Abstract
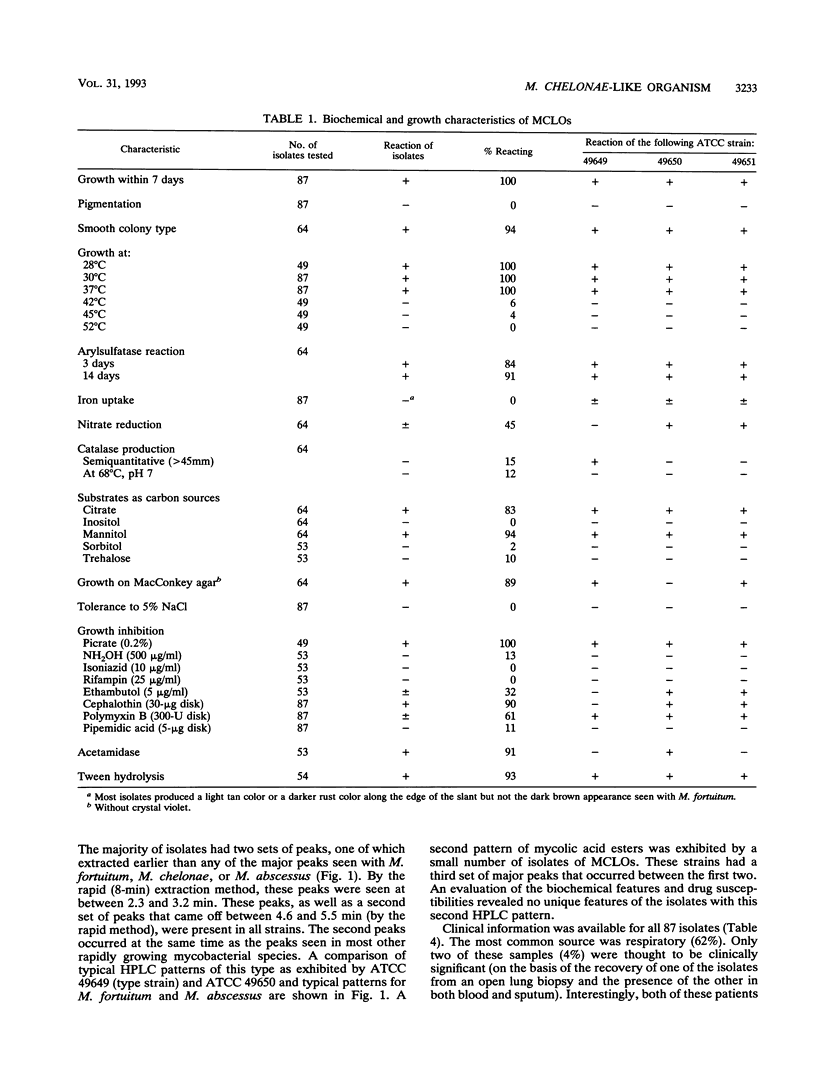
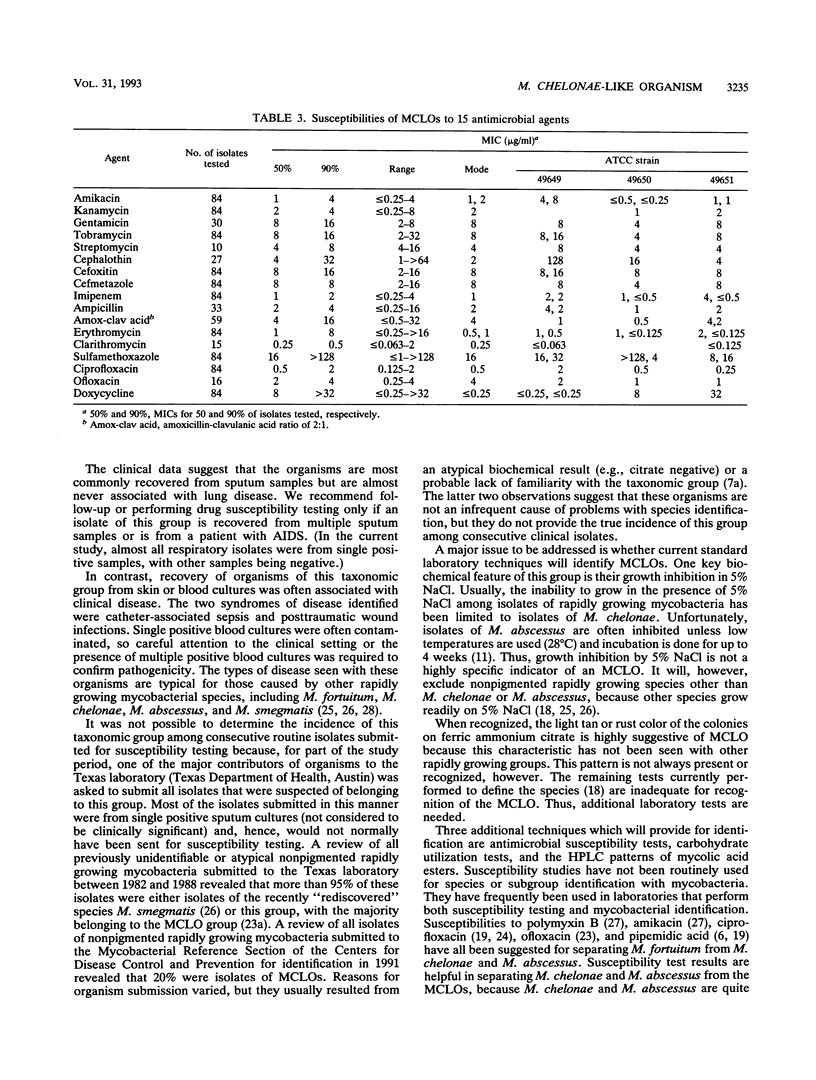
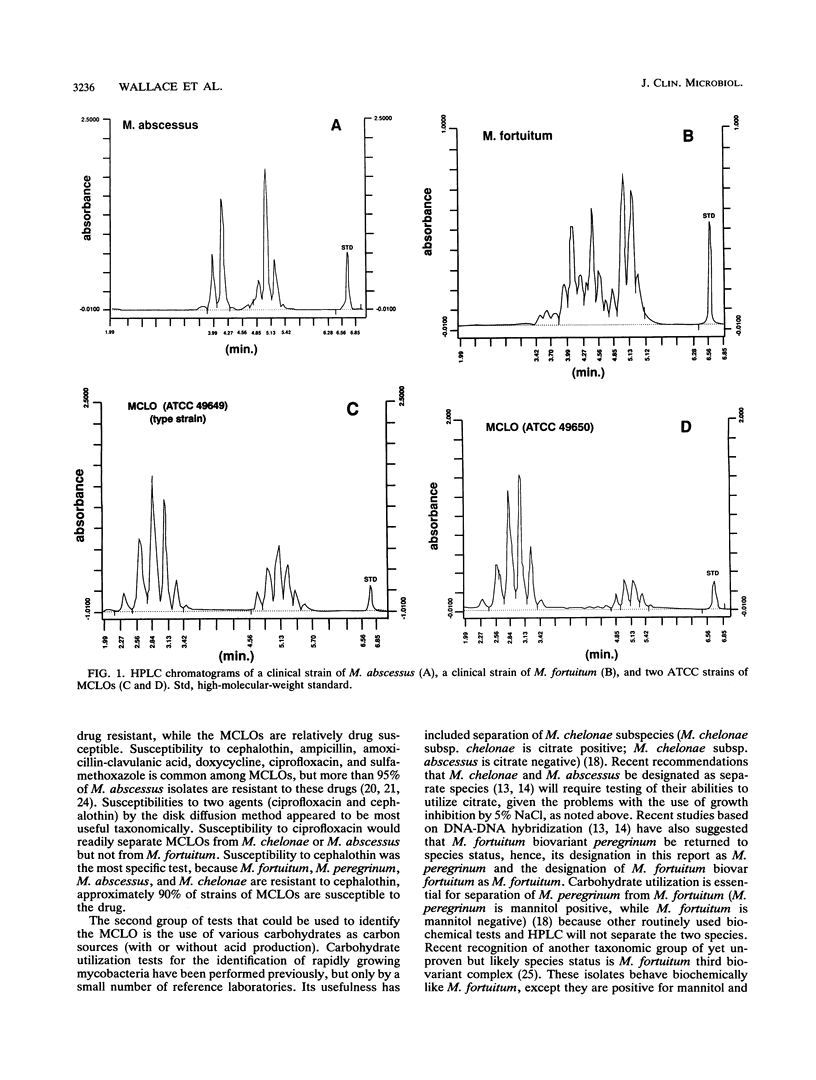
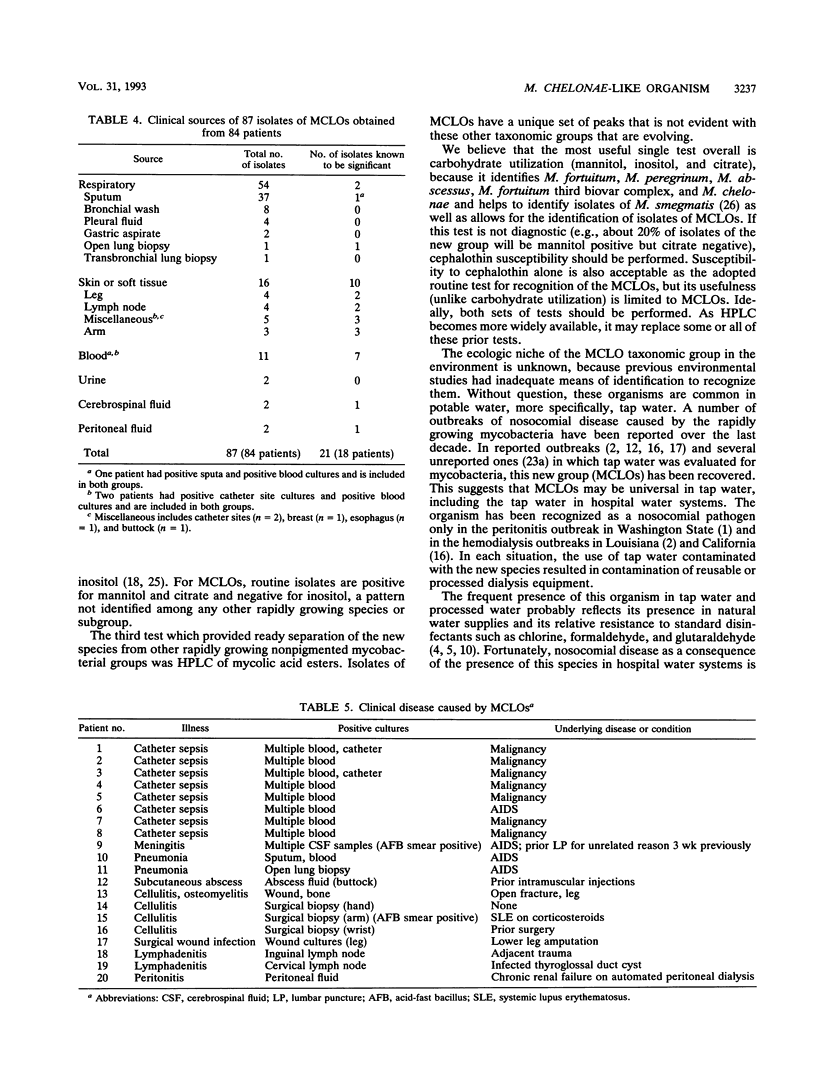
Mycobacterium chelonae-like organisms are nonpigmented rapidly growing mycobacteria whose clinical significance is unknown. We evaluated 87 sporadic isolates encountered in a clinical laboratory. Most isolates (62%) were respiratory; only 2 of 54 (4%) (both from patients with AIDS) were clinically significant. Among 33 nonrespiratory isolates, 20 of 33 (or 61%) were clinically significant. Clinical diseases included posttraumatic wound infections and catheter-related sepsis. Routine biochemical features included growth inhibition by 5% NaCl (100%), a smooth colony morphology (94%), positive 3-day arylsulfatase reaction (84%), no color or a light tan color on iron uptake (100%), and variable nitrate reduction (45%). Additional characteristics that helped to separate this group from M. chelonae and Mycobacterium abscessus were susceptibility to cephalothin (90%) and ciprofloxacin (100%), utilization of mannitol (94%) and citrate (83%) as carbon sources, and unique patterns of mycolic acid esters by high-performance liquid chromatography. This group was quite drug susceptible, with 100% of isolates inhibited by amikacin, imipenem, cefoxitin, cefmetazole, and the newer quinolones ciprofloxacin and ofloxacin. Three examples of this group, including a proposed type strain, have been deposited in the American Type Culture Collection.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Band J. D., Ward J. I., Fraser D. W., Peterson N. J., Silcox V. A., Good R. C., Ostroy P. R., Kennedy J. Peritonitis due to a mycobacterium chelonei-like organism associated with intermittent chronic peritoneal dialysis. J Infect Dis. 1982 Jan;145(1):9–17. doi: 10.1093/infdis/145.1.9. [DOI] [PubMed] [Google Scholar]
- Bolan G., Reingold A. L., Carson L. A., Silcox V. A., Woodley C. L., Hayes P. S., Hightower A. W., McFarland L., Brown J. W., 3rd, Petersen N. J. Infections with Mycobacterium chelonei in patients receiving dialysis and using processed hemodialyzers. J Infect Dis. 1985 Nov;152(5):1013–1019. doi: 10.1093/infdis/152.5.1013. [DOI] [PubMed] [Google Scholar]
- Brown B. A., Wallace R. J., Jr, Onyi G. O., De Rosas V., Wallace R. J., 3rd Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae-like organisms. Antimicrob Agents Chemother. 1992 Jan;36(1):180–184. doi: 10.1128/aac.36.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson L. A., Bland L. A., Cusick L. B., Favero M. S., Bolan G. A., Reingold A. L., Good R. C. Prevalence of nontuberculous mycobacteria in water supplies of hemodialysis centers. Appl Environ Microbiol. 1988 Dec;54(12):3122–3125. doi: 10.1128/aem.54.12.3122-3125.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson L. A., Cusick L. B., Bland L. A., Favero M. S. Efficacy of chemical dosing methods for isolating nontuberculous mycobacteria from water supplies of dialysis centers. Appl Environ Microbiol. 1988 Jul;54(7):1756–1760. doi: 10.1128/aem.54.7.1756-1760.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal M. J., Rodriguez F. C. Simple, new test for rapid differentiation of the Mycobacterium fortuitum complex. J Clin Microbiol. 1981 May;13(5):989–990. doi: 10.1128/jcm.13.5.989-990.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. H., Yates M. D., Uttley A. H. Differentiation of Mycobacterium chelonei from M. fortuitum by ciprofloxacin susceptibility. J Hyg (Lond) 1985 Dec;95(3):619–621. doi: 10.1017/s002217240006071x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd M. M., Silcox V. A., Jones W. D., Jr, Butler W. R., Kilburn J. O. Separation of Mycobacterium bovis BCG from Mycobacterium tuberculosis and Mycobacterium bovis by using high-performance liquid chromatography of mycolic acids. J Clin Microbiol. 1992 May;30(5):1327–1330. doi: 10.1128/jcm.30.5.1327-1330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrant G. O., Lambert M. A., Moss C. W. Gas-chromatographic analysis of mycolic acid cleavage products in mycobacteria. J Clin Microbiol. 1981 May;13(5):899–907. doi: 10.1128/jcm.13.5.899-907.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes P. S., McGiboney D. L., Band J. D., Feeley J. C. Resistance of Mycobacterium chelonei-like organisms to formaldehyde. Appl Environ Microbiol. 1982 Mar;43(3):722–724. doi: 10.1128/aem.43.3.722-724.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica G. P. Differential identification of mycobacteria. VII. Key features for identification of clinically significant mycobacteria. Am Rev Respir Dis. 1973 Jan;107(1):9–21. doi: 10.1164/arrd.1973.107.1.9. [DOI] [PubMed] [Google Scholar]
- Kuritsky J. N., Bullen M. G., Broome C. V., Silcox V. A., Good R. C., Wallace R. J., Jr Sternal wound infections and endocarditis due to organisms of the Mycobacterium fortuitum complex. Ann Intern Med. 1983 Jun;98(6):938–939. doi: 10.7326/0003-4819-98-6-938. [DOI] [PubMed] [Google Scholar]
- Kusunoki S., Ezaki T. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol. 1992 Apr;42(2):240–245. doi: 10.1099/00207713-42-2-240. [DOI] [PubMed] [Google Scholar]
- Lowry P. W., Beck-Sague C. M., Bland L. A., Aguero S. M., Arduino M. J., Minuth A. N., Murray R. A., Swenson J. M., Jarvis W. R. Mycobacterium chelonae infection among patients receiving high-flux dialysis in a hemodialysis clinic in California. J Infect Dis. 1990 Jan;161(1):85–90. doi: 10.1093/infdis/161.1.85. [DOI] [PubMed] [Google Scholar]
- Lowry P. W., Jarvis W. R., Oberle A. D., Bland L. A., Silberman R., Bocchini J. A., Jr, Dean H. D., Swenson J. M., Wallace R. J., Jr Mycobacterium chelonae causing otitis media in an ear-nose-and-throat practice. N Engl J Med. 1988 Oct 13;319(15):978–982. doi: 10.1056/NEJM198810133191504. [DOI] [PubMed] [Google Scholar]
- Lévy-Frébault V. V., Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992 Apr;42(2):315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- Silcox V. A., Good R. C., Floyd M. M. Identification of clinically significant Mycobacterium fortuitum complex isolates. J Clin Microbiol. 1981 Dec;14(6):686–691. doi: 10.1128/jcm.14.6.686-691.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele L. C., Wallace R. J., Jr Ability of ciprofloxacin but not pipemidic acid to differentiate all three biovariants of Mycobacterium fortuitum from Mycobacterium chelonae. J Clin Microbiol. 1987 Feb;25(2):456–457. doi: 10.1128/jcm.25.2.456-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson J. M., Thornsberry C., Silcox V. A. Rapidly growing mycobacteria: testing of susceptibility to 34 antimicrobial agents by broth microdilution. Antimicrob Agents Chemother. 1982 Aug;22(2):186–192. doi: 10.1128/aac.22.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson J. M., Wallace R. J., Jr, Silcox V. A., Thornsberry C. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob Agents Chemother. 1985 Dec;28(6):807–811. doi: 10.1128/aac.28.6.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamura M. In vitro antimycobacterial activity of a new antibacterial substance DL-8280--differentiation between some species of mycobacteria and related organisms by the DL-8280 susceptibility test. Microbiol Immunol. 1983;27(12):1129–1132. doi: 10.1111/j.1348-0421.1983.tb02933.x. [DOI] [PubMed] [Google Scholar]
- Wallace R. J., Jr, Bedsole G., Sumter G., Sanders C. V., Steele L. C., Brown B. A., Smith J., Graham D. R. Activities of ciprofloxacin and ofloxacin against rapidly growing mycobacteria with demonstration of acquired resistance following single-drug therapy. Antimicrob Agents Chemother. 1990 Jan;34(1):65–70. doi: 10.1128/aac.34.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., Jr, Brown B. A., Silcox V. A., Tsukamura M., Nash D. R., Steele L. C., Steingrube V. A., Smith J., Sumter G., Zhang Y. S. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J Infect Dis. 1991 Mar;163(3):598–603. doi: 10.1093/infdis/163.3.598. [DOI] [PubMed] [Google Scholar]
- Wallace R. J., Jr, Nash D. R., Tsukamura M., Blacklock Z. M., Silcox V. A. Human disease due to Mycobacterium smegmatis. J Infect Dis. 1988 Jul;158(1):52–59. doi: 10.1093/infdis/158.1.52. [DOI] [PubMed] [Google Scholar]
- Wallace R. J., Jr, Swenson J. M., Silcox V. A., Good R. C. Disk diffusion testing with polymyxin and amikacin for differentiation of Mycobacterium fortuitum and Mycobacterium chelonei. J Clin Microbiol. 1982 Dec;16(6):1003–1006. doi: 10.1128/jcm.16.6.1003-1006.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., Jr, Swenson J. M., Silcox V. A., Good R. C., Tschen J. A., Stone M. S. Spectrum of disease due to rapidly growing mycobacteria. Rev Infect Dis. 1983 Jul-Aug;5(4):657–679. doi: 10.1093/clinids/5.4.657. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wallace R. J., Jr, Steingrube V. A., Brown B. A., Nash R., Silcox A., Tsukamura M. Isoelectric focusing patterns of beta-lactamases in the rapidly growing mycobacteria. Tuber Lung Dis. 1992 Dec;73(6):337–344. doi: 10.1016/0962-8479(92)90037-k. [DOI] [PubMed] [Google Scholar]



