). The transcripts with peak expression during the M phase (6–10 h) were categorized into three classes based on their peak expression amplitudes: classes A (highest), B (intermediate), and C (lowest). Classes D and E represented transcripts that were preferentially expressed at the G2 (2–6 h) and G1 (10–14 h) phases, respectively. The S phase-specific transcripts showed their highest expression at 0 h, decreased during the G2 and M phases, and increased again toward the second S phase (14–16 h). These transcripts were classified into classes H, I, and J, depending on their oscillation patterns. The transcripts of classes F and G showed their highest expression at 2 and 0 h, respectively, but did not show increased expression in the second S or G2 phase in the synchronous cultures. This suggests that the expression of these transcripts was probably unrelated to the cell cycle and may have been induced by some stresses of the cells when they were washed for removal of aphidicolin or transferred into the new medium.
Figure 2.
Classification of cell cycle-regulated transcripts in synchronous cultures of BY-2 cells. Synchronization of BY-2 cells was performed twice independently in experiments A and B. In each experiment, cells were sampled at 2-h intervals after release from an aphidicolin block, and the total RNA extracted from each sample was used for microarray analysis. Among 16,896 ESTs in the microarray, 3,846 ESTs showed more than a 2-fold variation in transcript levels during the culture period in both experiments. These ESTs were further analyzed by self-organizing map clustering using data sets of the two independent experiments as described in “Materials and Methods.” These analyses yielded 883 cell cycle-regulated transcripts that were classified into 10 classes (A–J). The expression pattern of each transcript in experiment A is shown separately for each class. Similar expression profiles in each class were obtained in experiment B (data not shown). In each graph, the time points in the synchronous cultures are indicated in hours on the x axis and the median polishing-normalized relative expression levels are shown on the y axis in logarithmic scale.
We performed real-time RT-PCR analysis of arbitrarily selected transcripts representing each class in order to confirm the expression patterns determined by microarray experiments (Supplemental Fig. S3). All examined transcripts in this experiment showed expression patterns that were consistent with the results of the microarray analysis. Moreover, we evaluated whether our classification matched with previously identified cell cycle-regulated genes with known expression patterns. Hence, the G2/M phase-specific CYCB1 transcripts were present only in classes A and B, whereas the G2-specific CYCA1 and CDKB1 transcripts belonged to class D. The histone genes, which are markers for the S phase, were present only in the S phase-specific classes (H–J). Class I contained transcripts for CDC6, CYCA3, NtE2F, and nine histone transcripts, whereas transcripts for PCNA, MCM, and 23 histone transcripts were placed in class J. These patterns are generally consistent with those previously reported for cDNA-amplified fragment length polymorphism-based transcript profiles of synchronized BY-2 cells (Breyne et al., 2002). We concluded that our microarray experiments successfully defined and properly classified the cell cycle-regulated transcripts.
Many G2/M Phase-Specific Genes Are Up-Regulated by NtmybA2ΔC
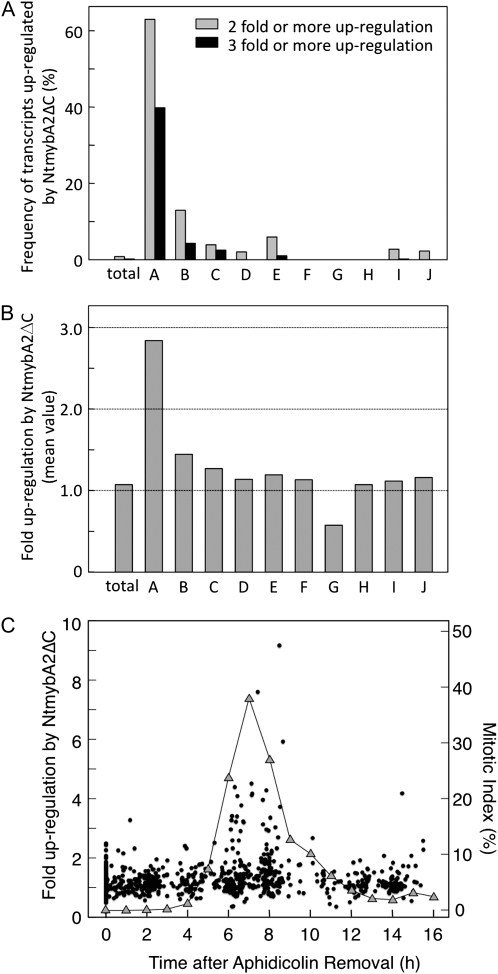
When our classification of the cell cycle-regulated transcripts was applied to the microarray data of 35S∷NtmybA2ΔC cells, it was clear that M phase-specific classes of transcripts were predominant in the group of transcripts that were up-regulated by NtmybA2ΔC. In Table I, there are 32 listed transcripts that were 3-fold or greater up-regulated in the 35S∷NtmybA2ΔC cells compared with the control cells transformed with the vector alone (Student's t test, P ≤ 0.05). Among them, 20 transcripts (63%) belonged to one of the three M phase-specific classes (A, B, or C), whereas these classes accounted for only 1.3% (214 transcripts) of the total clones in the microarray. Subsequently, we calculated the proportion of up-regulated transcripts in each of the 10 classes (A–J). Approximately 60% of the transcripts in class A increased more than 2-fold and nearly 40% increased more than 3-fold by overexpression of NtmybA2ΔC (Fig. 3A). The proportion of up-regulated transcripts in class B was lower than that in class A, but it was still substantially higher than that in the other classes (C–J). The ratio between expression signals in 35S∷NtmybA2ΔC and control cells was averaged for all transcripts in each class. As shown in Figure 3B, this value was much higher in class A (2.85) compared with the other classes, which varied around 1.0. Finally, for all cell cycle-regulated transcripts, the ratio between expression signals in 35S∷NtmybA2ΔC and control cells was plotted against their peak expression times, which were estimated from the expression data in the synchronous cultures (Fig. 3C). This analysis showed that the transcripts expressed maximally in the M phase (6–10 h) tended to be more strongly up-regulated by NtmybA2ΔC than those whose expression was maximal in the other phases during the cell cycle.
Table I.
Up-regulated transcripts in BY-2 cells overexpressing NtmybA2ΔC
cDNA clones with overlapping sequences are shown in the same row representing the data of both clones. In such cases, expression data from the two array spots were jointly used for statistical analysis to calculate fold change and P value.
| Fold Change | P Value | Gene ID | Phasea | Classb | Scorec | Annotation/Closest Homolog |
|---|---|---|---|---|---|---|
| 11.78 | 1.25E-04 | 39297 | M | A | 122 | Glycosyl hydrolase family 17 (Arabidopsis thaliana) |
| 7.18 | 2.20E-03 | 23145 | 328 | Patatin homolog (Nicotiana tabacum) | ||
| 7.03 | 1.01E-03 | 30077 | ||||
| 6.45 | 4.53E-03 | 29189/35265 | M | A/C | 95/84 | Thionin-like protein (Nicotiana tabacum) |
| 5.40 | 4.01E-03 | 26092/21549 | M | C | 73/72 | Protease inhibitor/seed storage/lipid transfer protein (LTP) family (Arabidopsis thaliana) |
| 5.04 | 4.80E-03 | 4484 | M | A | 36 | Microtubule-binding protein TAN1 (Arabidopsis thaliana) |
| 4.96 | 1.02E-03 | 27097 | ||||
| 4.79 | 8.19E-03 | 39367 | M | A | NACK1 kinesin (Nicotiana tabacum) | |
| 4.77 | 1.98E-03 | 27273/27065 | 113/104 | Unnamed protein product (Vitis vinifera) | ||
| 4.60 | 4.37E-03 | 1033 | M | A | 317 | B-type cyclin (Nicotiana tabacum) |
| 4.52 | 3.45E-03 | 28271 | M | A | ||
| 4.40 | 2.03E-03 | 5022 | M | A | 194 | Pentatricopeptide (PPR) repeat-containing protein (Arabidopsis thaliana) |
| 4.14 | 9.81E-06 | 6323 | M | A | 91.7 | Unnamed protein product (Vitis vinifera) |
| 4.12 | 7.69E-03 | 2455 | 70 | Putative cytochrome P450 (Oryza sativa, japonica group) | ||
| 4.07 | 1.98E-03 | 27045 | M | A | ||
| 3.92 | 3.69E-02 | 31040 | ||||
| 3.72 | 7.80E-04 | 6751 | M | B | 68.2 | Pectinesterase family protein (Brassica oleracea) |
| 3.53 | 2.38E-04 | 3201/2259 | S | I | 71.2/66 | Invertase/pectin methylesterase inhibitor family protein (Arabidopsis thaliana) |
| 3.43 | 2.32E-02 | 1005 | M | B | 384 | β-d-Glucan exohydrolase (Nicotiana tabacum) |
| 3.40 | 1.69E-03 | 1117 | M | A | 103 | Unnamed protein product (Vitis vinifera) |
| 3.37 | 9.60E-04 | 4820 | M | A | 46 | Hypothetical protein (Arabidopsis thaliana) |
| 3.33 | 6.93E-03 | 6658 | 70 | Auxin-induced (indole-3-acetic acid-induced) protein family (Arabidopsis thaliana) | ||
| 3.31 | 2.69E-04 | 24242 | M | B | 185 | Knolle (Capsicum annuum) |
| 3.29 | 1.64E-02 | 30283 | 53 | UP-9A (sulfur starvation-induced gene; Nicotiana tabacum) | ||
| 3.27 | 7.93E-03 | 10087 | M | A | ||
| 3.20 | 1.98E-02 | 2608 | 159 | Unnamed protein product (Vitis vinifera) | ||
| 3.19 | 2.02E-04 | 30017/24314 | M | B | 148/53.1 | Nonsymbiotic hemoglobin class 1 (Solanum lycopersicum) |
| 3.14 | 4.23E-03 | 22336 | M | A | 120 | Phytocyanin-like arabinogalactan protein (Gossypium hirsutum) |
| 3.11 | 7.73E-04 | 21354 | M | A | 216 | Cyclin-specific ubiquitin carrier protein, putative (Arabidopsis thaliana) |
| 3.10 | 5.51E-03 | 11337 | M | A | 106 | Unnamed protein product (Vitis vinifera) |
| 3.03 | 3.75E-03 | 6370 | 120 | Unnamed protein product (Vitis vinifera) | ||
| 3.02 | 1.79E-02 | 6468 | 78.2 | Gly-rich protein (Nicotiana tabacum) |
The cell cycle phase of peak expression for each cell cycle-regulated clone.
Classes representing defined patterns of cell cycle-regulated expression in the synchronized cultures.
Scores for the highest hit in the BLASTX search.
Figure 3.
M phase-specific classes of transcripts are preferentially and strongly up-regulated by NtmybA2ΔC overexpression. A, Frequency of the transcripts up-regulated by NtmybA2ΔC in each class. For each transcript, the ratio of the expression signal in 35S∷NtmybA2ΔC cells to that in control cells is averaged over three independent experiments. Frequencies of transcripts with this average expression ratio being more than 2 (gray bars) or 3 (black bars) are shown for each class (A–J). The frequency of such transcripts is also calculated in the total transcripts of the microarray (total). B, Magnitude of up-regulation by NtmybA2ΔC overexpression in each class. The expression ratio between the 35S∷NtmybA2ΔC and control cells for each transcript is averaged for each class (A–J). The averaged expression ratio is also calculated in the total transcripts of the microarray (total). C, Scatterplot showing correlation between the time of peak expression and the magnitude of up-regulation by 35S∷NtmybA2ΔC. For each cell cycle-regulated transcript, the expression ratio between 35S∷NtmybA2ΔC and control cells is plotted against the time of peak expression estimated from the expression data in synchronous cultures as described in “Materials and Methods” (black dots). The mitotic index for each time point is also plotted (gray triangles) for comparison.
Taken together, the combination of microarray data from the transgenic lines and those from the synchronized cultures demonstrated that overexpression of NtmybA2ΔC preferentially affects the transcript levels of M phase-specific genes in BY-2 cells. Furthermore, this analysis allowed us to identify novel target genes that are potentially regulated by NtmybA2.
Up-Regulated Genes in 35S∷NtmybA2ΔC Contain MSA Motifs
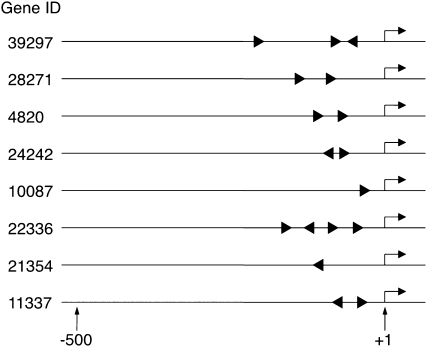
Our microarray analysis identified 20 M phase-specific transcripts that were up-regulated 3-fold or more in 35S∷NtmybA2ΔC cells (Table I). We hypothesized that these genes were regulated by NtmybA2 through direct binding to the MSA elements. In fact, this group of genes contained CYCB1;3 and NACK1, which were previously identified as target genes of NtmybA2 (Ito et al., 2001). To test if other genes in this group also have MSA motifs, we cloned upstream regions from eight representative genes (Gene ID/GenBank accession numbers are as follows: 4820/BP132633, 11337/BP526600, 10087/BP137396, 21354/BP530340, 22336/BP530697, 24242/BP531347, 28271/BP531840, and 39297/BP526919) by thermal asymmetric interlaced (TAIL)-PCR and determined the promoter sequences 0.9 to 1.9 kb upstream from the transcriptional start sites. The MSA elements contain a perfectly conserved characteristic core pentamer, AACGG, which is surrounded by less conserved nucleotide sequences (Ito, 2000). First, we searched for AACGG sequences in the forward and complementary strands in the eight promoter sequences and found 25 potential MSA core sites. By analyzing the sequences adjacent to the core motif, 18 of the 25 sequences matched the consensus MSA motif (TCYAACGGYYA) with identity scores of more than 70%. Figure 4 shows the positions and orientations of such putative MSA motifs in the promoter regions of the eight selected genes. All genes analyzed in this study contained one to four putative MSA motifs, most of which (17 of 18) were located proximal to the transcriptional start sites (−214 to −41). The presence of MSA-like motifs in these NtmybA2ΔC-responsive genes further supports our categorization of potential target genes of NtmybA2.
Figure 4.
Positions and orientations of MSA-like motifs in upstream regions of the genes up-regulated by NtmybA2ΔC. Each line shows the promoter region of the gene represented by a Gene ID, where arrows above the lines indicate transcriptional start sites. Numbers below the lines show nucleotide positions relative to transcriptional start sites (+1).
The MSA Element in NtE2C Is Functional in Vivo
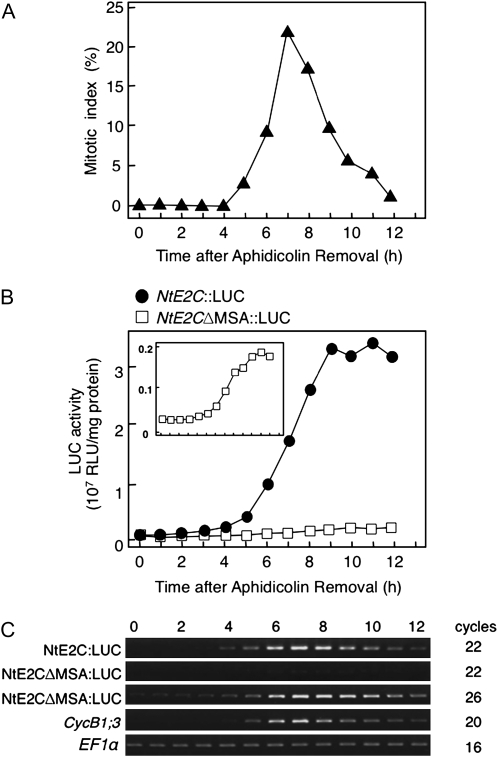
The gene corresponding to 21354/BP530340 was further analyzed to examine the in vivo function of its MSA motif. This transcript, designated NtE2C, encodes for the cyclin-specific ubiquitin carrier protein, also known as E2-C, which is involved in cyclin B degradation in animal systems (Zachariae and Nasmyth, 1999). In Arabidopsis, functional homologues are expressed in tissues with active cell division (Criqui et al., 2002). The G2/M phase-specific expression of NtE2C was confirmed by RNA gel blot analysis in the synchronized BY-2 cells (Supplemental Fig. S4). To analyze the cell cycle-regulated activity of the NtE2C promoter, the 1.0-kb promoter region was fused to the luciferase (LUC) reporter gene, and this construct (NtE2C∷LUC) was introduced into BY-2 cells. The resulting transgenic BY-2 cells were synchronized by the aphidicolin method. The peak mitotic index was observed 7 h after release from the aphidicolin block (Fig. 5A); the LUC activity of the NtE2C∷LUC cells rapidly increased from 6 to 9 h and then remained relatively constant (Fig. 5B). Similar changes in LUC activity were observed in our previous experiments, in which the LUC gene was fused to the CYCB1;3 and NACK1 promoters (Ito et al., 1998, 2001). Semiquantitative RT-PCR analysis of NtE2C∷LUC cells showed that LUC mRNA levels changed dramatically during the cell cycle, with maximal levels at 7 to 8 h (Fig. 5C). Therefore, we concluded that the 1.0-kb NtE2C promoter, which contains a single MSA motif, is sufficient for cell cycle-regulated transcription in BY-2 cells.
Figure 5.
Functional analysis of the MSA element in the upstream region of the NtE2C gene. The LUC reporter gene was fused to either the wild-type NtE2C promoter (NtE2C∷LUC) or its mutant form lacking the MSA elements (NtE2CΔMSA∷LUC). BY-2 cells were transformed with these constructs and synchronized by aphidicolin treatment. After release from the aphidicolin block, the cells were sampled at 1-h intervals during culture. A, Change in the mitotic index in the synchronized culture. Mitotic index of the NtE2C∷LUC cells is shown. The NtE2CΔMSA∷LUC cells showed a similar pattern (data not shown). B, Change in LUC enzyme activity in the synchronized cultures. Protein extracts were prepared from each sample, and LUC enzyme activities were determined. The inset shows the change in the LUC activities of NtE2CΔMSA∷LUC cells with an expanded scale. RLU, Relative light units. C, Semiquantitative RT-PCR analysis of LUC mRNA levels in the synchronized culture. Total RNA was extracted from each sample and used for the RT-PCR analysis of LUC, CYCB1;3, and EF1α mRNAs. The EF1α mRNA was analyzed as a control that shows constitutive expression during the cell cycle. After electrophoresis in the presence of ethidium bromide, the gels were photographed under UV light illumination. Numbers above the gel images show time in hours after aphidicolin removal, and those on the right indicate numbers of PCR amplification cycles.
To evaluate the significance of the MSA element, we introduced a point mutation in the core sequence of the MSA to create NtE2CΔMSA and fused it to the LUC gene. When this construct (NtE2CΔMSA∷LUC) was introduced into the BY-2 cells, LUC activity was about 20 times lower than that of the wild-type promoter, but surprisingly, the NtE2CΔMSA∷LUC cells still showed G2/M phase-specific increase in LUC activity in the synchronized cultures (Fig. 5B, inset). Similar results were obtained by semiquantitative RT-PCR analysis (Fig. 5C). Mutation of the MSA motif dramatically reduced LUC mRNA levels, although these levels still oscillated in the cell cycle, peaking at the M phase. This indicates that most of the transcriptional activity from the NtE2C promoter depended on the single MSA element; however, it could also contain an additional cis-element that acts as a weak G2/M phase-specific enhancer.
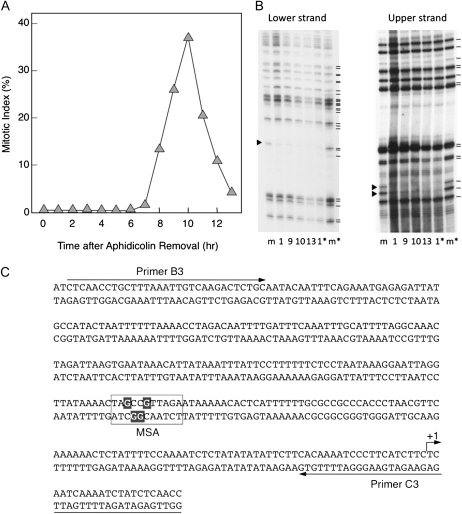
Subsequently, we analyzed the in vivo occupancy of the MSA element in the NtE2C promoter. In vivo footprinting experiments were designed to allow the analysis of both strands in the region between −296 and +22, which contains the MSA motif (Fig. 6C). As shown in Figure 6, B and C, the G residues in the MSA core sequence (AACGG) are strongly protected on both forward and complementary strands. No other site of clear protection was observed in the promoter region tested. The G residues in the MSA motif were constantly protected throughout the cell cycle in the synchronized cultures of BY-2 cells (Fig. 6, A and B). This suggests that some nuclear factors, most likely the c-Myb-like factors (NtmybA1, NtmybA2, and/or NtmybB), specifically bind to the MSA element in vivo, and such binding constantly occurs throughout the cell cycle.
Figure 6.
In vivo footprinting analysis of the NtE2C promoter during the cell cycle. A, Change in the mitotic index during the synchronized cultures of BY-2 cells. The cells were synchronized by aphidicolin treatment. After release from the aphidicolin block, the cells were sampled at 1-h intervals during the culture period to measure the mitotic index. B, Cells at the indicated times after aphidicolin removal were treated in vivo with DMS, and ligation-mediated PCR amplification was performed as described in “Materials and Methods.” As a control, ligation-mediated PCR was performed on in vitro DMS-treated genomic DNA (lane m). Signals representing G residues are shown by black bars, and the protected G residues are indicated by arrowheads. Asterisks indicate the results of the independent experiment. C, Protected residues in in vivo footprinting. The nucleotide sequences of the NtE2C promoter are shown for both strands. The MSA-like motif is boxed, and the protected G residues are shown with a black background. Positions of the end-labeled primers used for the ligation-mediated PCR are shown by arrows (primers B3 and C3).
Possible Autoregulation of NtmybA1 and NtmybA2 Genes
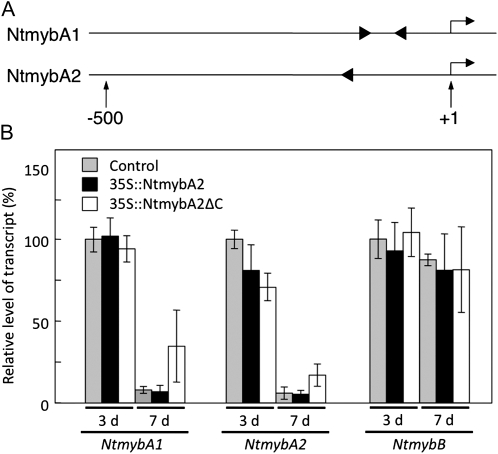
NtmybA1 and NtmyA2 are expressed in a cell cycle-regulated manner that slightly precedes the expression of CYCB1;3 (Ito et al., 2001). To examine if these Myb genes also have MSA elements in the promoter regions, we isolated their genomic fragments by TAIL-PCR and sequenced the 2.5-kb (NtmybA1) and 2.1-kb (NtmybA2) upstream regions. The upstream region of NtmybA1 contained two putative MSA motifs that contained the AACGG core motif and fitted the 11-bp consensus MSA sequence with an identity score of more than 70%, whereas that of NtmybA2 contained only one such motif (Fig. 7A). These motifs were located close to the transcriptional start sites, similar to their positions in other G2/M phase-specific genes (−74 and −127 in NtmybA1 and −148 in NtmybA2). RT-PCR analysis further showed that NtmybA2ΔC overexpression significantly increased NtmybA1 and NtmybA2 mRNA levels in cells during the stationary phase (Fig. 7B); however, there was no such change in the NtmybB mRNA levels. This suggests that NtmybA2, and possibly NtmybA1, activated its own transcription by binding to the MSA elements.
Figure 7.
Possible autoregulation of NtmybA2. A, Positions and orientations of the MSA-like motifs in the upstream regions of NtmybA1 and NtmybA2. Arrows above the lines indicate transcriptional start sites; numbers below the lines show the nucleotide positions relative to the transcriptional start sites (+1). B, Up-regulation of NtmybA1 and NtmybA2 genes in the 35S∷NtmybA2ΔC cells. The transgenic BY-2 cells carrying the vector alone (Control), 35S∷NtmybA2, and 35S∷NtmybA2ΔC were sampled on day 3 (logarithmic phase) and day 7 (stationary phase) after subculture. Total RNA was extracted from each sample, and real-time RT-PCR analysis was performed to quantify the transcript abundance of NtmybA1, NtmybA2, and NtmybB. The results of real-time PCR analysis are shown after normalization to EF1α mRNA expression. The relative transcript levels were averaged over the three biological replicates and are shown with the sds (error bars).
DISCUSSION
Coregulation of G2/M Phase-Specific Genes by NtmybA2
The MSA element has been identified as a cis-regulatory element that is necessary and sufficient for the G2/M phase-specific transcription of the CYCB1 gene in BY-2 cells (Ito et al., 1998). These elements are commonly present in the G2/M phase-specific genes of different plant species, including alfalfa (Medicago sativa), bell pepper (Capsicum annuum), tobacco, soybean (Glycine max), periwinkle (Vinca minor), and Arabidopsis (Ito, 2000; Schantz et al., 2005; Zhiponova et al., 2006). In tobacco, the R1R2R3-Myb transcription factor, NtmybA2, binds to the MSA motif in vitro, and its overexpression activates MSA-containing promoters in transient expression assays in BY-2 protoplasts (Ito et al., 2001). These findings suggested that many G2/M phase-specific genes might be regulated by a common mechanism that involves interaction between the MSA elements and NtmybA2 in tobacco. The present study provided direct evidence supporting this hypothesis. First, microarray analysis showed that the M-specific classes of transcripts were much more frequently and strongly up-regulated by NtmybA2ΔC overexpression than those expressed at other phases of the cell cycle. Second, promoter sequences of the strongly up-regulated genes contained motifs that are highly homologous to the MSA consensus sequence. Third, the single MSA-like motif present in one of the up-regulated genes, NtE2C, was functional in vivo. Mutation of this motif resulted in a dramatic reduction of NtE2C promoter activity in BY-2 cells. Taken together, our results provide strong evidence that NtmybA2, and possibly NtmybA1, directly and positively regulates the transcription of many G2/M phase-specific genes in tobacco.
Differential Effects of NtmybA2ΔC on G2/M Phase-Specific Genes
We found that the effect of NtmybA2ΔC overexpression varied significantly depending on the individual genes of M-specific classes that were defined by our microarray analysis. One possibility is that not all G2/M phase-specific genes may be directly targeted by NtmybA2. We noticed a strong tendency for the genes up-regulated by NtmybA2ΔC to show sharp expression peaks at the M phase in synchronous cultures. Among the transcripts of the M phase-specific classes, those in class A, which showed their expression peaks with the greatest sharpness, were most frequently and strongly up-regulated by NtmybA2ΔC overexpression (Figs. 2 and 3). We speculate that most of the class B and C transcripts may not be directly regulated by NtmybA2. Our data also showed that overexpression of the truncated hyperactive from of NtmybA2 did not equally affect the target genes. There was substantial variation in the effects of NtmybA2ΔC on the expression levels of individual class A genes that were shown to have MSA-like motifs. The ratio of transcript levels in the 35S∷NtmybA2ΔC and control cells varied greatly, ranging from 3.10 (Gene ID 11337) to 11.8 (Gene ID 39297). We could not find a direct correlation between the number of MSA motifs in the promoter regions and the effect of NtmybA2ΔC overexpression. Thus, gene 22336, which has four MSA-like motifs, was less affected by NtmybA2ΔC compared with gene 10087, which has a single MSA motif. These findings are consistent with our previous study in Arabidopsis plants with simultaneous mutations in MYB3R1 and MYB3R4 genes, which are highly homologous to NtmybA2. In this double mutant, several G2/M phase-specific genes were down-regulated, but the reduction in transcript levels varied significantly depending on the individual genes, irrespective of the number of MSA-like motifs. For example, the KN and CYCB1;1 genes both contained three MSA-like motifs; however, the former was down-regulated by 3.3-fold in the double mutant, whereas the latter showed no such down-regulation at all (Haga et al., 2007). Nevertheless, it has been reported that the MSA elements are essential for CYCB1;1 expression in Arabidopsis (Li et al., 2005). Our hypothesis is that Arabidopsis and tobacco may express some other MSA-binding activators and that the contribution of NtmybA2 and its homologues may vary depending on the individual target genes. Another possibility is that some other cis-acting elements might act positively on the transcription of some G2/M phase-specific genes, which could also explain the differential effects of NtmbyA2C overexpression. Indeed, the presence of additional cis-elements was suggested by our analysis of NtE2C promoters, which showed residual transcriptional activity at the G2/M phase in the absence of the MSA element (Fig. 5). In the light of this result, it is noteworthy that the NtE2C promoter contains the GCCCR motif, which could enhance the transcriptional activity of the CYCB1;1 gene (Li et al., 2005).
Stable Transformants Versus Transient Assays
We found clear differences in the effects of NtmybA2 overexpression in the two different experimental systems: stable overexpression in transformed BY-2 cells (this study) and transient overexpression in BY-2 protoplasts transfected with a plasmid containing 35S∷NtmybA2 (Ito et al., 2001). In the earlier experiments, overexpression of the full-length NtmybA2 strongly activated LUC reporter gene expression from plasmids containing CYCB1;3∷LUC or NACK1∷LUC. The transcription of the LUC gene was further enhanced by C-terminal truncation of NtmybA2 (Araki et al., 2004). Conversely, this study showed that CYCB1;3 and NACK1 genes were up-regulated only in 35S∷NtmybA2ΔC stably transformed cells but not in 35S∷NtmybA2 cells. This discrepancy may be explained by the difference in the state of the cells (intact cells versus protoplasts) or levels of expression of NtmybA2 between the two experimental systems. Another possible explanation is that increased NtmybA2 expression may activate target promoters when they are present in naked DNA molecules but not when they are present in a native chromatin configuration. Our speculation is that there might be some other rate-limiting steps in addition to the increased expression of NtmybA2 for the activation of target promoters in the native chromatin structure. Indeed, it has been reported that chromatin modification mediated by HISTONE MONOUBIQUITINATION1 positively regulates the expression of many G2/M phase-specific genes in Arabidopsis (Fleury et al., 2007). Our results suggest that the C-terminal region of NtmybA2 inhibits its own activity for transcriptional activation of the endogenous genes and that relieving this inhibitory effect might enable up-regulation of target genes in native chromatin structure. The CDK/CYCB1-mediated phosphorylation of NtmybA2 is one possible mechanism for relieving this inhibitory effect (Araki et al., 2004).
Positive Feedback Regulation of G2/M Phase-Specific Transcription
We showed that NtmybA1 and NtmybA2 contain MSA motifs in their promoter regions. These genes were up-regulated by NtmybA2ΔC overexpression in BY-2 cells during the stationary phase, as was observed for other G2/M phase-specific genes such as NtKN and CYCB1;3. Our data suggest that the NtmybA2 gene itself may be one of the downstream targets of NtmybA2. Therefore, we can postulate the existence of an autoregulatory loop in which NtmybA2 activates its own transcription. In general, the autoregulatory control provides a very sensitive means to control the accumulation of rate-limiting products. Similar autoregulation is also known for the E2F transcription factor in animal cells, which acts as a rate-limiting factor during the G1-to-S transition (Johnson et al., 1994). In this autoregulation, transcription of the E2F1 gene is activated by E2F activity, which comprises its own products. Moreover, we previously suggested another mode of positive feedback control for the activation of NtmybA2 activity, in which CYCB1 transcription is activated by NtmybA2, which in turn is activated by a CDK in a complex with CYCB1 (Araki et al., 2004). These positive feedback controls would allow an initial modest accumulation of NtmybA2 activity to rapidly amplify NtmybA2 expression, thus resulting in a burst of NtmybA2 activity. Such mechanisms might help to explain the observed sharp increase in the levels of the G2/M phase-specific transcripts during the cell cycle, which might further explain the rapid and irreversible entry into mitosis. The existence of multiple pathways of positive feedback regulation might provide robustness to the mechanisms that ensure the transcription of a specific set of genes at the correct time during the cell cycle.
In summary, we propose that NtmybA2 positively coregulates many G2/M phase-specific genes by binding to the MSA elements in tobacco and that its activity might be controlled by multiple mechanisms of positive feedback regulation. Such mechanisms might provide a sensitive switch that allows a rapid increase in the set of transcripts at the G2-to-M transition.
MATERIALS AND METHODS
Construction of Plasmids
For cloning of the cauliflower mosaic virus 35S promoter into a binary vector, the KpnI/EcoRV fragment of pJIT60 (Guerineau and Mullineaux, 1993) was blunt ended by T4 DNA polymerase and cloned into the EcoRI/HindIII interval of pPZP211 (Hajdukiewicz et al., 1994). The resulting plasmid, pPZP211-35S, was used to construct 35S∷NmybA2 and 35S∷NtmybA2ΔC. The pGAD10 plasmid containing the full-length NtmybA2 cDNA was isolated previously (Ito et al., 2001); this plasmid was cut by SalI, and the excised cDNA fragment was cloned into the SalI site of pPZP211-35S to create 211-35S∷NtmbyA2. Similarly, the 211-35S∷NtmbyA2ΔC construct was created by inserting the SalI fragment of pJIT-NtmybA2Δ630 (Araki et al., 2004) into the SalI site of pPZP211-35S.
The promoter region of NtE2C was amplified from BY-2 genomic DNA by PCR using the sense primer 5′-AACTGCAGGAGTAGCAGAGACAAAGTGCCAA-3′ containing a PstI site and the antisense primer 5′-AAGTCGACTTGTAGAACAGAAAAAGGAGACGT-3′ containing the SalI site. The amplified fragment was cut with PstI and SalI and cloned into the PstI/SalI site of pPZP211-LUC (Kono et al., 2003) to create 211-NtE2C∷LUC. The NtE2C promoter was mutagenized at the MSA motif with the QuikChange multi site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions using the mutagenic primer 5′-GGTTATAAAACTAGCCAATAGAATAAAAACACTC-3′. The mutated NtE2C promoter, in which CCGTT in the MSA motif was changed to CCAAT, was cloned into the PstI/SalI site of pPZP211-LUC to create 211-NtE2CΔMSA∷LUC.
Cells, Transformation, and Synchronization
Maintenance, synchronization, and Agrobacterium tumefaciens-mediated transformation of tobacco (Nicotiana tabacum) BY-2 cells were performed as described previously (Ito et al., 1998). DNA synthesis and mitotic index determination were described previously (Genschik et al., 1998).
Expression Analysis
Extraction of total RNA, cDNA synthesis, and real-time RT-PCR analysis were performed as described previously (Haga et al., 2007). For semiquantitative PCR analysis, ExTaq polymerase (TaKaRa) was used. Amplification was performed with cycles of 93°C for 30 s, 56°C for 1 min, and 72°C for 2 min. The PCR products were electrophoresed on 1.0% agarose gels in the presence of ethidium bromide (0.5 mg L−1). RNA gel blot analysis was performed as described previously (Ito et al., 1997). The primer sequences used for real-time RT-PCR and semiquantitative PCR analyses are available upon request.
Microarray Analysis
Tobacco microarrays were prepared by spotting PCR-amplified cDNA fragments from BY-2 cells onto microarray slides (Amersham Biosciences) as described previously (Gális et al., 2006). Sequences of cDNA clones that provided quality sequence during single-run sequencing can be retrieved from http://www.ncbi.nlm.nih.gov/sites/entrez after the addition of prefix “BY” to designated clone numbers (e.g. BY4820).
Probe Labeling and Microarray Hybridization
One microgram of mRNA was labeled using SuperScript II reverse transcriptase (Invitrogen), random 9-mer primers, poly(A)-specific poly(dT) primers, and Cy-3/Cy-5-labeled dCTP nucleotides (Amersham Biosciences) for the first-strand cDNA synthesis. The slides with UV cross-link-immobilized DNA were prehybridized for 1 h at 55°C in prehybridization buffer (1% [w/v] BSA, 5× SSC, and 0.1% [w/v] SDS). Hybridization of microarrays with Cy-5-labeled sample cDNA and the Cy-3-labeled vector sequence used as a signal normalization factor was performed in ExpressHyb hybridization solution (Clontech) at 60°C for 4 h as described previously (Gális et al., 2006). The slides were washed in SSC/SDS buffer solutions at 55°C (final wash in 0.1× SSC and 0.1% [w/v] SDS), and the Cy-5 (sample signal) and Cy-3 (normalization signal) values were obtained by scanning with a GenePix 4000A microarray scanner (Molecular Dynamics). The scanned images were analyzed with the MicroArray Suite program (Scananalytics, BD Biosciences).
Normalization and Processing of Microarray Data
Duplicate spot signals for each cDNA EST on the microarray slides from two independent biological replicates of the cell cycle experiment (A and B) and three biological replicates of the overexpression experiment (A, B, and C) were processed using GeneSpring software (Agilent Technologies) as follows: (1) each average sample value of the duplicated spots (Cy-5-labeled cDNA) was normalized relative to the average Cy-3-labeled vector control signal (per gene normalization), and (2) median polishing (per gene and per chip) normalization was determined by the implemented default method of the GeneSpring software program. Normalized values for the cell cycle experiment were further filtered as described in detail in Supplemental Figure S2. The NtmybA2/control and NtmybA2ΔC/control data spots, with average normalized signal ratios at least 1.5-fold higher (or lower for suppressed clones) than the corresponding spot in the control cells, were selected independently for experiments A, B, and C. Only signals that simultaneously increased or decreased in all three biological replicates of the experiment were considered significant and selected for further analysis. Furthermore, the data points that did not fulfill the statistical criteria for arithmetic average from three replicated signal ratios (Student's t test, P > 0.05) were excluded from the analysis. In order to validate the microarray data by an independent method, 10 genes were selected from the cell cycle kinetic experiment and the target sequences were amplified by PCR using specific primers and template cDNA from synchronized BY-2 cells (Supplemental Fig. S3).
The peak expression time in synchronized cultures was estimated for each cell cycle-regulated gene using microarray data from experiment A. We first selected genes that showed highest signal intensity at time points between 2 and 14 h. The expression signals of the highest time point and 2 h before and after the highest time point were used to establish the second-order regression curve. The time point of the vertex of the curve was defined as the estimated time of the peak expression and used to create Figure 3C.
LUC Assay
LUC assays were performed as described previously (Ito et al., 1998).
In Vivo Genomic Footprinting
In vivo footprinting was done as described previously (Chabouté et al., 2000). About 20 mL of BY-2 cell suspensions was incubated with 0.5% to 1% (v/v) dimethyl sulfate (DMS). After 2 min of incubation at 25°C, the reaction was stopped by adding 500 mL of ice-cold water. The cells were intensively washed with ice-cold water, collected on a filter, and frozen in liquid nitrogen. DNA samples were prepared with the Nucleon Phytopure plant DNA extraction kit (Amersham Biosciences) or the Plant DNAzol reagent (Invitrogen). The DNA samples were further purified by RNaseA treatment and extraction with phenol/chloroform. DNA was cleaved at the level of DMS-modified residues with 1 m piperidine at 95°C for 30 min, lyophilized thrice, and precipitated. The cleaved DNA was then amplified by ligation-mediated PCR. Two sets of three overlapping primers were used to separately analyze the lower and upper strands of the NtE2C promoter. The asymmetric linkers used for ligation had the canonical sequences 5′-GCGGTGACCCGGGGAGATCTGAATTC-3′ and 5′-GAATTCAGATC-3′. The primers used were 5′-CGACCTTAAGCATAAAACTCGAT-3′ and 5′-CGACCTTAAGCATAAAACTCGAT-3′ for the first PCR, 5′-TTTATATAATTGATCTCAACCTGCTTTAAAT-3′ and 5′-GAGAGAAGGGTTGAGATAGATTTTGATTGAG-3′ for the second PCR, and 5′-CTCAACCTGCTTTAAATTGTCAAGACTCTGC-3′ and 5′-GGTTGAGATAGATTTTGATTGAGAAGATGAAGGGATTTTGTG-3′ for the third PCR. Primers for the third PCR were end labeled with [γ-32P]ATP by T4 DNA polynucleotide kinase. The amplified products were analyzed on standard sequencing gels, and the gels were dried and autoradiographed.
Flow Cytometric Analysis
Samples for flow cytometric analysis were prepared using the Cystain UV Precise P reagent kit (Partec). About 0.1 g fresh weight of BY-2 cells was collected on filter paper, frozen in liquid nitrogen, and stored at −80°C. The samples were then chopped using a razor blade in 0.5 mL of nuclei extraction buffer. After filtration through a 30-μm-mesh nylon sieve, 2 mL of staining solution containing 4′,6-diamidino-2-phenylindole was added. The samples were then analyzed with a PAS flow cytometer (Partec).
Other Nucleic Acid Procedures
Nucleotide sequences were determined using an ABI 3130xl genetic analyzer and the BigDye terminator cycle sequencing kit version 3.1 (Applied Biosystems). The TAIL-PCR procedure was carried out as described previously (Liu et al., 1995). Sequences of arbitrary degenerate primers and gene-specific primers for NtE2C, NtmybA1, and NtmybA2 are available upon request. Transcription start sites were determined by sequencing PCR products of 5′ RACE, which was performed using the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer's instructions.
Sequence data for the promoter regions in Figure 4 can be found in the GenBank/EMBL data libraries under accession numbers AB472848 to AB472855. Microarray data from this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE14250 (synchronization experiment) and GSE14251 (overexpression experiment).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cell cycle progression in synchronized cultures of BY-2 cells.
Supplemental Figure S2. Flow chart showing the processing of microarray data for cell cycle-regulated transcripts.
Supplemental Figure S3. Real-time RT-PCR analysis of the representative transcripts in each of the cell cycle-regulated classes.
Supplemental Figure S4. Expression pattern of NtE2C in synchronized BY-2 cells.
Supplemental Table S1. Normalized expression data for transgenic BY-2 cells carrying 35S∷NtmybA2, 35S∷NtmybA2ΔC, and the vector alone.
Supplemental Table S2. Summary of genes up- or down-regulated in 35S∷NtmybA2ΔC cells.
Supplemental Table S3. Normalized expression data during the cell cycle in synchronized cultures of BY-2 cells.
Supplemental Table S4. Summary of cell cycle-regulated genes in BY-2 cells.
Supplementary Material
Acknowledgments
We thank Tomoko Narisawa and Mami Sasaki for helping in the microarray analysis, Miki Yoshioka, Kanako Komatsu, Yuka Sako, and Hiro Iguchi for technical assistance, and Yasunori Machida for helpful discussions.
This work was supported by Grants-in-Aid from the Japan Society for the Promotion of Science (grant no. 20061013) and from the Ministry of Education, Culture, Sports, Science, and Technology (grant no. 19570034) and by the Yamada Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masaki Ito (masakito@agr.nagoya-u.ac.jp).
The online version of this article contains Web-only data.
References
- Araki S, Ito M, Soyano T, Nishihama R, Machida Y (2004) Mitotic cyclins stimulate the activity of c-Myb-like factors for transactivation of G2/M phase-specific genes in tobacco. J Biol Chem 279 32979–32988 [DOI] [PubMed] [Google Scholar]
- Breeden LL (2003) Periodic transcription: a cycle within a cycle. Curr Biol 13 31–38 [DOI] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G, et al (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Sekine M, Philipps G, Chaubet-Gigot N (2000) Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 12 1987–2000 [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ (2001) Transcriptional regulation and function during the human cell cycle. Nat Genet 27 48–54 [DOI] [PubMed] [Google Scholar]
- Criqui MC, de Almeida Engler J, Camasses A, Capron A, Parmentier Y, Inzé D, Genschik P (2002) Molecular characterization of plant ubiquitin-conjugating enzymes belonging to the UbcP4/E2-C/UBCx/UbcH10 gene family. Plant Physiol 130 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8 655–665 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Joubès J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6 536–543 [DOI] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális I, Simek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H, Matsuoka K (2006) A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J 46 573–592 [DOI] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J (1998) Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 10 2063–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR (2007) Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev 21 2880–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau F, Mullineaux P (1993) Plant transformation and expression vectors. In RRD Croy, ed, Plant Molecular Biology Labfax. Bios Scientific Publishers, Oxford, pp 121–147
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voss U, Jürgens G, et al (2007) R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134 1101–1110 [DOI] [PubMed] [Google Scholar]
- Ito M (2000) Factors controlling cyclin B expression. Plant Mol Biol 43 677–690 [DOI] [PubMed] [Google Scholar]
- Ito M (2005) Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res 118 61–69 [DOI] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13 1891–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Criqui MC, Sakabe M, Ohno T, Hata S, Kouchi H, Hashimoto J, Fukuda H, Komamine A, Watanabe A (1997) Cell-cycle-regulated transcription of A- and B-type plant cyclin genes in synchronous cultures. Plant J 11 983–992 [DOI] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR (1994) Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev 8 1514–1525 [DOI] [PubMed] [Google Scholar]
- Kono A, Umeda-Hara C, Lee J, Ito M, Uchimiya H, Umeda M (2003) Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol 132 1315–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whitter RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Demura T, Gális I, Horiguchi T, Sasaki M, Tashiro G, Fukuda H (2004) A comprehensive gene expression analysis toward the understanding of growth and differentiation of tobacco BY-2 cells. Plant Cell Physiol 45 1280–1289 [DOI] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41 546–566 [DOI] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S (1992) Tobacco BY2 cell line as the “HeLa” cell in cell biology of higher plants. Int Rev Cytol 132 1–30 [Google Scholar]
- Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B, Hauser S, Gaubatz S (2007) The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schantz ML, Jamet E, Guitton AE, Schantz R, Houlné G (2005) Functional analysis of the bell pepper KNOLLE gene (cakn) promoter region in tobacco plants and in synchronized BY2 cells. Plant Sci 169 155–163 [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inzé D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K (1999) Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev 13 2039–2058 [DOI] [PubMed] [Google Scholar]
- Zhiponova MK, Pettkó-Szandtner A, Stelkovics E, Neer Z, Bottka S, Krenács T, Dudits D, Fehér A, Szilák L (2006) Mitosis-specific promoter of the alfalfa cyclin-dependent kinase gene (Medsa;CDKB2;1) is activated by wounding and ethylene in a non-cell division-dependent manner. Plant Physiol 140 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.