Abstract
HIV-1 latency in resting CD4+ T cells represents a major barrier to virus eradication in patients on highly active antiretroviral therapy (HAART). Eliminating the latent HIV-1 reservoir may require the reactivation of viral gene expression in latently infected cells. Most approaches for reactivating latent HIV-1 require nonspecific T cell activation, which has potential toxicity. To identify factors for reactivating latent HIV-1 without inducing global T cell activation, we performed a previously undescribed unbiased screen for genes that could activate transcription from the HIV-1 LTR in an NF-κB-independent manner, and isolated an alternatively spliced form of the transcription factor Ets-1, ΔVII-Ets-1. ΔVII-Ets-1 activated HIV-1 transcription through 2 conserved regions in the LTR, and reactivated latent HIV-1 in cells from patients on HAART without causing significant T cell activation. Our results highlight the therapeutic potential of cellular factors for the reactivation of latent HIV-1 and provide an efficient approach for their identification.
Keywords: antiretroviral therapy, ΔVII-Ets-1, expression cloning, long terminal repeat, viral reservoir
Advances in antiretroviral therapy have dramatically reduced mortality among patients with HIV-1 infection (1). However, there is still no therapeutic regimen to cure chronic HIV-1 infection. Although highly active antiretroviral therapy (HAART) can suppress plasma viral load to undetectable levels, viremia rebounds within weeks after discontinuation of HAART. The major barrier to eradication of HIV-1 infection is the existence of viral reservoirs. Among them, the best characterized is a small pool of latently-infected resting memory CD4+ T cells harboring an integrated provirus (2–4). Previous studies have demonstrated the stability of this latent reservoir in patients on HAART (5). The half-life of this reservoir was estimated to be >44 months. At this rate of decay, it is expected to take >60 years to purge HIV-1 from infected patients on HAART. Thus, this reservoir necessitates the lifetime use of HAART, and strategies are needed for eradication of latently infected cells (6, 7).
Recently, reactivation of latent virus has gained wide interest as a potential strategy to eradicate the viral reservoirs (8–11). It is assumed that latently infected cells can be killed either by immune attack or direct viral cytopathic effects after reactivation of latent HIV-1. A reactivation strategy, along with simultaneous efficient suppression of viral spread by HAART, might reduce and ultimately eliminate the latent reservoirs (6, 7). Although logical, this approach has practical limitations. Because signals that cause T cell activation also activate HIV-1 replication, some studies have focused on strategies to induce some level of T cell activation as a means of reactivating latent HIV-1 (10, 11). Unfortunately, the potential toxicity of such nonspecific T cell activation has severely complicated this approach (10, 11). For example, patients treated with agonistic anti-CD3 monoclonal antibody and IL-2 suffered from severe side effects, transient renal failure, and seizure. An ideal reactivation strategy for virus eradication might allow activation of HIV-1 without inducing global T cell activation.
The HIV-1 provirus responds to various extracellular stimuli, including T cell activation signals and some proinflammatory cytokines (12–14). The HIV-1 promoter, located within the U3 region of the LTR, contains an array of cis-acting transcription factor binding sites (15). The interaction between these diverse signals and the various binding sites in the LTR forms a complex regulatory network. In particular, the host transcription factor NF-κB is important for activating HIV-1 gene expression through 2 conserved κB sites in the core enhancer region of the HIV-1 LTR (12, 13). However, HIV-1 can replicate in the absence of κB sites in the LTR (16), consistent with the existence of NF-κB-independent pathways in the activation of HIV-1 (17, 18). NF-κB also has a critical role in innate and adaptive immune responses, and regulates genes that have important roles during T cell activation (19). Because of the central role of NF-κB in T cell activation, we reasoned that to find genes that could uncouple the activation of latent HIV-1 from T cell activation it would be desirable to identify factors that could activate the HIV-1 LTR in an NF-κB-independent manner.
To systematically search for NF-κB-independent pathways for the activation of HIV-1, we performed an expression cloning screen using a reporter containing mutated NF-κB sites in the enhancer region of the HIV-1 LTR. By screening a human splenocyte cDNA expression library, we isolated an alternatively spliced form of the Ets-1 transcription factor, ΔVII-Ets-1. ΔVII-Ets-1 was able to activate the NF-κB site-mutated HIV-1 LTR without stimulating T cell activation and could activate latent HIV-1 from resting CD4+ T cells isolated from patients on HAART. Our results identify a cellular factor that can reactivate latent HIV-1 without inducing T cell activation, and illustrate the potential of this expression cloning strategy to yield novel approaches for eradicating latent reservoirs of HIV-1.
Results
Expression Cloning Screen to Identify NF-κB-Independent Pathways for the Reactivation of Latent HIV-1.
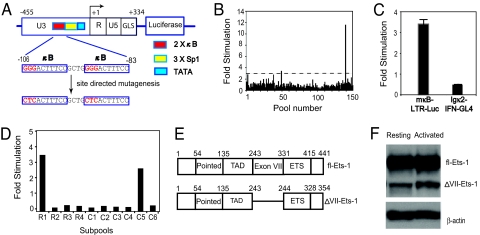
To facilitate the identification of NF-κB-independent pathways that could activate the HIV-1 LTR, we generated a luciferase reporter, mκB-LTR-Luc, which contains the HIV-1 LTR from reference strain NL4-3 with mutated κB sites within the core enhancer region (−106 to −83) (Fig. 1A) that have been shown to abolish the activity of NF-κB on the HIV-1 LTR (13). We then screened a human splenocyte cDNA expression library for the ability to stimulate the mκB-LTR-Luc reporter. To maximize the number of the cDNAs that could be assayed, we generated cDNA pools with ≈100 cDNAs per pool (20). Each pool was transfected into HEK293T cells with the mκB-LTR-Luc reporter and pCSK-lacZ, an NF-κB-independent β-galactosidase expression vector used for normalizing transfection efficiency and extract recovery. A pool was considered positive if it activated the mκB-LTR-Luc reporter by 3-fold or more, as compared with an empty expression vector. In all, 477 pools were screened (Fig. 1B), and 7 positive pools were identified.
Fig. 1.
Strategy for expression cloning and isolation of ΔVII-Ets-1. (A) The structure of wt-LTR-Luc and mκB-LTR-Luc reporters. The wt-LTR-Luc reporter contains the 5′ LTR and the untranscribed region of Gag (GLS) derived from NL4-3 upstream of the luciferase gene. Two tandem NF-κB sites (2x κB), 3 Sp1 sites (3x Sp1) and the TATA box (TATA) in the U3 region are shown as boxes with indicated colors. The NF-κB-binding sites were mutated as indicated. Numbering above the boxed regions is relative to the transcription start site (+1), based on the sequence of the reference strain HIVHXB2. (B) Example of primary screening. Pooled cDNAs were transfected into HEK293T cells with the mκB-LTR-Luc reporter and the pCSK-lacZ control vector, and the fold stimulation was determined as described in Materials and Methods. Only 150 out of 477 pools screened are shown. (C) HEK293T cells were cotransfected with pool 50, Igκ2-IFN-GL4 and pCSK-lacZ. The fold stimulation of pool 50 is shown relative to reference control pcDNA3.1(−). (D) Identification of the positive clone from pool 50 in a 4 row (R) × 6 column (C) matrix of subpools. The well in R 1 and C 5 contained the positive clone. (E) The functional domains of fl-Ets-1 and ΔVII-Ets-1. The exon VII, originally designated in alignment with chicken c-ets gene, is actually exon 6 in the human ETS1 gene (22). (F) Western blot of fl-Ets-1 and ΔVII-Ets-1 in 10 μg of lysates of primary resting CD4+ T cells and CD4+ T cells activated with 2.5 μg/mL anti-CD3 and 1 μg/mL anti-CD28 antibodies for 3 days. The indicated β-actin levels serve as loading control for the T cell lysates.
To verify that each positive pool activated the HIV-1 LTR in an NF-κB-independent manner, we tested each pool for its ability to activate the NF-κB-responsive reporter Igκ2-IFN-GL4, which contains 2 NF-κB sites upstream of the IFN-β minimal promoter and the luciferase gene (20). Four of the seven positive pools did not activate the Igκ2-IFN-GL4 reporter. For example, although pool 50 activated the mκB-LTR-Luc reporter by 3.4-fold, pool 50 did not activate the Igκ2-IFN-GL4 reporter at all (Fig. 1C). We purified the activity of 1 pool (pool 50) for further study.
Isolation of Human ΔVII-Ets-1 from a Human Splenocyte cDNA Library.
To identify the cDNA responsible for the activity of pool 50, DNA from pool 50 was used to retransform bacteria, and individual colonies were amplified in wells of 24-well plates. Each 24-well plate was screened as a subpool after preparing DNA from pooled aliquots from each well. Once a 24-well plate was found to exhibit the activity present in the original pool 50, a conceptual matrix (4 row × 6 column) was used to identify the coordinates of the positive clone within the 24-well plate (Fig. 1D). Sequencing revealed that this clone was a previously described spliced variant of the transcription factor Ets-1, ΔVII-Ets-1.
Ets-1 is a member of Ets family of transcription factors that share an 85-aa conserved DNA binding domain, the ETS domain (21, 22). Ets-1 is highly expressed in T cells, and has an essential role in their proliferation and survival (23). Two isoforms of Ets-1, full-length (fl)-Ets-1 and ΔVII-Ets-1, which lacks the region encoded by exon VII, can be detected in T cells; however, fl-Ets-1 is expressed at a much higher level (21, 22). The functional domains of Ets-1 are shown in Fig. 1E. Previous studies have revealed that exon VII encodes an autoinhibitory domain that regulates the DNA binding activity of Ets-1 (24). The C-terminal portion of the exon VII-encoding region contains helices that inhibit the DNA-binding domain in concert with helices encoded C-terminal to the DNA-binding domain of Ets-1 (ETS). The N-terminal portion of the exon VII domain is phosphorylated in a calcium-responsive manner, and phosphorylation in this region further inhibits DNA binding and transcriptional activation (25). Because ΔVII-Ets-1 lacks the exon VII-encoded region, its DNA-binding affinity is significantly enhanced. The role of fl-Ets-1 on HIV-1 gene expression has been studied before (26, 27). Full-length Ets-1 binds upstream to the core enhancer of HIV-1 LTR, and stimulates viral gene expression by interacting with other transcription factors, such as NF-κB, NFAT, and USF-1. Because HIV-1 replication is closely linked to the activation state of the T cell, we determined the expression of fl-Ets-1 and ΔVII-Ets-1 in resting and activated CD4+ T cells. Both fl-Ets-1 and ΔVII-Ets-1 were readily detectable in resting and activated CD4+ T cells; however, the expression of ΔVII-Ets-1 was less than that of fl-Ets-1 (Fig. 1F).
Specific Activation of the HIV-1 LTR by ΔVII-Ets-1.
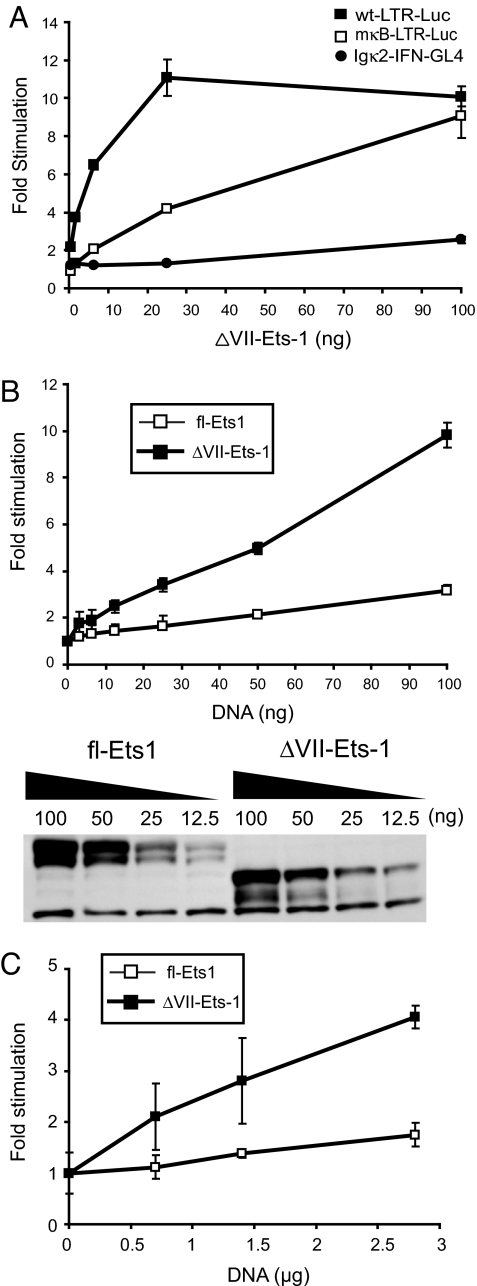
We verified that ΔVII-Ets-1 was responsible for the activity displayed by pool 50. Overexpression of ΔVII-Ets-1 in HEK293T cells activated the wild-type HIV-1 LTR (wt-LTR-Luc) and the LTR with mutated NF-κB sites (mκB-LTR-Luc), but did not activate the NF-κB-dependent reporter (Igκ2-IFN-GL4) (Fig. 2A). Thus, ΔVII-Ets-1 was able to specifically activate the HIV-1 LTR without activating NF-κB. Compared with its effect on mκB-LTR-Luc, ΔVII-Ets-1 showed a higher level of induction on wt-LTR-Luc, suggesting that, although ΔVII-Ets-1 did not require NF-κB for transcriptional activity, it does act cooperatively with NF-κB in the context of the HIV-1 LTR. This finding is consistent with a previous report demonstrating the cooperative activity between Ets-1 and NF-κB on the HIV-1 LTR (26). When directly compared in this assay, ΔVII-Ets-1 showed a greater ability than fl-Ets-1 to activate the mκB-LTR-Luc reporter at comparable expression levels (Fig. 2B). To confirm that ΔVII-Ets-1 could activate the HIV-1 LTR in T cells, we also assessed its ability to activate the mκB-LTR-Luc in the Jurkat T cell line. Overexpression of ΔVII-Ets-1 in Jurkat T cells did induce the mκB-LTR-Luc reporter, but to a lower degree than in HEK293T cells (Fig. 2C). As in HEK293T cells, ΔVII-Ets-1 displayed more activity than fl-Ets-1 in Jurkat T cells (Fig. 2C).
Fig. 2.
ΔVII-Ets-1 specifically activates the HIV-1 LTR. (A) Titration of ΔVII-Ets-1 expression vector in the presence of 0.5 ng of the control vector TK-RLuc plus 1 of 3 different reporters: 4 ng wt-LTR-Luc, 4 ng mκB-LTR-Luc, or 10 ng Igκ2-IFN-GL4 in HEK293T cells. The fold stimulation is shown normalized to that observed with the reference control vector pmax-Empty. Data are means ± SD of triplicate transfections, and represent 2 independent experiments. (B) Titration of fl-Ets-1 and ΔVII-Ets-1 expression vectors in the presence of 4 ng mκB-LTR-Luc and 0.5 ng pTK-RLuc in HEK293T cells. (Upper) Fold stimulation, normalized to that observed with pmax-Empty. Data are means ± SD of triplicate transfections, and represent 2 independent experiments. (Lower) Western blot analysis of the relative expression level of fl-Ets1 and ΔVII-Ets-1 using the lysates analyzed in the Upper. (C) Titration of fl-Ets-1 and ΔVII-Ets-1 expression vectors in the presence of 100 ng mκB-LTR-Luc and 50 ng pTK-RLuc in Jurkat T cells. The fold stimulation is shown normalized to that observed with pmax-Empty. Data are means ± SD of triplicate transfections, and represent 2 independent experiments.
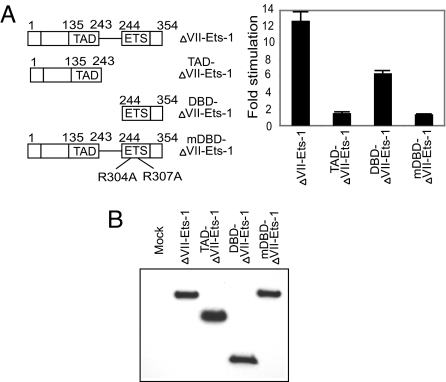
The DNA-Binding Domain of ΔVII-Ets-1 Is Essential for Activation of HIV-1 LTR.
ΔVII-Ets-1 contains an N-terminal transactivation domain and C-terminal DNA-binding domain (Fig. 3A). To investigate the functional domains that are responsible for the activation of mκB-LTR-Luc, we generated 3 FLAG-tagged mutants: TAD-ΔVII-Ets-1 (transactivation domain of ΔVII-Ets-1), DBD-ΔVII-Ets-1 (DNA-binding domain of ΔVII-Ets-1), and mDBD-ΔVII-Ets-1 (R304A, R307A mutant of ΔVII-Ets-1). R304 and R307 of ΔVII-Ets-1 are equivalent to R391 and R394 of fl-Ets-1. These 2 arginine residues are critical for the DNA binding of Ets-1 (28). Cotransfection of mκB-LTR-Luc and each of these variants revealed that TAD-ΔVII-Ets-1 alone could not activate the mutant HIV-1 LTR, whereas DBD-ΔVII-Ets-1 alone had a moderate effect (Fig. 3A). Also, mutation of the 2 essential arginine residues (R304 and R307) in mDBD-ΔVII-Ets-1 abolished the stimulatory effect of ΔVII-Ets-1, indicating the necessity of the DBD-ΔVII-Ets-1 in activation of the HIV-1 LTR (Fig. 3A). The expression of these ΔVII-Ets-1 variants in HEK293T cells was confirmed by Western blotting with anti-FLAG antibodies (Fig. 3B). These results demonstrate the essential role of DNA binding in the activation of the HIV-1 LTR by ΔVII-Ets-1.
Fig. 3.
Mapping the functional domains of ΔVII-Ets-1. (A) Cotransfection of HEK293T cells with 100 ng of FLAG-tagged ΔVII-Ets-1, TAD-ΔVII-Ets-1, DBD-ΔVII-Ets-1, or mDBD-ΔVII-Ets-1 with 4 ng of mκB-LTR-Luc and 0.5 ng of TK-RLuc. (Left) Displayed are the constructs assayed. (Right) Fold stimulation is shown normalized to that observed with pmax-Empty. Data are means ± SD of triplicate transfections, and represent 3 independent experiments. (B) Western blot analysis of the relative expression level of FLAG-tagged ΔVII-Ets-1 variants using the identical HEK293T cell lysates analyzed in A.
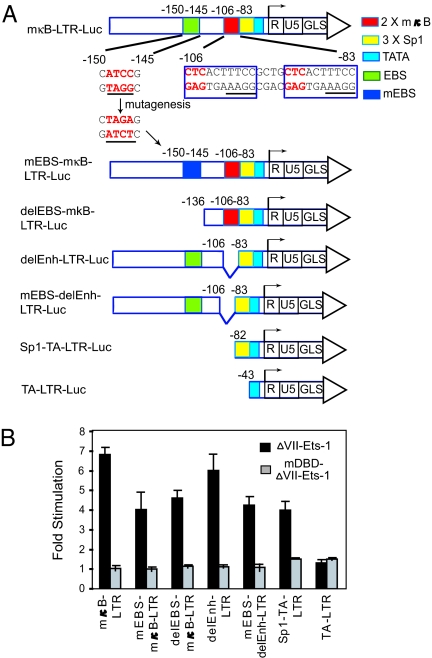
Mapping of the ΔVII-Ets-1-Responsive Elements in HIV-1 LTR.
We assayed a panel of LTR mutant and deletion constructs to determine which DNA elements in the LTR are required for ΔVII-Ets-1-mediated activation (Fig. 4A). Previous studies revealed that Ets-1 can bind to the Ets-binding site (EBS) in the distal region (−150 to −145) of the LTR (27). Also, there are another 2 EBSs that bind the Ets family transcription factor GABPα within the HIV-1 core enhancer (−106 to −83) (29). These 2 EBSs are located in the 3′ half of κB sites with the core sequence 5′-GGAA-3′ (Fig. 4A). Mutation or deletion of the EBS in the distal region (−150 to −145) (mEBS-mκB-LTR-Luc and delEBS-mκB-LTR-Luc) reduced ΔVII-Ets-1-mediated activation by ≈40%. However, deletion of the enhancer region of LTR (delEnh-LTR-Luc), which contains binding sites for NF-κB, NFAT and GABPα, did not significantly reduce ΔVII-Ets-1-mediated activation, either in the context of mκB-LTR-Luc or mEBS-LTR-Luc, indicating that the 2 defined EBSs in this core enhancer region are not required for ΔVII-Ets-1 activity. A construct containing only the 3 Sp1 binding sites and the TATA box (Sp1-TA-LTR-Luc) displayed ≈60% of the activity of the mκB-LTR-Luc. Deletion of the Sp1-binding sites in the context of Sp1-TA-LTR-Luc destroyed the activity of ΔVII-Ets-1, indicating that this region is as important for ΔVII-Ets-1 responsiveness as is the EBS between −150 and −145 (Fig. 4B). Interestingly, the ΔVII-Ets-1 mutant that could not bind DNA (mDBD-ΔVII-Ets-1) did not display any activity on any of these reporter constructs, indicating that the DNA-binding activity of ΔVII-Ets-1 is required for transactivation through the elements between −150 and −145 and between −82 and −44.
Fig. 4.
Mapping the ΔVII-Ets-1-responsive elements in the HIV-1 LTR. (A) Reporter constructs with truncations or point mutations of the HIV-1 LTR. Numbering indicates the nucleotide positions relative to the transcription start site (+1). The underlined sequences within the κB sites indicate putative EBSs. (B) Cotransfection of HEK293T cells with 0.5 ng TK-RLuc and either 70 ng of ΔVII-Ets-1-expression vector, mDBD-ΔVII-Ets-1, or pmax-Empty with the indicated reporters. The fold stimulation normalized to that observed with pmax-Empty is shown. Data are means ± SD of triplicate transfections, and represent 2 independent experiments.
Overexpression of ΔVII-Ets-1 Does Not Induce Full Activation of Primary CD4+ T Cells.
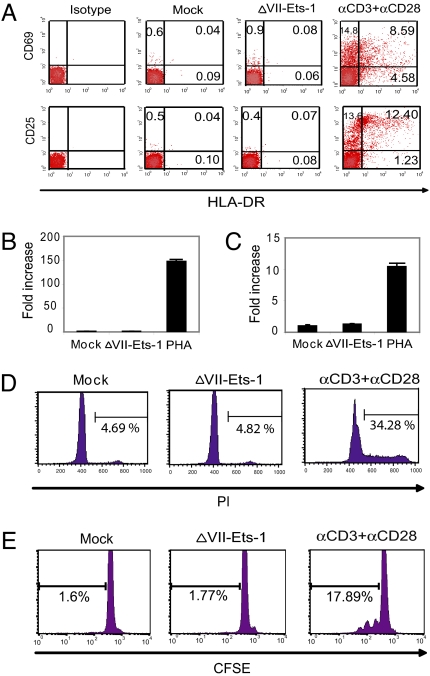
Because our goal was to find a way to reactivate latent HIV-1 without inducing global T cell activation, we next determined whether overexpression of ΔVII-Ets-1 could activate resting CD4+ T cells. We isolated resting CD4+ T cells with high purity from HIV-1 negative donors, and then transfected the cells with either a mock vector or the ΔVII-Ets-1-expression vector under conditions that achieved 50–70% transfection of cells. Overexpression of ΔVII-Ets-1 in resting CD4+ T cells did not cause the up-regulation of the T cell activation markers CD69, CD25, or HLA-DR, when compared with the mock vector. In contrast, very strong up-regulation of these proteins was observed in cells activated with agonistic anti-CD3 and anti-CD28 antibodies (Fig. 5A). We also compared the expression of IFN-γ and IL-2 in these cells. The cells transfected with either mock vector or ΔVII-Ets-1 had very low levels of mRNA for IFN-γ and IL-2, whereas the transcripts of IFN-γ and IL-2 in PHA-activated cells were significantly up-regulated by 147- and 10-fold, respectively (Fig. 5 B and C). We first assessed cell proliferation by staining the cells with propidium iodide. A low fraction of cells transfected with either mock vector or ΔVII-Ets-1 had entered S phase (<5%), whereas 34% of cells costimulated by anti-CD3 and anti-CD28 antibodies entered S phase (Fig. 5D). To independently assess proliferation, we stained resting CD4+ T cells with CFSE before transfection; <2% of cells transfected with either mock vector or ΔVII-Ets-1 proliferated, whereas almost 18% cells treated with anti-CD3 and anti-CD28 for 3 days proliferated (Fig. 5E). These data indicate that overexpression of ΔVII-Ets-1 does not induce significant T cell activation.
Fig. 5.
Overexpression of ΔVII-Ets-1 in primary resting CD4+ T cells does not induce significant activation of T cells. (A) Expression of activation markers on ΔVII-Ets-1-transfected primary resting CD4+ T cells or CD4+ T cells activated with 2.5 μg/mL anti-CD3 and 1 μg/mL anti-CD28 for 72 h; 5 μg of ΔVII-Ets-1-expression vector was transfected into 3.5 × 106 primary resting CD4+ T cells by nucleofection. ΔVII-Ets-1-transfected cells were analyzed for surface expression of CD25, HLA-DR, and CD69 by flow cytometry 72 h after transfection. IFN-γ (B) and IL-2 (C) transcripts from primary CD4+ T cells 72 h after transfection or after 72 h of PHA treatment were quantified by real-time RT-PCR, and were normalized to ubiquitin mRNA levels. The fold stimulation was normalized to that observed with the reference control vector pmax-Empty. Data are means ± SD of triplicate measurements, and represent 2 independent experiments. (D) ΔVII-Ets-1-transfected cells, pmax-Empty-transfected cells, and anti-CD3/anti-CD28-treated cells were stained with propidium iodide, and analyzed for cell proliferation by flow cytometry on day 3 after nucleofection or antibody treatment. (E) Primary CD4+ T cells were stained with 1 μM CFSE and then transfected with ΔVII-Ets-1 expression vector or pmax-Empty or activated with anti-CD3 and anti-CD28, and analyzed for cell proliferation by flow cytometry on day 3 after nucleofection or antibody treatment.
Ability of ΔVII-Ets-1 Overexpression to Stimulate Virus Production by Resting CD4+ T Cells from HIV-1-Infected Patients on HAART.
The results of our transient reporter assay demonstrated that ΔVII-Ets-1 can activate the HIV-1 LTR without the involvement of NF-κB. We extended this result by determining whether overexpression of ΔVII-Ets-1 could reactivate HIV-1 from the latent reservoir in patients on HAART. We isolated highly purified resting CD4+ T cells (>99%) from 6 different patients whose viral loads were below the limit of detection (<50 copies/mL of HIV-1 RNA), and then transfected these T cells with either a GFP-expressing or empty expression vector to serve as a negative control, a ΔVII-Ets-1-expression vector, or a positive control expression vector encoding HIV-1 Tat, a strong transactivator of the LTR that induces high levels of virus production when overexpressed in latently infected cells (30, 31).
Overexpression of ΔVII-Ets-1 resulted in an increase of virus release from 9- to 560-fold, indicating that overexpression of ΔVII-Ets-1 alone is sufficient to cause virus production in these resting CD4+ T cells of patients on HAART (Table 1). This stimulation was comparable with that achieved with the positive control Tat-expression vector. The transfection efficiency was estimated to be 45 to 70% based on the frequency of GFP+ cells among cells transfected with the GFP-expression vector. For 3 patients, we also tested whether fl-Ets-1 expression could activate virus production. Full-length Ets-1 expression resulted in a measurable activation of virus, but the effect was weaker than that observed with either ΔVII-Ets-1 or Tat, consistent with the weaker ability of fl-Ets-1 to activate transcription from the HIV-1 LTR (Fig. 2).
Table 1.
Induction of virus production from primary CD4+ T cells isolated from patients on HAART by transfection of ΔVII-Ets-1, fl-Ets-1, or Tat
| Control | ΔVII-Ets-1 | fl-Ets-1 | Tat | |
|---|---|---|---|---|
| Patient 1 | 128 | 1206 (9.4) | ND | ND |
| Patient 2 | <50 | 778 (>15.6) | ND | 815 (>16.3) |
| Patient 3 | <50 | 28006 (>560.1) | ND | 17353 (>347.1) |
| Patient 4 | <50 | 1132 (>22.6) | 112 (>2.2) | 745 (>14.9) |
| Patient 5 | <50 | 643 (>12.9) | 85 (>1.7) | 821 (>16.4) |
| Patient 6 | <50 | 788 (>15.8) | 220 (>4.4) | 851 (>17.0) |
The number in each cell represents the copy number of viral RNA released in the culture supernatant. The number in parentheses is the fold increase, as compared with the reference control vector pmax-GFP (patients 1–3) or pmaxEmpty (patients 4–6). ND, not determined.
Discussion
The reactivation of latent HIV-1 provirus is an essential prerequisite for the eradication of HIV-1 infection. We have demonstrated that an unbiased screen for activators of LTR-dependent transcription is a straightforward approach toward the identification of activities that can reactivate latent HIV-1. We isolated ΔVII-Ets-1 as an activity that was sufficient to induce transcription from the HIV-1 LTR. When introduced into latently infected CD4+ T cells from HIV-1-positive patients, ΔVII-Ets-1 could activate HIV-1 production without inducing T cell activation.
The HIV-1 LTR integrates the function of a multitude of transcription factors, some of which respond to signaling pathways that are central to cell activation, proliferation, and survival. Our results indicate that it is possible to use only one of the factors that contributes to LTR regulation to activate latent HIV-1 provirus without the need to activate multiple transcription factors or pathways that may be toxic. Other regulators of LTR transcription may provide a similar opportunity, and should be explored.
To focus our screen on activities that would be less likely to induce T cell activation, we used a mutant LTR that could not respond to NF-κB. This approach removed from our analysis the high frequency of genes that can activate NF-κB when overexpressed (20), and targeted the detection of genes that function through elements of the LTR distinct from the NF-κB-binding sites. However, it is still possible that some activators of NF-κB may be particularly efficient at activating latent HIV-1 without inducing T cell activation.
Full-length Ets-1 has previously been shown to participate in LTR regulation in concert with other transcription factors, like NF-κB, NFAT, and USF-1 (26, 27), by binding to a conserved Ets-1-binding site (−150 to −145) in the distal region of the LTR (32). Our results demonstrate that overexpression of the alternatively spliced form of Ets-1, ΔVII-Ets-1, is sufficient both for LTR transcriptional activation and activation of latent HIV-1. The activation of the LTR by ΔVII-Ets-1 required its DNA-binding activity, and proceeded through 2 conserved regions of the LTR, the region containing the Ets-1-binding site (−150 to −145) and the region of the LTR containing the Sp1 binding sites (3x Sp1) (−82 to −44). Although Ets-1 has not previously been reported to bind between −82 and −44, it is possible that ΔVII-Ets-1 can bind to a previously unidentified EBS in this region. Alternatively, ΔVII-Ets-1 may activate a cellular gene that in turn acts in this region to induce transcription from the HIV-1 LTR. Further studies are necessary to resolve these possibilities. ΔVII-Ets-1 is an isoform that is not subject to the regulation imposed by the autoinhibitory domain that is encoded by exon VII, or by phosphorylation sites also encoded by exon VII that negatively regulate Ets-1 DNA binding and transcriptional activity. Our data indicate that ΔVII-Ets-1 is a more potent activator of LTR-dependent transcription than fl-Ets-1, and this enhanced activity correlates with a greater ability to activate HIV-1 production. Therefore, the ΔVII-Ets-1 isoform may be uniquely suited to being active without the need for other factors or pathways that may be required for the activity of fl-Ets-1. The ability of ΔVII-Ets-1 to reactivate latent HIV-1 suggests that potential therapies might be aimed at increasing the levels of expression of this isoform of Ets-1 in latently infected cells. Alternatively, potential therapies might involve the conversion of fl-Ets-1 from an inhibited to an active transcription factor.
Our screen was conducted with only 1 cDNA expression library, and screening was well below saturation. Therefore, it is likely that our approach can be expanded and lead to the isolation of further interesting genes with desirable therapeutic possibilities. Our results prove that the screen for activators of LTR transcription is a good surrogate screen for genes that can activate latent HIV-1. It will allow efficient, inexpensive screening of more cDNA libraries, and can be adapted easily for the screening of small molecules.
Materials and Methods
Patients.
Resting CD4+ T cells were isolated from 6 HIV-1-infected adults who had been continuously receiving HAART for at least 6 months and had suppression of viremia to <50 copies/mL.
Primary and Secondary Screening.
We adapted the strategy of Pomerantz et al. (20). A human splenocyte cDNA library was divided into pools of 100 cDNAs per pool. HEK293T cells were plated at 9 × 104 cells per well in 24-well dishes, and transfected 24 h later with a total of 356 ng of DNA, including 2 ng of pCSK-LacZ, 4 ng of mκB-LTR-Luc, and 350 ng of pool cDNA by using the calcium phosphate method. The reference control transfection contained 350 ng pcDNA3.1(−) (Invitrogen). At 40–48 h after transfection, the cells were lysed in 100 μL of passive lysis buffer (Promega) at room temperature, and centrifuged at 13,000 × g at room temperature for 5 min to pellet debris. We used 20 and 10 μL of lysate, respectively, to assay luciferase and β-Gal activities, by using the luciferase assay system (Promega), a chemiluminescent β-Gal reporter gene assay (Roche), and luminometer (Central LB 960; Berthold) in accordance with the manufacturers' instructions. Fold stimulation was calculated for each sample by dividing the luciferase activity, normalized to the β-Gal activity, by that observed in the empty vector control sample. Positive pools were considered NF-κB-independent if their activity with the Igκ2-IFN-GL4 reporter was <30% of that observed with the mκB-LTR-Luc reporter, or if they stimulated the Igκ2-IFN-GL4 reporter <1.5-fold.
Transient Transactivation Assay in HEK293T Cells and Jurkat T Cells.
In Figs. 23–4, we used TK-RLuc (pGL4.74; Promega) as an internal control instead of pCSK-lacZ; 20 μL of lysates were analyzed by using the Dual-luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Fold stimulation was calculated by comparing observed activities with that achieved with the vector pmax-Empty. Jurkat T cells were grown in RPMI medium 1640 with GlutaMax-I (Invitrogen) supplemented with 10% FBS (Gemini Bio) and 100 U/mL each of penicillin and streptomycin. On the day of transfection, 5 × 105 cells were plated in 2 mL in each well of a 6-well plate before incubation with DNA-Fugene 6 (Roche) complexes using a total of 2.95 μg of DNA and 9 μL of Fugene 6. Transfections included 100 ng mκB-LTR-Luc, 50 ng TK-RLuc, and up to 2.8 μg pmax-ΔVII-Ets-1; 40–48 h after transfection, cells were lysed in 150 μL of passive lysis buffer (Promega) for 15 min at room temperature. Debris was removed by centrifugation at 14,000 × g for 5 min in a microcentrifuge at room temperature, and 40 μL of lysates were analyzed as described above.
Cell Lysates, Western Blots, and Antibodies.
Primary CD4+ T cells or HEK293T cells were lysed in RIPA buffer (50 mM Tris, pH 7.5/150 mM NaCl/10 mM EDTA/1% Nonidet P-40/0.1% SDS) plus protease inhibitors (Sigma). Debris was removed by centrifugation for 20 min at 13,000 × g in a microcentrifuge at 4 °C. Anti-Ets-1 (sc-350; Santa Cruz), anti-β-actin (Sigma), and anti-FLAG-HRP (Sigma) were used at dilutions of 1:400, 1:2,500, and 1:3,000, respectively.
RNA Isolation and Real-Time RT-PCR.
Total cellular RNA was isolated by using RNeasy Mini Kit (Qiagen) and RT reactions were performed by using SuperScript III Reverse Transcriptase (Invitrogen) with random primers (Invitrogen). Expression of IFN-γ or IL-2 transcripts was measured by using TaqMan Gene Expression Assay products on an ABI 7300 Real Time PCR System. The ubiquitin mRNA was measured by RT-PCR with SYBR Green PCR Master Mix (Applied Biosystems). All control reactions with no template or without the addition of RT were negative.
T Cell Purification and Transfections, RNA Isolation, and Real-Time RT-PCR for Viral Load.
Resting CD4+ T cells from patients on HAART were isolated through a 2-step purification procedure as previously described (2). Transfections were performed by using an Amaxa Nucleofector. Purified CD4+ T cells (2.5–4 × 106) were resuspended in 100 μL of Nucleofector solution, transfected by using program U-014, then cultured in 2 mL of RPMI medium 1640 with GlutaMax-I (Invitrogen) plus 10% FBS (Gemini Bio) and 100 units/mL each of penicillin and streptomycin. For each patient, the number of cells used was identical for each condition. For each transfection, 3.5–5.0 μg of pmax-GFP, pmax-Empty, pmax-ΔVII-Ets-1, pmax-fl-Ets-1, or pcDNA-Tat-86 was transfected. GFP expression served as a negative control and an indicator of transfection efficiency based on the frequency of GFP-positive cells assayed by flow cytometry. To measure the copy number of released virus, supernatants of transfected resting CD4+ T cells were collected 72 h after nucleofection. RNA isolation and real-time RT-PCR were done by using Amplicor Ultrasensitive for HIV-1 Kit (Roche) following the manufacturer's instructions.
Further details regarding the construction of reporter constructs and expression vectors are available in supporting information (SI) Materials and Methods.
Supplementary Material
Acknowledgments.
We thank H. Zhang for help in cell sorting, the laboratory of Dr. Thomas C. Quinn for measuring viral load, and Drs. J. Blankson and A. Shen for critical comments on this manuscript. This work was supported by National Institutes of Health Grant AI043222 (to R.F.S.). J.L.P is a Rita Allen Foundation Scholar and a recipient of a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809536106/DCSupplemental.
References
- 1.Palella FJ, Jr, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 3.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 6.Geeraert L, Kraus G, Pomerantz RJ. Hide-and-Seek: The Challenge of Viral Persistence in HIV-1 Infection. Annu Rev Med. 2008;59:487–501. doi: 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- 7.Margolis DM, Archin NM. Attacking HIV provirus: Therapeutic strategies to disrupt persistent infection. Infect Disord Drug Targets. 2006;6:369–376. doi: 10.2174/187152606779025824. [DOI] [PubMed] [Google Scholar]
- 8.Brooks DG, et al. Molecular characterization, reactivation, and depletion of latent HIV. Immunity. 2003;19:413–423. doi: 10.1016/s1074-7613(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 9.Lehrman G, et al. Depletion of latent HIV-1 infection in vivo: A proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prins JM, et al. Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1-infected patients on potent antiretroviral therapy. AIDS. 1999;13:2405–2410. doi: 10.1097/00002030-199912030-00012. [DOI] [PubMed] [Google Scholar]
- 11.van Praag RM, et al. OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion. J Clin Immunol. 2001;21:218–226. doi: 10.1023/a:1011091300321. [DOI] [PubMed] [Google Scholar]
- 12.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 14.Tong-Starksen SE, Luciw PA, Peterlin BM. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc Natl Acad Sci USA. 1987;84:6845–6849. doi: 10.1073/pnas.84.19.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BK, Feinberg MB, Baltimore D. The kappaB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J Virol. 1997;71:5495–5504. doi: 10.1128/jvi.71.7.5495-5504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoni BA, Rabson AB, Kinter A, Bodkin M, Poli G. NF-kappa B-dependent and -independent pathways of HIV activation in a chronically infected T cell line. Virology. 1994;202:684–694. doi: 10.1006/viro.1994.1390. [DOI] [PubMed] [Google Scholar]
- 18.Leonard J, et al. The NF-kappa B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989;63:4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 2002;21:5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorcyk CL, Watson DK, Mavrothalassitis GJ, Papas TS. The human ETS1 gene: Genomic structure, promoter characterization and alternative splicing. Oncogene. 1991;6:523–532. [PubMed] [Google Scholar]
- 23.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 24.Jonsen MD, Petersen JM, Xu QP, Graves BJ. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowley DO, Graves BJ. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 2000;14:366–376. [PMC free article] [PubMed] [Google Scholar]
- 26.Bassuk AG, Anandappa RT, Leiden JM. Physical interactions between Ets and NF-kappaB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieweke MH, Tekotte H, Jarosch U, Graf T. Cooperative interaction of ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J. 1998;17:1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 29.Flory E, Hoffmeyer A, Smola U, Rapp UR, Bruder JT. Raf-1 kinase targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin X, et al. Transcriptional profiles of latent human immunodeficiency virus in infected individuals: Effects of Tat on the host and reservoir. J Virol. 2003;77:8227–8236. doi: 10.1128/JVI.77.15.8227-8236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Estable MC, et al. Human immunodeficiency virus type 1 long terminal repeat variants from 42 patients representing all stages of infection display a wide range of sequence polymorphism and transcription activity. J Virol. 1996;70:4053–4062. doi: 10.1128/jvi.70.6.4053-4062.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.