Abstract
Autophagy is an intracellular process in which a portion of cytoplasm is transported into vacuoles for recycling. Physiological roles of autophagy in plants include recycling nutrients during senescence, sustaining life during starvation, and the formation of central digestive vacuoles. The regulation of autophagy and the formation of autophagosomes, spherical double membrane structures containing cytoplasm moving toward vacuoles, are poorly understood. HVA22 is a gene originally cloned from barley (Hordeum vulgare), which is highly induced by abscisic acid and environmental stress. Homologs of HVA22 include Yop1 in yeast, TB2/DP1 in human, and AtHVA22a to -e in Arabidopsis (Arabidopsis thaliana). Reverse genetics followed by a cell biology approach were employed to study the function of HVA22 homologs. The AtHVA22d RNA interference (RNAi) Arabidopsis plants produced small siliques with reduced seed yield. This phenotype cosegregated with the RNAi transgene. Causes of the reduced seed yield include short filaments, defective carpels, and dysfunctional pollen grains. Enhanced autophagy was observed in the filament cells. The number of autophagosomes in root tips of RNAi plants was also increased dramatically. The yop1 deletion mutant of Saccharomyces cerevisiae was used to verify our hypothesis that HVA22 homologs are suppressors of autophagy. Autophagy activity of this mutant during nitrogen starvation increased in 5 min and reached a plateau after 2 h, with about 80% of cells showing autophagy, while the wild-type cells exhibited low levels of autophagy following 8 h of nitrogen starvation. We conclude that HVA22 homologs function as suppressors of autophagy in both plants and yeast. Potential mechanisms of this suppression and the roles of abscisic acid-induced HVA22 expression in vegetative and reproductive tissues are discussed.
Abscisic acid (ABA) regulates the expression of hundreds of genes in a plant (Seki et al., 2002). The specific functions of many of these genes are still not clear, but they are assumed to be related to the physiological roles of ABA, such as promoting seed maturation, inhibiting seed germination, and conferring tolerance to environmental stress. HVA22, a gene isolated from barley (Hordeum vulgare) aleurone cells treated with ABA, encodes a small peptide of 130 amino acids without an obvious functional motif (Shen et al., 1993). Expression of HVA22 is dramatically up-regulated by ABA not only in aleurone cells but also in vegetative tissues (Shen et al., 2001). Homologs of HVA22 have been identified in diverse eukaryotes, including plants, fungi (named Yop1 in Saccharomyces cerevisiae), mammals (named TB2 or DP1 in human), flies, and worms, but not in any prokaryotes. Expression of yeast Yop1 is also up-regulated by stress (Shen et al., 2001). Studies of the physiological role of Yop1 protein revealed that Yop1p is a Yip1p-interacting protein that is an essential protein regulating a critical step in the Rab GTPase-mediated membrane transport in S. cerevisiae (Calero et al., 2001). In another study, synthetic enhancement mutant screening was performed and SEY1 was identified as a Yop1-dependent complementation gene (Brands and Ho, 2002). The yeast gene SEY1 is a homolog of the Arabidopsis (Arabidopsis thaliana) gene RHD3, whose function is to facilitate vesicle transport in root hairs. Mutation of RHD3 causes vesicle accumulation at the tips of root hairs and the formation of short and wavy root hairs (Schiefelbein and Somerville, 1990; Galway et al., 1997). Due to the apparent vesicle accumulation in the yeast yop1/sey1 double mutant, it was suggested that Yop1 protein is involved in vesicular trafficking.
To obtain more insight into the role of HVA22, a functional analysis of HVA22 homologs was carried out in Arabidopsis. Five HVA22 homologs were identified in Arabidopsis, named AtHVA22a to -e. Expression of these five genes is differentially up-regulated by ABA and environmental stress, except for AtHVA22c, which is nearly unaffected by these treatments. Expression levels of AtHVA22 genes vary in different organs; generally, fast-growing organs such as flowers and inflorescence stems have higher expression levels than slow-growing organs such as mature rosette and cauline leaves. Among the five, expression of AtHVA22d is most tightly regulated by ABA in vegetative tissues (Chen et al., 2002). In this work, reverse genetics followed by a cell biology approach were used to study the function of HVA22 homologs. The results we obtained suggest that the role of the HVA22 homologs is to regulate autophagy in a negative manner in both Arabidopsis and S. cerevisiae.
Autophagy is a cytoplasmic recycling mechanism by which a portion of cytoplasm is transported into vacuoles (lysosomes in animal cells) and degraded therein. The regained nutrients are either mobilized to different parts of the organism or used in the same cell. Aspects of autophagy, including physiological and molecular, have been described in eukaryotes (Yorimitsu and Klionsky, 2005). Recent studies of autophagy in plants have been largely based on the knowledge obtained from S. cerevisiae autophagy mutants (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Klionsky and Ohsumi, 1999). More than 30 S. cerevisiae autophagy genes have been identified and are involved in different aspects of autophagy, such as regulation, autophagosome formation, and degradation of autophagic vesicles in vacuoles (Bassham et al., 2006). One gene involved in the regulation, encoding a Ser/Thr protein kinase called Tor1, functions as an autophagy suppressor in S. cerevisiae and mammalian cells (Noda and Ohsumi, 1998; De Virgilio and Loewith, 2006; Inoki and Guan, 2006). Its negative regulatory role is executed by inhibiting Atg1p-Atg13p complex formation (Kamada et al., 2000), although the underlying mechanism is still not clear. Atg1p is also a Ser/Thr protein kinase, and Atg13p is an activator for Atg1p activity. When Tor1p activity is suppressed chemically or S. cerevisiae is under starvation, Atg13p is dephosphorylated and binds to Atg1p. This binding activates Atg1p activity and leads to autophagy. Although the Atg1p-Atg13p complex is essential for autophagy, it is not sufficient for this process. An additional component, Atg17p, is also required in the Atg1p-Atg13p complex to carry out autophagy (Kabeya et al., 2005). How the Atg1p-Atg13p-Atg17p complex promotes the formation of autophagosomes, and the relation between Tor1p and the Atg1p-Atg13p-Atg17p complex, still need to be elucidated. The immediate substrate(s) of Tor1p in the cytoplasm has not been identified; nevertheless, Tor1p does not seem to be involved in the membrane/lipid trafficking directly. Therefore, it is possible that another autophagy regulation mechanism(s) might exist that controls membrane trafficking and thus the formation of autophagosomes.
Autophagosome formation is the central part of macroautophagy. Similar to microautophagy, which engulfs cytoplasm into vacuoles nonselectively, macroautophagy nonselectively enwraps cytoplasm using a double membrane, thereby creating autophagosomes. When autophagosomes fuse with vacuoles, the outer membrane integrates into the vacuole membrane and the cytoplasm surrounded by the inner membrane is released into the vacuole lumen. Although many proteins have been identified that are associated with the preautophagosomal structures (Suzuki et al., 2001) and autophagosomes (Yorimitsu and Klionsky, 2005), the mechanisms of preautophagosomal structure formation, expansion into autophagosomes, and the following targeting and fusion with vacuoles are still poorly understood. Most of the S. cerevisiae autophagy genes have homologs in Arabidopsis and other eukaryotes, suggesting that autophagy is a well-conserved process. Yet, the origin of the autophagosome membrane is still controversial. Studies on induced autophagy in rat liver showed that the autophagosome membrane arises from the endoplasmic reticulum (ER; Dunn, 1990). On the other hand, the origin of the autophagosome membrane in S. cerevisiae is still under debate (Noda et al., 2002; Yorimitsu and Klionsky, 2005).
Here, we report the results from studies on AtHVA22 genes in Arabidopsis and Yop1 in S. cerevisiae. Our results reveal that these HVA22 homologs negatively regulate autophagy in both Arabidopsis and yeast.
RESULTS
Selection and Phenotyping of AtHVA22d RNAi Transgenic Plants
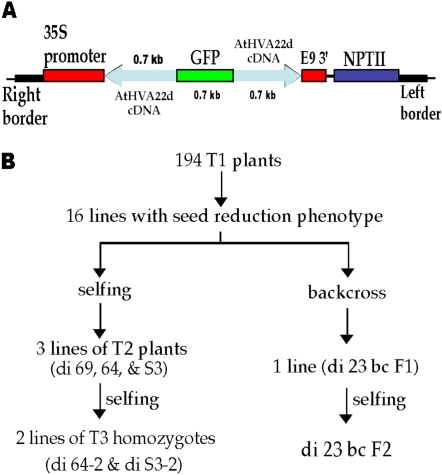
AtHVA22d cDNA, including parts of its 5′ and 3′ untranslated regions, was used to make the RNAi construct for Arabidopsis transformation (Fig. 1A). After transformation, T1 seeds were harvested and germinated on agar plates containing kanamycin for transgenic plant selection. One hundred ninety-four lines were selected and grown in soil for phenotyping and allowed to set T2 seeds. Among these plants, 16 lines displayed a promising phenotype of aberrant siliques in which no or few seeds were produced. The selection scheme is shown in Figure 1B. Among the 16 lines, some were able to produce seeds by self-pollination. Those that could not set seeds by self-pollination either perished or were rescued by backcrossing with wild-type ecotype Columbia (Col-0) pollen. Among the surviving lines, four showed stably heritable phenotypes, and these lines were used in this study. Among the four lines, di 23 bc F2 was derived from a backcross as follows. This line was generated by backcrossing di 23 T1 with Col-0 pollen to produce di 23 bc F1. The F1 seeds were germinated on agar plates containing kanamycin, and the surviving seedlings (about 10% survived) were transferred to soil. The di 23 bc F2 seeds were harvested from one of these di 23 bc F1 plants that exhibited a heritable phenotype. Because homozygous di 23 bc F3 could not be identified (see below for explanation), this line was retained as a segregating F2 population for this study. Another line, di 69 T2, resulting from self-pollination, was also retained as T2 segregates because homozygous T3 could not be identified. The transgenic-nontransgenic ratios of the F2 and the T2 populations were both about 1:1 as genotyped by PCR (see below) and based on kanamycin selection. This non-Mendelian segregation of di 23 bc F2 and di 69 T2 and the low kanamycin resistance ratio of di 23 bc F1 seeds suggest that lethality was associated with the action of the AtHVA22d RNAi transgene. Two other lines, di S3-2 and di 64-2, were T3 homozygous, and their phenotype was weaker than that of the aforementioned F2 and T2 lines. Their nontransformed T3 segregates were di S3-5 and di 64-5, respectively.
Figure 1.
AtHVA22d RNAi construct for Arabidopsis transformation and the selection scheme for RNAi transgenic plants. A, Two AtHVA22d cDNA fragments (protein-coding region plus part of 5′ and 3′ untranslated regions) were inserted into pMON 10098 to flank a GFP cDNA fragment in either sense or antisense orientation for Agrobacterium-mediated transformation. The size of each DNA fragment is shown above or below the fragment, and the orientation of the AtHVA22d cDNA fragments is indicated by the arrows. This construct is driven by the cauliflower mosaic virus 35S promoter, and the terminator is E9 (pea rbcs-E9 terminator). The NPTII (for neomycin phosphotransferase II) gene is the antibiotic selection marker. B, After transformation, 194 T1 lines were selected by kanamycin resistance. Among the 194 T1 lines, 16 displayed severe seed reduction in siliques. These plants were either backcrossed to wild-type Arabidopsis Col-0 or allowed to self-pollinate. Only one line showed a stable phenotype in the backcrossed progeny, and this line had difficulties in producing homozygous F3 plants. Three T2 lines showed a stable phenotype in the self-pollination group, and two of these three lines were able to produce T3 homozygotes. [See online article for color version of this figure.]
Cross-Suppression of the Expression of the AtHVA22 Gene Family by the RNAi Transgene
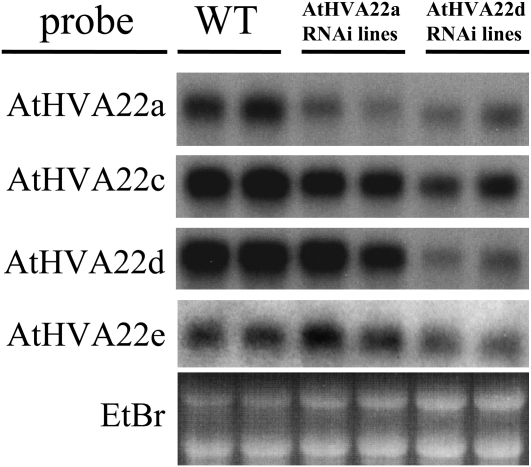
AtHVA22a and AtHVA22d RNAi transgenic plants were generated initially to compare their phenotypes. The phenotype of AtHVA22a T1 RNAi transgenics was similar to that of AtHVA22d T1 RNAi plants, although weaker. To examine the cosuppression effectiveness of the two RNAi transgenes, two T1 plants were arbitrarily selected from AtHVA22a and AtHVA22d RNAi plants for northern-blot analysis. As shown in Figure 2, expression of AtHVA22a was suppressed significantly but not for the other three homologs (AtHVA22c, -d, and -e) in AtHVA22a RNAi plants. In AtHVA22d RNAi plants, however, apparent suppression was observed for the four genes (AtHVA22a, c, d, and e) whose transcripts could be detected by northern-blot analysis. Expression of AtHVA22b was too low to detect (Chen et al., 2002). Based on the phenotype of the transgenic plants and the effectiveness of the RNAi transgenes, AtHVA22d RNAi plants were focused on for the gene functional study in this work.
Figure 2.
Effectiveness of AtHVA22a and AtHVA22d RNAi transgenes on the expression of the AtHVA22 gene family in Arabidopsis. Northern-blot analysis was performed with two arbitrarily selected transgenic plants from AtHVA22a or AtHVA22d T1 RNAi transformants to examine the RNAi suppression on AtHVA22 gene family expression. The phenotype of AtHVA22a RNAi plants was similar to that of AtHVA22d RNAi plants, although weaker. Different degrees of suppression on AtHVA22 gene family expression were observed among the AtHVA22a and AtHVA22d RNAi lines, and the suppression effect of AtHVA22d RNAi was stronger than that of AtHVA22a RNAi. The level of expression of AtHVA22b was too low to be detected. EtBr, Ethidium bromide; WT, wild type.
The Seed Reduction Phenotype Cosegregated with the RNAi Transgene
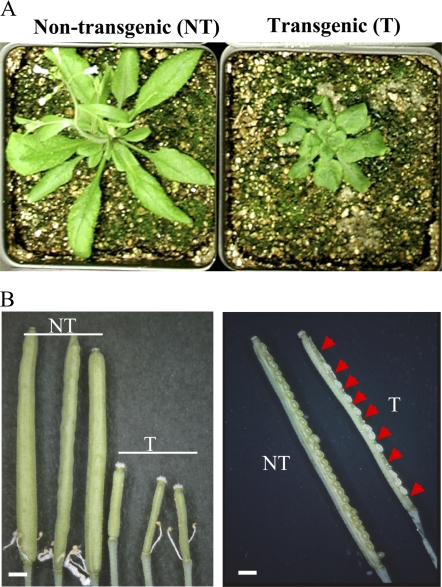
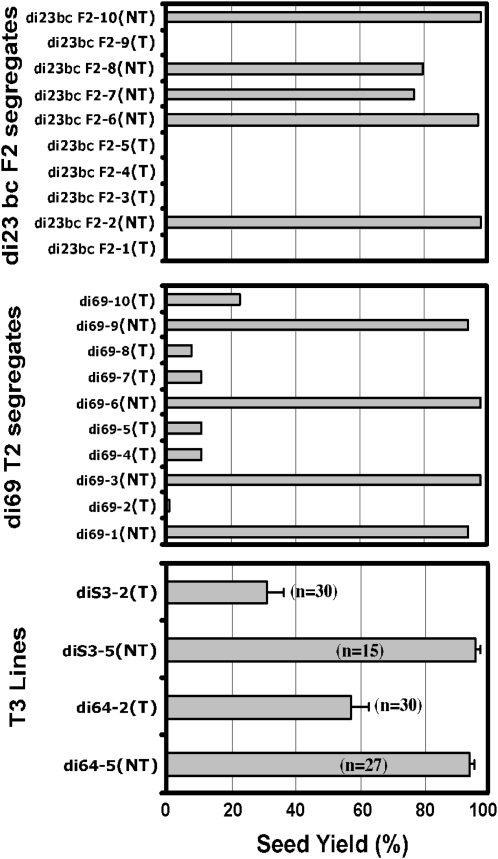
In addition to slower growth rate (Fig. 3A), these AtHVA22d RNAi plants produced smaller siliques containing no or fewer mature seeds compared with the wild-type plants, as shown in Figure 3B. To examine whether this phenotype was due to artificial selection or caused by the action of the RNAi transgene, a statistical analysis was carried out with the F2, T2, and T3 populations to determine whether this phenotype cosegregated with the RNAi transgene. Plants from these populations were genotyped by either amplifying the GFP DNA fragment in the transgene (for di 23 bc F2 and di 69 T2; Fig. 1) using PCR or by examining their kanamycin resistance (for di S3-2, di S3-5, di 64-2, and di 64-5). The fourth to sixth siliques, numbering from the base to the apex on the primary inflorescence, were dissected to count the number of successful and unsuccessful seeds. The results show that the AtHVA22d RNAi transgene cosegregates with the seed reduction phenotype of these plants (Fig. 4). This indicates that the seed reduction phenotype is indeed caused by the RNAi transgene.
Figure 3.
Slow growth rate, small siliques, and seed reduction of the AtHVA22d RNAi transgenic Arabidopsis. A, The growth rate of AtHVA22d RNAi transgenic plants (T; at right) was slower than that of the nontransformed segregates (NT; at left), as shown in the photographs of 4-week-old plants grown under long-day conditions. B, The AtHVA22d RNAi transgenic plants (T) produced smaller siliques compared with those of the nontransformed segregates (NT). There were no seeds in some very small siliques compared with those of the nontransformed segregates (left). Some siliques of the AtHVA22d RNAi plants produced fewer seeds (right). The arrowheads indicate the positions of unsuccessful seeds. Bars = 0.1 cm.
Figure 4.
Cosegregation of the seed reduction phenotype and the AtHVA22d RNAi transgene. Ten plants from the di 23 bc F2 (top) or the di 69 T2 (middle) population were genotyped by PCR using the GFP DNA fragment as a marker. T denotes transgenic and NT denotes nontransformed segregates. The fourth to sixth siliques (from the base to apex) on the primary inflorescence were dissected to count the numbers of successful and unsuccessful seeds. Siliques of the T3 homozygous populations of di S3-2 (T), di S3-5 (NT), di 64-2 (T), and di 64-5 (NT) were examined in the same manner (bottom). These results indicate that the seed reduction phenotype cosegregated with the AtHVA22d RNAi transgene. Error bars in the bottom panel represent se.
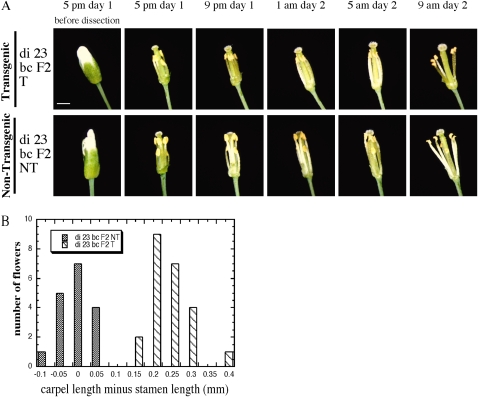
Stamen Development of the AtHVA22d RNAi Arabidopsis Was Impaired
To determine the cause of seed reduction in AtHVA22d RNAi siliques, flowers of these RNAi plants that were grown in long-day growth conditions were dissected. Intriguingly, the stamens of the RNAi plants were not long enough to pollinate. As shown in Figure 5A, growth of the stamens one night before flower opening was documented every 4 h from 5 pm on day 1 to 9 am on day 2. The stamens of the nontransformed segregate (di 23 bc F2 NT) elongated substantially from 5 pm to 9 pm on day 1, allowing the length of these stamens to match that of the carpel and permit self-pollination (Fig. 5A, bottom row). The growth of the stamens in the RNAi transgenic plants (di 23 bc F2 T), however, was impaired, and they were still shorter than the carpel when the flower was open (Fig. 5A, top row). To further investigate the stamen length defect, the latest open flowers of di 23 bc F2 T and NT at the midreproductive stage of the plants were dissected and the lengths of the carpels and stamens were measured. The carpels of di 23 bc F2 T were about 0.2 to 0.25 mm longer than the stamens of the same flower on average, while carpels of di 23 bc F2 NT were about the same length as their corresponding stamens (Fig. 5B). To be sure that the length difference between the carpels and stamens in the di 23 bc F2 T flowers was caused by short stamens rather than by long carpels, the lengths of carpels of di 23 bc F2 T and NT were compared, and those of those stamens were compared as well. The carpel length of the RNAi and control plants was 2.49 ± 0.031 mm (n = 23; mean ± se) and 2.74 ± 0.027 mm (n = 17), respectively; the stamen length of the RNAi and the control plants was 2.25 ± 0.036 mm (n = 23) and 2.75 ± 0.025 mm (n = 17), respectively. This indicates that the presence of the AtHVA22d RNAi transgene results in a length decrease in both stamens and carpels, but the effect on the stamens is greater, causing reduction in self-pollination in RNAi transgenic plants.
Figure 5.
Impaired filament development in AtHVA22d RNAi plants. A, Flowers of the di 23 bc F2 T and NT plants were dissected and photographed at 4-h intervals. The filaments of the nontransformed segregate (di 23 bc F2 NT) grew significantly from 5 to 9 pm on day 1, and the stamens reached a length comparable to that of the carpel (bottom row). The filaments of the RNAi plants (di 23 bc F2 T) did not grow to the same extent, which limited self-pollination in these plants (top row). Bar = 1 mm. B, Statistics of length difference between carpels and stamens of lines di 23 bc F2 T and NT. Lengths of the carpel and the longest stamen of each flower were measured using a dissection microscope. Twenty-four di 23 bc F2 T and 17 di 23 bc F2 NT flowers were measured. The flowers sampled were the latest open ones on each inflorescence at the midreproductive stage.
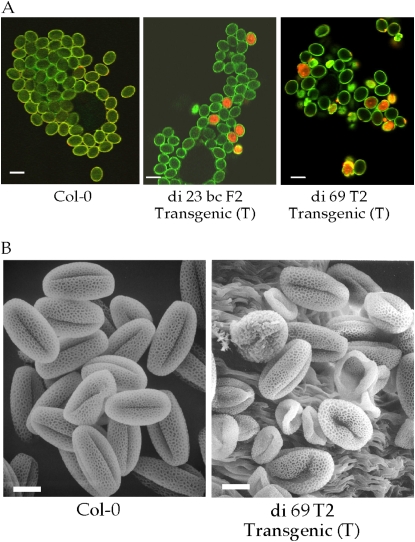
Pollen Development Was Also Affected by the AtHVA22d RNAi Transgene
We were unable to rescue some T1 transgenic plants using their pollen to fertilize wild-type plants. This led us to examine the pollen grains of these perishing T1 plants and the surviving transgenics. The majority of the pollen grains of these perishing T1 plants were deformed. Pollen grains of di 23 bc F2 T, di 69 T2 T, and wild-type Col-0 were stained with propidium iodide to examine their viability. Propidium iodide is a dead cell stain that emits light in the yellow-red region when it binds to nucleic acid and is excited by blue light. As shown in Figure 6A, about 10% to 20% of pollen grains from the RNAi plants examined by confocal laser scanning microscopy absorbed the stain, indicating that they were dead. Since di 23 bc F2 T and di 69 T2 T are not homozygous lines, the actual percentage of dead pollen grains among those containing the RNAi transgene is expected to be higher. Many abnormal pollen grains were observed in the di 69 T2 T line (Fig. 6A), suggesting that proper expression of the AtHVA22 gene family is required for pollen development. Scanning electron microscopy revealed that many pollen grains from di 69 T2 T plants were collapsed (Fig. 6B).
Figure 6.
Dead and abnormal pollen grains of lines di 23 bc F2 T and di 69 T2 T. A, Pollen grains were stained by the fluorescent dye propidium iodide to examine their viability using laser scanning confocal microscopy. Red staining indicates nonviable pollen. Staining of pollen grains of lines di 23 bc F2 T, di 69 T2 T, and wild-type Col-0 showed that a portion of the pollen of the RNAi plants was either dead or abnormal. The green color is autofluorescence of the cell wall of pollen grains. Bars = 25 μm. B, Pollen grains of di 69 T2 T were examined by scanning electron microscopy compared with wild-type Col-0 pollen. Bars = 10 μm.
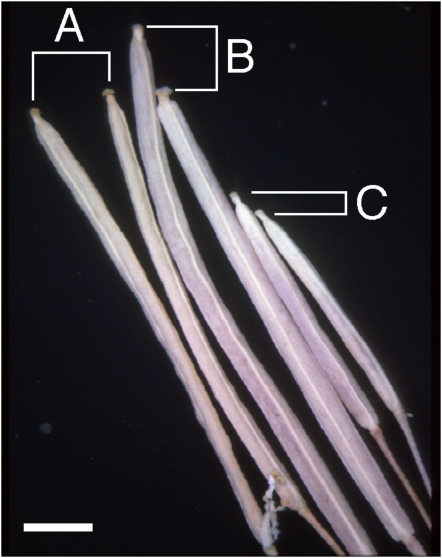
Carpels of the AtHVA22d RNAi Flowers Were Also Defective
Since the RNAi plants produced fewer seeds than the controls and the average length of the transgenic carpels was shorter than that of nontransformed segregates, this raised the question of a potential developmental problem associated with the female parts of the flowers of the RNAi plants. To examine whether development of the female parts was impaired, reciprocal crosses were carried out between the RNAi plants and wild-type Col-0. As shown in Figure 7, siliques from di 69 T2 T carpels fertilized with Col-0 pollen (C) were much smaller than the siliques from Col-0 self-pollination (A) or Col-0 carpels fertilized with di 69 T2 T pollen (B). Seventeen seeds were produced by six siliques resulting from Col-0 pollinating di 69 T2 T carpels, which was much fewer than the approximately 60 seeds produced by a normal Col-0 silique. This indicates that the AtHVA22d RNAi transgene affected not only the development of stamens and pollen but also the development of female parts of the transgenic plants. This conclusion is supported by the observation that Col-0 pollinating the RNAi lines produced a lower than expected number of seeds able to survive kanamycin selection: 10% of di 23 T1-derived seeds, 25% from di 23 bc F2 T, and 6% from di 69 T2 T plants. These ratios are markedly lower than the 50% (assuming that they were single-locus inserted transgenic plants) that would be expected from these hemizygous plants in the absence of any fertility defects.
Figure 7.
Effect of defective carpels on the short siliques of AtHVA22d RNAi plants. Siliques resulting from wild-type Col-0 self-pollination (A), di 69 T2 T (a hemizygote of the RNAi transgenic) pollinating Col-0 carpels (B), and Col-0 pollinating di 69 T2 T carpels (C) were compared to show that carpels of the RNAi plants are defective. Bar = 0.2 cm. [See online article for color version of this figure.]
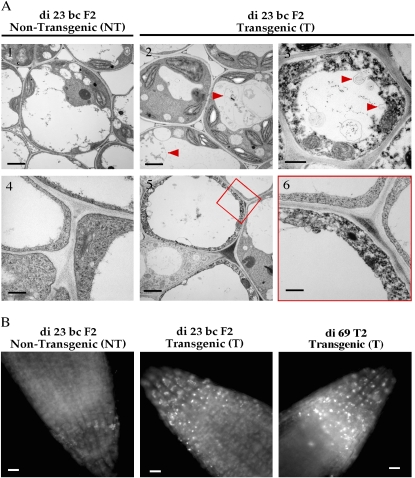
Autophagy Was Enhanced in the AtHVA22d RNAi Plants
The shortened stamens of the RNAi Arabidopsis lines prompted us to investigate the subcellular structure of the filament cells. Images obtained with transmission electron microscopy showed vesicles and membranes accumulated in the vacuoles of many filament cells (Fig. 8A2). Invagination of cytoplasm into vacuoles was also detected (Fig. 8A3). Abnormal electron-dense particles in the cytoplasm and thinning of cytoplasm were also observed (Fig. 8A, 3, 5 and 6). These events were not seen in the filament cells of the control plants (Fig. 8A, 1 and 4). These observations led to our hypothesis that autophagy is enhanced in the RNAi plants, resulting in impaired development of some cell types. To more directly examine levels of autophagy activity in these plants, roots of the RNAi plants and wild-type Col-0 were stained with the autophagosome-specific fluorescent dye monodansylcadaverine (Contento et al., 2005). Under nonstarvation conditions (half-strength Murashige and Skoog [MS] medium supplemented with 2% Suc), autophagosomes were not detected (data not shown) in the roots of either the control or the RNAi plants through 10 d of germination. In medium lacking Suc (half-strength MS salts only), autophagosomes were detected in the root tips of the RNAi seedlings 4 d after the start of germination (Fig. 8B). In contrast, autophagosomes were only detected 8 d after germination in the control seedlings. These observations support the hypothesis that autophagy activity is enhanced in the RNAi plants.
Figure 8.
Enhanced autophagy in the AtHVA22d RNAi Arabidopsis. A, Transmission electron microscopy revealed overactive autophagy in the AtHVA22d RNAi plants. Membrane and vesicle deposition in vacuoles (image 2) and invagination of cytoplasm into a vacuole (image 3) were detected in the filament cells of di 23 bc F2 T, as indicated by the arrowheads. As a consequence of overactive autophagy, thinning and abnormal electron density of cytoplasm were observed (images 5 and 6). Bars = 2 μm in images 1, 2, 3, and 5 and 0.4 μm in images 4 and 6. B, Autophagosomes were detected in the root tips of di 23 bc F2 T (center) and di 69 T2 T (right) stained by the fluorescent dye monodansylcadaverine after germination on sugar-free medium, compared with the root tip of nontransformed segregate di 23 bc F2 NT (left). Bars = 10 mm. [See online article for color version of this figure.]
Autophagy Was Enhanced in the yop1 Deletion Mutant of S. cerevisiae
If it was true that autophagy was enhanced in AtHVA22d RNAi plants, we expected that the yop1 deletion mutant in S. cerevisiae would have a similar phenotype, because Yop1 is the only HVA22 homolog in S. cerevisiae. In S. cerevisiae, the membrane protein Atg8p has been used as an autophagy molecular marker because it is associated with autophagosomes and transported into vacuoles when autophagy occurs (Kirisako et al., 1999; Ichimura et al., 2000). Occurrence of autophagy can be detected by expressing GFP-Atg8ΔRp and using antibodies to detect the dissociation of GFP and Atg8ΔRp on western blots, because the linker between GFP and Atg8ΔRp is cleaved first when GFP-Atg8ΔRp is transported into vacuoles (Yorimitsu et al., 2006; Klionsky et al., 2008). As shown in Figure 9, a low-level autophagy activity had occurred even before the start of nitrogen starvation stress in the yop1 deletion mutant, and more pronounced autophagy activity was observed right after the onset of nitrogen starvation. In contrast, it took 1 h of nitrogen starvation to trigger any detectable autophagy activity in wild-type yeast.
Figure 9.
Enhanced autophagy in the yop1 deletion mutant of S. cerevisiae. Wild-type and yop1 (HVA22 homolog in yeast) deletion mutant plants were transformed with GFP-Atg8ΔR (an autophagy molecular marker) to monitor the occurrence of autophagy using western-blot analysis. Atg8p is an autophagosomal membrane protein, and the deletion of its C-terminal Arg is to facilitate its lipidation in order to insert the protein into the autophagosomal membrane. GFP-Atg8ΔR protein resides in the cytoplasm under normal conditions. Upon the occurrence of autophagy, GFP-Atg8ΔRp is transported into vacuoles and degraded therein. The linker between the two protein domains is attacked first, thus leaving GFP from Atg8ΔRp. The occurrence of autophagy can be monitored by detecting GFP using antibody. The results indicate that the occurrence of autophagy in the yop1 deletion mutant was earlier than in the wild-type yeast after nitrogen starvation.
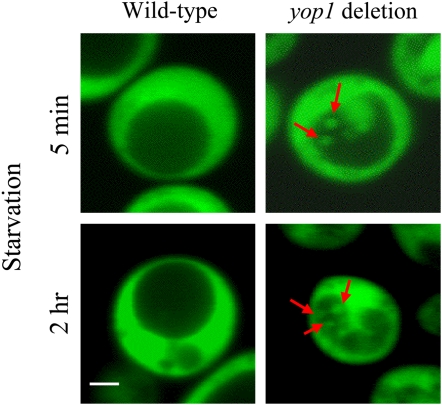
To verify the autophagy detected by western-blot analysis, a cytosolic GFP was overexpressed in yeast to label the distribution of its cytoplasm. Laser scanning confocal microscopy was used to monitor autophagy by monitoring the distribution of GFP-labeled cytoplasm. Little difference was discovered between the wild-type yeast and the yop1 deletion mutant under nonstarvation conditions. However, a difference emerged rapidly when the two strains were transferred to a nitrogen starvation medium. In the yop1 deletion mutant, GFP-labeled cytoplasm started to appear in the vacuoles in as soon as 5 min under nitrogen starvation (Fig. 10). Although this phenomenon was observed only in a low percentage of cells initially, cells displaying autophagy increased to over 80% after 2 h of the starvation. In contrast, autophagy in the wild-type cells was much less obvious through 8 h of nitrogen starvation. For more details of our monitoring of autophagy in yeast, see http://biology4.wustl.edu/autophagy.
Figure 10.
Autophagy enhancement in the yop1 deletion mutant of S. cerevisiae. Wild-type yeast and the yop1 (HVA22 homolog in yeast) deletion mutant cells were transformed with cytosolic GFP to label the distribution of cytoplasm. These cells were starved by transferring to nitrogen-deficient medium, and autophagy was monitored using laser scanning confocal microscopy. Enhanced autophagy was detected as rapidly as 5 min after the start of nitrogen starvation. Red arrows indicate cytoplasm engulfed in the vacuole. Bar = 1 μm. For more details, see Supplemental Data S1.
DISCUSSION
HVA22 was one of the many stress-induced proteins in plants whose function had not been fully explored. The presence of HVA22 homologs among diverse eukaryotes provided us the opportunity to investigate its function in both Arabidopsis and yeast. Taking advantage of convenient features in genetics and cell biology in these organisms, we have been able to reveal that HVA22 functions as an autophagy suppressor in both systems. We have demonstrated that autophagy is enhanced in both AtHVA22d RNAi transgenic Arabidopsis lines and the yop1 deletion mutant of S. cerevisiae. This suggests that HVA22 homologs regulate autophagy in a negative manner. A recent study showed that Yop1p and TB2/DP1 (all are HVA22 homologs) are integral proteins in tubular ER membranes with three cytosolic domains (N terminus, central, and C terminus) separated by two hydrophobic hairpins inserted into the outer leaflet of the ER lipid bilayer and that the insertion per se is required to maintain the shape of tubular ER (Voeltz et al., 2006). Our findings coupled with this work suggest the following two possible cellular roles for HVA22 homologs in autophagy. First, ER membrane with HVA22 homologs inserted could be restrained from forming preautophagosomal structures, because the shape of the ER membrane is constrained by the presence of the protein, leading to negative autophagy regulation. Second, it is possible that HVA22 homologs prevent preautophagosomal structure formation by interacting with an unknown protein(s) that contributes to negative regulation of autophagy. The topology of Yop1p and TB2/DP1 in the ER membrane suggests that these proteins execute multiple functions in cells. In addition to maintaining the shape of tubular ER, the studies of Calero et al. (2001) and Brands and Ho (2002) showed that Yop1p is involved in vesicular trafficking and that the N terminus is responsible for this activity. However, the most conserved part of Yop1p homologs is located in the central cytosolic domain. It is reasonable to assume that this central cytosolic domain is responsible for a cellular mechanism that has yet to be discovered. As for the C-terminal domain, it is very diverse among these homologs; thus, presumably this domain is not responsible for the conserved autophagy.
The initial attempt in this work was to use RNAi to study the role of the AtHVA22d gene particularly, because AtHVA22d was the most responsive to ABA and stress treatments. Although T-DNA insertion mutants are available for the AtHVA22d gene, phenotypes we have observed in the AtHVA22d mutants are much milder than the RNAi lines (Supplemental Data S2). As shown in Figure 2, the RNAi lines have reduced transcript levels for all members of the AtHVA22 gene family. Therefore, the phenotypes described in this paper are most likely related to knocking down of the expression of all members of AtHVA22 family.
HVA22 is barely detectable and highly inducible by ABA in barley aleurone cells and young vegetative tissues (Shen et al., 2001). During seed germination of cereals, aleurone cells perform de novo synthesis of hydrolytic enzymes that are secreted into the endosperm to degrade this nutrient reservoir, and GAs are required for this process. Concomitant with this enzyme synthesis, multiple small vacuoles in aleurone cells fuse into a big central vacuole accompanied by diminishing of cytoplasm (Zentella et al., 2002), which is a sign of cytoplasm degradation during the process that eventually leads to the death of aleurone cells. The recycled materials from the degraded cytoplasm are used to synthesize these secretory hydrolytic enzymes. When treated with ABA, HVA22 expression is induced and vacuolation and seed germination are arrested. Based on these phenomena and the negative role of HVA22 homologs in autophagy, it is highly plausible that the arrest of seed germination by ABA is mediated, at least in part, by suppression of autophagy via HVA22. Indeed, it was recently demonstrated by Guo and Ho (2008) that overexpression of HVA22 alone is sufficient to suppress the formation of large digestive vacuoles. In vegetative tissues of plants under environmental stresses such as high salinity, drought, and low temperature, ABA levels increase and anabolism is greatly reduced due to the reduced efficiency of photosynthesis. The physiological role of HVA22 during environmental stress is possibly a part of a protective mechanism used to suppress unnecessary catabolism while anabolic activities are reduced.
Autophagy is less active in fast-growing cells (Stephan and Herman, 2006). This is conceivable because anabolism has to be greater than catabolism in order for cells to grow and divide. In fast-growing tissues or organs such as inflorescence stems and flowers of Arabidopsis, expression levels of AtHVA22 genes are much higher than in slow-growing mature leaves (Chen et al., 2002). Thus, it is not surprising that the development of filaments, carpels, and pollen grains was impaired more severely than in other cell types in the RNAi Arabidopsis lines whose autophagy was overactive. It is intriguing that two recent reports have suggested that the Arabidopsis homolog of the yeast autophagy gene ATG6/VSP30 is essential for pollen germination (Fujiki et al., 2007; Qin et al., 2007), while our work indicates that knocking down HVA22 expression, which results in enhanced autophagy, leads to defects in pollen development. Since mutations in other Arabidopsis autophagy genes did not show pollen germination defects, it is possible that the pollen germination defect in AtATG6 mutants was caused by its role in vesicular trafficking rather than in autophagy (Fujiki et al., 2007). On the other hand, based on our results, it appears that autophagy is an important process in pollen development and, hence, needs to be properly regulated.
Many of the T1 RNAi plants in this study showing severe phenotypes, including dwarf and bushy stature, perished because they could not set seeds by either self-pollination or crossing to wild-type plants. This suggests that strong suppression of the AtHVA22 gene family is lethal. This view is supported by the non-Mendelian segregation of di 23 bc F2 and di 69 T2 and the difficulties in identifying di 23 bc F3 and di 69 T3 homologous transgenic RNAi plants. These phenomena are well explained by our observation of overactive autophagy in these RNAi plants: strong suppression of the AtHVA22 gene family causes overactive autophagy that leads to self-destruction of the transgenic cells.
The severity of phenotype is expected to be correlated with the degree of gene expression knockdown in transgenic plants if low degrees of gene knockdown cause an easily quantifiable phenotype and lethality is not associated with high levels of knockdown. In our study, we obtained 16 lines of transgenic plants with obvious and stable phenotypes among the original 194 T1 plants. This indicates that minor levels of knockdown did not lead to an apparent phenotype. This notion is supported by the fact that single-gene T-DNA insertion lines (Supplemental Data S2) also did not show any phenotype. If it takes a certain threshold level of knockdown in order to cause a phenotype, the correlation between phenotype severity and degree of knockdown may not be observed. Moreover, lethality in this study had caused the loss of many lines with severe phenotypes. This problem further cuts down the analyzable range of phenotype versus gene expression knockdown.
Phenotyping based on bias had been the greatest concern since the beginning of this study. We eventually investigated four transgenic lines with inheritable and stable phenotypes, two strong ones and two weak ones. The best way in this case to exclude the possibility of artificial selection was to conduct a phenotype-versus-transgene cosegregation analysis. Although this approach may not be flawless, we believe it to be the most convincing way to correlate the presence of an RNAi-generating transgene with the appearance of a phenotype. In addition, our hypothesis obtained from Arabidopsis was confirmed in yeast.
The negative regulation mechanism executed by AtHVA22 and Yop1p might be downstream of a starvation sensor, as shown by the production of autophagosomes in the root tips of the RNAi plants and the yop1 deletion yeast only in starvation conditions. However, the relation between AtHVA22/Yop1p and Tor1p, as well as the Atg1p-Atg13p complex, is not clear. We hypothesize that AtHVA22/Yop1p controls the lipid flow from the ER to the preautophagosomal structures by either stabilizing the ER membrane or by recruiting a negative autophagy regulator(s) that interacts with their central cytosolic domain. This would place their activity upstream of the Atg1p-Atg13p complex. Since Tor1p inactivation enhances autophagy in nonstarvation conditions and its substrate has yet to be identified, it is difficult to determine whether Tor1p may act in the same pathway as AtHVA22/Yop1p. Clearly, further studies are required to clarify the regulation of autophagy.
MATERIALS AND METHODS
RNAi Construct
The GFP cDNA fragment was amplified by PCR using primers 5′-ACGAATTCGTGGAGGAGGTATTCTAGATC-3′ and 5′-ATGAGCTCATCCATGCCATGTGTAATCCC-3′. This cDNA fragment was inserted into the multicloning sites of the Arabidopsis (Arabidopsis thaliana) transformation vector pMON 10098. The AtHVA22d cDNA fragment was amplified by PCR using primers 5′-AAGAATTCGAGCTCTTTACACAGACTTGTCGCTCT-3′ and 5′-TCTCTAGAGGATCCGAAATACGCGTGGAAGATTAG-3′. This amplified AtHVA22d cDNA fragment contained the coding region and parts of the 5′ and 3′ untranslated regions. There were two enzyme digestion sites at each end of this cDNA fragment, which were used for the sense or antisense orientation insertion between the GFP cDNA fragment and the terminator or the promoter, respectively.
Plants, Growth Conditions, and Transformation
Arabidopsis ecotype Col-0 was grown on a germination mix in a growth chamber as described by Chen et al. (2002). Briefly, these plants were grown in 16/8-h day/night cycles set at 22°C under 150 μmol m−2 s−1 light intensity. Plants were transformed using Agrobacterium tumefaciens (Bechtold and Pelletier, 1998).
RNA Isolation and Northern-Blot Analysis
Total RNA isolation and northern-blot analysis were carried out based on the procedures described by Chen et al. (2002) using a guanidine method, formaldehyde gel electrophoresis, and 32P-labeled DNA probes. Ten micrograms of total RNA was separated on the formaldehyde gel, and the gel was stained by ethidium bromide to verify equal loading of RNA quantities in each lane.
Small-Scale Genomic DNA Isolation and Genotyping of the Transformants
Small-scale genomic DNA isolation was carried out with a modified mini-prep procedure (Cocciolone and Cone, 1993). Two fresh leaves were ground in 0.5 mL of urea buffer (7 m urea, 0.3 m NaCl, 20 mm EDTA, 34 mm N-lauroyl sarcosine, and 50 mm Tris-HCl, pH 8) with mortar and pestle. The extract was then transferred to a microtube and incubated at 65°C for 10 min. After the incubation, the extract was gently mixed with 0.4 mL of phenol:chloroform (1:1) for 5 min. After centrifuging for 5 min at room temperature, 0.4 mL of the supernatant was transferred to a new microtube. The supernatant was mixed with an equal volume of isopropanol and allowed to rest for 5 min at room temperature. After centrifuging the new tube for 5 min at room temperature, the supernatant was discarded and the pellet was rinsed with 70% ethanol twice. The pellet was dried in air and dissolved in 50 μL of Tris-EDTA buffer containing RNase. Genotyping was carried out using PCR to amplify the GFP DNA fragment with the following two primers, 5′-GGAGAAGAACTTTTCACTGGA-3′ and 5′-ATCCATGCCATGTGTAATCC-3′.
Pollen Viability Staining and Imaging
Mature pollen was spread on glass slides and stained with 1 mg mL−1 propidium iodide (Sigma P4170) in distilled water. The absorption spectrum of propidium iodide bound to DNA is between 450 and 600 nm, and the emission is in the yellow-red region (550–700 nm). The samples were examined with 488-nm excitation wavelength using a Leica laser scanning confocal microscope. The emission spectrum was collected with two channels: one in the green region for the autofluorescence from the cell wall of pollen grains, the other in the yellow-red region for the propidium iodide emission.
Scanning and Transmission Electron Microscopy
Mature anthers were attached onto specimen mounts for scanning electron microscopy and allowed to dry on a slide warmer. The materials were then sputter coated with gold and imaged with a Hitachi S-450 scanning electron microscope. Stamens were fixed with 2.5% glutaraldehyde in 100 mm sodium phosphate buffer, pH 7.4, containing a trace amount of Photo-Flo 200 (Kodak) as a wetting reagent. The samples were postfixed with 2% (w/v) osmium tetroxide in water, dehydrated, and embedded in Polybed 812 resin (Polysciences). Thin sections were stained with 2% uranyl acetate followed by 0.38 mm lead citrate. The samples were imaged with a Hitachi H-600 transmission electron microscope.
Autophagosome Staining
Autophagosome staining in root tips was carried out using a modified method of Contento et al. (2005). Seeds were germinated on agar plates containing half-strength MS salts (pH 5.7) and grown vertically. Roots were harvested and stained at the stages indicated in “Results” with 100 mm monodansylcadaverine (Sigma D4008) in 50 mm potassium phosphate buffer (pH 7.4) containing 0.01% L77 for 10 min, followed by three rinses in the phosphate buffer. Root tips were imaged with the Nikon inverted fluorescence microscope TE2000-S equipped with the Cool Snap ES digital camera (Photometrics).
Western-Blot Analysis of Yeast Autophagy
Wild-type Saccharomyces cerevisiae (strain w303) and its yop1 deletion mutant (Brands and Ho, 2002) were transformed with the plasmid pRS416-GFP-Atg8ΔR, in which the construct was driven by a copper-inducible promoter (kindly provided Dr. Wei-Pang Huang, National Taiwan University), and grown in the synthetic dropout medium (−Ura) with 2% Glc until stationary phase. The C-terminal Arg of Atg8p is deleted, which is necessary for the lipidation processing in order to insert this protein into an autophagosomal membrane (Ichimura et al., 2000). Five hundred microliters of each culture was transferred to 250 mL of the synthetic dropout medium (−Ura) with 2% Gal and 25 μm Cu2+ and grown in an orbital shaker incubator at 200 rpm at 30°C for 22 h. Cells were harvested by centrifugation at 3,750 rpm at room temperature for 2 min and resuspended in the synthetic medium with 2% Gal and 25 μm Cu2+ without ammonium sulfate and amino acids. Phenylmethylsulfonyl fluoride (PMSF) was added to the nitrogen-starved cells to 1 mm when the cultures were sampled at the times indicated. Cells were broken by glass beads using a beadbeater. Ten micrograms of protein from each sample was separated by 8% SDS-PAGE and transferred onto the Immobilon-P membrane (PVDF; Millipore) in 25 mm Tris, 192 mm Gly, and 10% methanol (pH 8.3) using a Bio-Rad protein transfer tank. The membrane was incubated in TBST buffer (137 mm NaCl, 20 mm Tris-HCl [pH 7.6], and 0.05% Tween 20) containing 5% nonfat milk for 1 h at room temperature. Washed with TBST buffer for 5 min, the membrane was incubated in TBST containing 3% nonfat milk and a primary antibody against GFP (mouse monoclonal antibody; Santa Cruz Biotechnology) for at least 1 h. The membrane was washed three times (5 min each) with TBST and incubated in TBST containing a secondary antibody (goat anti-mouse IgG-horseradish peroxidase; Santa Cruz Biotechnology) for 1 h. Chemiluminescence imaging was carried out based on the procedure of Yang and Widmann (2001).
Microscopic Observation of Yeast Autophagy
Wild-type yeast and the yop1 (HVA22 homolog in yeast) deletion mutant cells were transformed with cytosolic GFP driven by a Gal-inducible promoter to label the distribution of cytoplasm. These cells were grown in the synthetic dropout medium (−Trp) containing 2% Gal for 44 h to late log phase and then starved by transferring to nitrogen-depleted medium (without ammonium sulfate and amino acids) containing 2% Gal and 1 mm PMSF, and autophagy was monitored using laser scanning confocal microscopy. More details of this procedure are given in the Supplemental Data S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data S1. Monitoring autophagy in S. cerevisiae with the laser scanning confocal microscopy is available at http://biology4.wustl.edu/autophagy.
Supplemental Data S2. Confirmed Arabidopsis AtHVA22 T-DNA insertion mutants showing occasional and variable morphological phenotypes.
Supplementary Material
Acknowledgments
We thank Drs. Craig Pikaard and Eric Richards for helpful discussions and Mr. Mike Veith and Dr. Tatsuhiko Noguchi for assistance with electron and fluorescence microscopy, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tuan-Hua David Ho (tho@sinica.edu.tw).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K (2006) Autophagy in development and stress responses of plants. Autophagy 2 2–11 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82 259–266 [DOI] [PubMed] [Google Scholar]
- Brands A, Ho TH (2002) Function of a plant stress-induced gene, HVA22: synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol 130 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Whittaker GR, Collins RN (2001) Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J Biol Chem 276 12100–12112 [DOI] [PubMed] [Google Scholar]
- Chen CN, Chu CC, Zentella R, Pan SM, Ho TH (2002) AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol Biol 49 633–644 [DOI] [PubMed] [Google Scholar]
- Cocciolone SM, Cone KC (1993) Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42 598–608 [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R (2006) The TOR signalling network from yeast to man. Int J Biochem Cell Biol 38 1476–1481 [DOI] [PubMed] [Google Scholar]
- Dunn WA Jr (1990) Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol 110 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y (2007) An Arabidopsis homolog of yeast ATG/VPS30 is essential for pollen germination. Plant Physiol 143 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Heckman JW Jr, Schiefelbein JW (1997) Growth and ultrastructure of Arabidopsis root hairs: the rhd3 mutation alters vacuole enlargement and tip growth. Planta 201 209–218 [DOI] [PubMed] [Google Scholar]
- Guo WJ, Ho THD (2008) An ABA-induced protein HVA22 inhibits GA-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al (2000) A ubiquitin-like system mediates protein lipidation. Nature 408 488–492 [DOI] [PubMed] [Google Scholar]
- Inoki K, Guan KL (2006) Complexity of the TOR signaling network. Trends Cell Biol 16 206–212 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 16 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryoytes. Autophagy 4 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol 15 1–32 [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273 3963–3966 [DOI] [PubMed] [Google Scholar]
- Noda T, Suzuki K, Ohsumi Y (2002) Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol 12 231–235 [DOI] [PubMed] [Google Scholar]
- Qin G, Ma Z, Zhang L, Xing S, Hou X, Deng J, Liu J, Chen Z, Qu L, Gu H (2007) Arabidopsis AtBECLIN1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res 17 249–263 [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2 282–291 [DOI] [PubMed] [Google Scholar]
- Shen Q, Chen CN, Brands A, Pan SM, Ho THD (2001) The stress- and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol Biol 45 327–340 [DOI] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho THD (1993) Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J Biol Chem 268 23652–23660 [PubMed] [Google Scholar]
- Stephan JS, Herman PK (2006) The regulation of autophagy in eukaryotic cells. Autophagy 2 146–148 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH (1994) Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349 275–280 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333 169–174 [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124 573–586 [DOI] [PubMed] [Google Scholar]
- Yang JY, Widmann C (2001) Antiapoptotic signaling generated by caspase-induced cleavage of RasGAP. Mol Cell Biol 21 5346–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self-eating. Cell Death Differ (Suppl 2) 12 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ (2006) Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Yamauchi D, Ho THD (2002) Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14 2289–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.