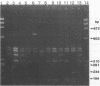
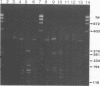
Abstract
Two methods, based on analysis of the polymerase chain reaction-amplified 16S rRNA gene by restriction enzyme analysis (REA) or direct cycle sequencing, were developed for rapid identification of mycobacteria isolated from animals and were compared to traditional phenotypic typing. BACTEC 7H12 cultures of the specimens were examined for "cording," and specific polymerase chain reaction amplification was performed to identify the presence of tubercle complex mycobacteria. Combined results of separate REAs with HhaI, MspI, MboI, and ThaI differentiated 12 of 15 mycobacterial species tested. HhaI, MspI, and ThaI restriction enzyme profiles differentiated Actinobacillus species from mycobacterial species. Mycobacterium bovis could not be differentiated from M. bovis BCG or Mycobacterium tuberculosis. Similarly, Mycobacterium avium and Mycobacterium paratuberculosis could not be distinguished from each other by REA but were differentiated by cycle sequencing. Compared with traditional typing, both methods allowed rapid and more accurate identification of acid-fast organisms recovered from 21 specimens of bovine and badger origin. Two groups of isolates were not typed definitively by either molecular method. One group of four isolates may constitute a new species phylogenetically very closely related to Mycobacterium simiae. The remaining unidentified isolates (three badger and one bovine) had identical restriction enzyme profiles and shared 100% nucleotide identify over the sequenced signature region. This nucleotide sequence most closely resembled the data base sequence of Mycobacterium senegalense.
Full text
PDF


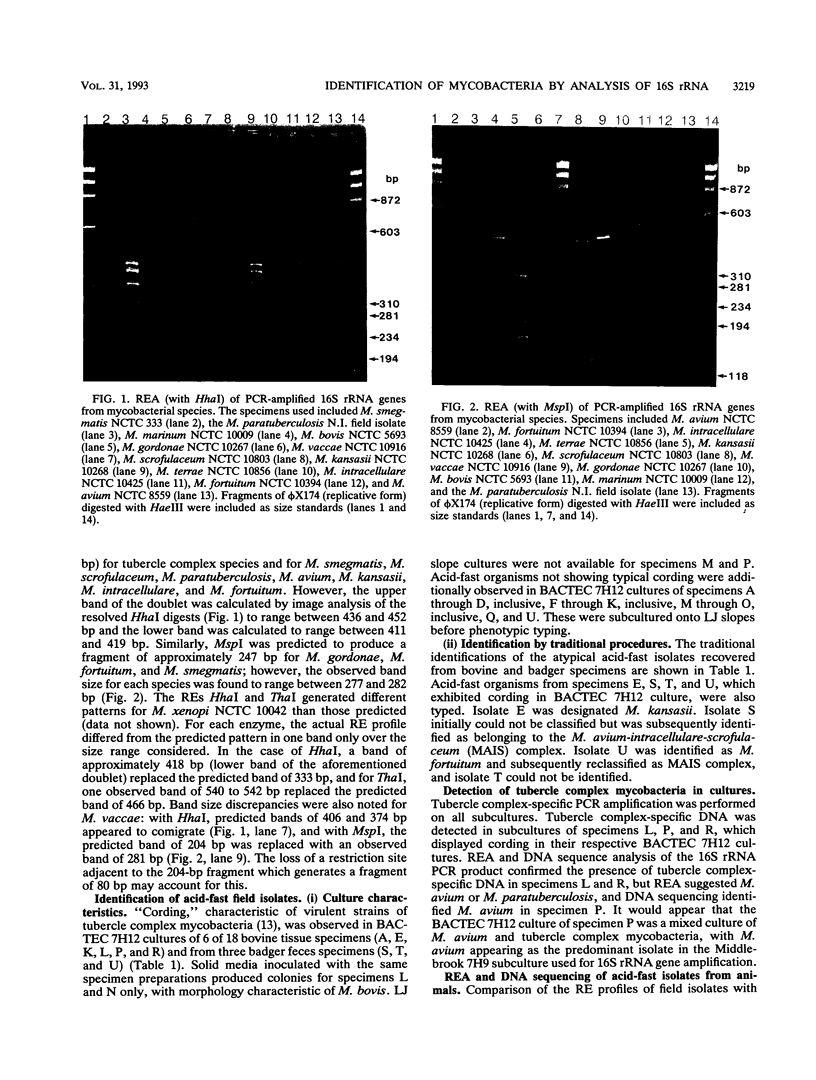



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Böddinghaus B., Rogall T., Flohr T., Blöcker H., Böttger E. C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990 Aug;28(8):1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böddinghaus B., Wolters J., Heikens W., Böttger E. C. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol Lett. 1990 Jul;58(2):197–203. doi: 10.1111/j.1574-6968.1990.tb13978.x. [DOI] [PubMed] [Google Scholar]
- Edwards U., Rogall T., Blöcker H., Emde M., Böttger E. C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989 Oct 11;17(19):7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenach K. D., Cave M. D., Bates J. H., Crawford J. T. Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis. J Infect Dis. 1990 May;161(5):977–981. doi: 10.1093/infdis/161.5.977. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Wisotzkey J. D., Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992 Jan;42(1):166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- Gilkerson S. W., Moss M., Cuthrell F. Microculture morphology of mycobacteria. J Bacteriol. 1966 Apr;91(4):1652–1654. doi: 10.1128/jb.91.4.1652-1654.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner P., Böttger E. C. Microheterogeneity within rRNA of Mycobacterium gordonae. J Clin Microbiol. 1992 Apr;30(4):1049–1050. doi: 10.1128/jcm.30.4.1049-1050.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica G. P., Baess I., Gordon R. E., Jenkins P. A., Kwapinski J. B., McDurmont C., Pattyn S. R., Saito H., Silcox V., Stanford J. L. A co-operative numerical analysis of rapidly growing mycobacteria. J Gen Microbiol. 1972 Nov;73(1):55–70. doi: 10.1099/00221287-73-1-55. [DOI] [PubMed] [Google Scholar]
- Kusaka T., Mori T. Pyrolysis gas chromatography-mass spectrometry of mycobacterial mycolic acid methyl esters and its application to the identification of Mycobacterium leprae. J Gen Microbiol. 1986 Dec;132(12):3403–3406. doi: 10.1099/00221287-132-12-3403. [DOI] [PubMed] [Google Scholar]
- Kusunoki S., Ezaki T., Tamesada M., Hatanaka Y., Asano K., Hashimoto Y., Yabuuchi E. Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 Mycobacterium species. J Clin Microbiol. 1991 Aug;29(8):1596–1603. doi: 10.1128/jcm.29.8.1596-1603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquin M., Ausina V., López Calahorra F., Belda F., García Barceló M., Celma C., Prats G. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J Clin Microbiol. 1991 Jan;29(1):120–130. doi: 10.1128/jcm.29.1.120-130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan M. L., Kazda J. F., McGill K., Quinn P. J., Cook B. M. Sensitisation of cattle to bovine and avian tuberculins with Mycobacterium cookii. Vet Rec. 1991 Oct 26;129(17):383–383. doi: 10.1136/vr.129.17.383-a. [DOI] [PubMed] [Google Scholar]
- Pitulle C., Dorsch M., Kazda J., Wolters J., Stackebrandt E. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol. 1992 Jul;42(3):337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- Plikaytis B. B., Plikaytis B. D., Yakrus M. A., Butler W. R., Woodley C. L., Silcox V. A., Shinnick T. M. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J Clin Microbiol. 1992 Jul;30(7):1815–1822. doi: 10.1128/jcm.30.7.1815-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHMOND L., CUMMINGS M. M. An evaluation of methods of testing the virulence of acid-fast bacilli. Am Rev Tuberc. 1950 Dec;62(6):632–637. doi: 10.1164/art.1950.62.6.632. [DOI] [PubMed] [Google Scholar]
- Rogall T., Flohr T., Böttger E. C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990 Sep;136(9):1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- Rogall T., Wolters J., Flohr T., Böttger E. C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990 Oct;40(4):323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- Telenti A., Marchesi F., Balz M., Bally F., Böttger E. C., Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993 Feb;31(2):175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagupsky P. V., Kaminski D. A., Palmer K. M., Nolte F. S. Cord formation in BACTEC 7H12 medium for rapid, presumptive identification of Mycobacterium tuberculosis complex. J Clin Microbiol. 1990 Jun;28(6):1451–1453. doi: 10.1128/jcm.28.6.1451-1453.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]