Abstract
Evaluation of eating quality in early breeding generations of rice is critical to developing varieties with better palatability. This paper reports DNA markers associated with eating quality of temperate japonica rice and an evaluation method aided by multiple regression analysis. A total of 30 markers comprising STSs, SNPs, and SSRs were tested for their association with palatability using 22 temperate japonica varieties with different palatability values. Eating quality-related traits of the 22 varieties were also measured. Of the 30 markers, 18 were found to be significantly associated with palatability and, consequently, a model regression equation with an R2 value of 0.99 was formulated to estimate the palatability by the marker data set. Validation of the model equation using selected breeding lines indicated that the marker set and the equation are highly applicable to evaluation of the palatability of cooked rice in temperate japonica varieties.
Keywords: Rice, eating quality, palatability, marker, regression analysis
Introduction
Rice (Oryza sativa L.) is the staple food for half of the world’s population. The eating quality of rice is increasingly important to meet the market demand. Therefore, one of the major goals in a breeding program is to develop rice varieties of better eating quality to satisfy the requirements of both the food industry and consumers. Even though indica rice varieties are popular worldwide, consumers in northeastern Asian countries such as Korea, Japan, northern China, and Taiwan prefer japonica rice, mainly due to its moderate elasticity and stickiness.
The eating quality of rice is a complex trait involving many physicochemical properties, and thus it has been challenging to accurately evaluate eating quality for selection in rice-breeding programs. Some key physicochemical properties affecting the eating quality are amylose content (AC) (1), pasting properties (PP) (2), gel consistency (GC), gelatinization temperature (GT) (3), and protein content (PC) (4). Good eating quality is also associated with stickiness, sweet flavor, glossiness of the cooked rice, and palatability. Palatability, the trait directly related to rice eating quality, is determined by aroma, appearance, taste, and texture (4). In addition to genetic determinants, such as genes involved in the synthesis of starch and protein, rice eating quality is also largely affected by environmental factors, cultural practices, and postharvest practices such as air temperature during ripening, the amount of fertilizer, irrigation management, grain-drying after harvest, and cooking methods (5).
In breeding programs, accurate evaluation of eating quality in early generations is critical. A sensory test by trained panels is the most appropriate evaluation method. However, because this method both requires a large amount of rice per sample and allows the evaluation of only a few samples per day, the sensory test is more efficient when performed at a later stage when selected lines are homozygous (6). Moreover, the results of sensory evaluation are sometimes not consistent even for the same sample, presumably due to the physical and emotional condition of members of the panel or subtle differences in sample preparation. Recently, an instrument for evaluating the palatability value of rice has been developed and used for line selection in breeding programs (7, 8). However, it also requires a large amount of rice per sample, and thus the palatability test using this instrument is usually performed only for advanced breeding generations.
A number of genetic studies on eating quality traits have been conducted. These have revealed that some rice physicochemical properties such as AC, GT, GC, and pasting viscosity are controlled by one to three major genes with one or more modifiers. The enzymes involved in starch biosynthesis, such as starch branching enzyme (SBE), starch synthase (SS), and granule bound starch synthase (GBSS) contribute greatly to the variation of starch physicochemical properties and thus eating quality (2). Major genes and/or quantitative trait loci (QTLs) associated with eating quality (9), PC (10), and palatability (9, 11) such as Wx (waxy gene) and alk (starch synthase II) (3) have been reported. In addition, interaction among these genes along with others may govern rice grain physicochemical properties, which in turn determine the eating quality of cooked rice. Collectively, the genetic complexity of eating quality, as well as the difficulty in accurate evaluation of eating quality at early breeding generations, has constrained the development of rice varieties with high eating quality.
To complement the physicochemical analyses and sensory tests available to evaluate eating quality, DNA marker-based approaches have been developed. These methods offer the additional advantages of screening at early breeding generations as well as simplicity and accuracy. Markers based on the Polymerase Chain Reaction (PCR) have been tested for quality evaluation of rice varieties (12). Recently, sequence-tagged site (STS) primers developed from random amplified polymorphic DNA (RAPD) analysis were able to differentiate rice varieties according to their palatability (12, 13). Several functional markers have also been developed to distinguish the physicochemical properties of rice, especially the effect of the waxy locus on PP (14), that of SBE on starch viscosity (15), and those of AC (2) and starch synthase IIa (SSIIa) on GT (3). Additional gene-tagged markers have also been developed from starch-synthesizing genes (2, 14, 15). Despite the recent progress in developing markers and identification of QTLs associated with eating quality, a marker-assisted breeding (MAB) system for better eating quality has not been established.
In this study, our aim was to develop DNA markers associated with eating quality and to formulate a marker-based evaluation and prediction method of eating quality of cooked rice in japonica varieties.
Materials and Methods
Plant Materials and DNA Extraction
A total of 22 japonica rice varieties, mostly bred in Korea, were evaluated for palatability. These consisted of 2 varieties from Japan (Koshihikari and Hitomebore), 1 variety from China (Hexi41), and 19 varieties from Korea (Gopum, Ilpum, Samgwang, Chucheong, Dongjin, Sinkeumo, Hwaseong, Hwacheong, Dobong, Samnam, Palkong, Baekjinju1, Seonong4, Onnuri, Manmi, Giho, Geuman, Nakdong, and Samdeok). These varieties were chosen because they represent diverse palatability scores among japonica rice (National Institute of Crop Science (NICS), personal communication). Varieties were grown using conventional cultural practices at the experimental farm of Seoul National University, Suwon, in 2006. In addition, 32 japonica breeding lines along with three check varieties grown in a regional yield trial plot in 2007 at NICS were also used for validation of the marker set developed in this study. For physicochemical analysis, rice grains were dried to 15% moisture content. All rice varieties and lines were grown in a light- and temperature-controlled greenhouse until the tillering stage, at which point tissue was collected for DNA extraction. Genomic DNA was extracted using the modified cetyltrimethylammonium bromide (CTAB) method based on the protocol of Murray and Thompson (16).
Previously Reported Markers
Twenty previously reported STS primer pairs developed from RAPD (12, 13, 17) were tested in this study. To identify the amplicon sequences, the amplified bands were excised, purified, TA cloned, and sequenced. In a homology search, each cloned sequence was compared against all sequences in the nonredundant databases using the BLASTX and BLASTN programs (http://www.ncbi.nlm.nih.gov/BLAST and www.gramene.org). However, only one of the six markers, SSIIa from Bao et al. (3), was polymorphic among the 22 rice varieties and was actually used in this study. The sequences and banding patterns of markers previously reported are presented in Table 1 and Figure 1, respectively.
Table 1. Previously Reported Markers for the Evaluation of Rice Eating Quality and Their Chromosomal Locations.
| primer sequence |
||||
|---|---|---|---|---|
| PCR marker | marker type | chra | forward (5′−3′) | reverse (5′−3′) |
| Ohtsubo et al. (13,17) Ohtsubo and Nakamura (12) | ||||
| A6 | STS | 7 | CCAGCTGTACGCCTGTACTAC | CCAGCTGTACGTCTTCCCCAGC |
| A7 | STS | 12 | TGCCTCGCACCAGAAATAG | TGCCTCGCACCATGAG |
| B1 | STS | 11 | GTTTCGCTCCTACAGTAATTAAGGG | GTTTCGCTCCCATGCAATCT |
| B43 | STS | 9 | GGCCGGCATGACTCAC | ACTGGCCGGCATCAAGAC |
| F6 | STS | 4 | ACCACTCCATATATATCATCCAAAG | ACCACTCCATATCACCACAAGG |
| G4 | STS | 1 | GAGACCGATATGCGATTC | GTGGTGTTTAGATCCAGAGACTTA |
| G22 | STS | 9 | CTCACTCAAATTTACAGTGCATTTTCTTG | AGGGCCATGATACAAGACTCTGT |
| G28 | STS | 1 | GGCGGTCGTTCTGCGAT | GGAGAATCCCACAGTAAGTTTTTCTTTG |
| J6 | STS | 11 | GTCGGAGTGGTCAGACCG | GTCGGAGTGGATGGAGTAGC |
| M2CG | STS | 8 | ACAACGCCTCCGATGA | ACAACGCCTCCGACAACAAGAT |
| M11 | STS | 6 | GTCCACTGTGACCACAACAT | GTCCACTGTGGGGATTGTTC |
| P5 | STS | 10 | ACAACGGTCCGTCCTTGCTT | ACAACGGTCCAACAGATACTTTTGA |
| S13 | STS | 1 | GTCGTTCCTGTGGTTAGGACAGGGT | GTCGTTCCTGCTGGTGTCTCAGAT |
| T16 | STS | 12 | GGTGAACGCTGTAGTTGGAATATA | GGTGAACGCTCAGATTTAAATATAAT |
| WK9 | STS | 9 | CCCGCAGTTAGATGCACCATT | CCGCAGTTAGATCAAGTGGC |
| E30 | STS | 1 | TACCTGGTTGATGTATACAGATCTGGTT | ATCCCTCGATCCCTCTAGCATTAT |
| B7 | STS | 2 | CAGGTGTGGGTTACAAGGATGA | CAGGTGGTTCACGGCCTTT |
| G49A | STS | 11 | AATCCAGACATGAAATTTATATGCAGATA | AATCCAGACATGTTGTCCTCAATTTTTG |
| G81 | STS | 6 | TACCTGAACCAGCAAGCATGCGCG | TACCTGAACCAGTATAATCTTTG |
| P3 | STS | 5 | AACGGGCCAAAAACGGAGGT | AACGGGCCAACGCAG |
| Bao et al. (2, 3) | ||||
| Wx (SNP) | dCAPS/AccI | 6 | CTTTGTCTATCTCAAGACAC | TTTCCAGCCCAACACCTTAC |
| SS1 (SSR) | SSR | 6 | GATCCGTTTTTGCTGTGCCC | CCTCCTCTCCGCCGATCCTG |
| SBE1 (SSR) | SSR | 6 | ATTTCTTTGGCCACAGGCGA | CCCAGATTCGGAACAAGAAC |
| SBE1 (STS) | STS | 6 | GAGTTGAGTTGCGTCAGATC | AATGAGGTTGCTTGCTGCTG |
| SBE3 (SNP) | dCAPS/SpeI | 2 | GTCTTGGACTCAGATGCTGGACTC | ATGTATAACTGGCAGTTCGAACGG |
| SSIIa | SNP | 6 | F7: CTGGATCACTTCAAGCTGTACGAC | R1: GCCGGCCGTGCAGATCTTAAC |
| F22: CAAGGAGAGCTGGAGGGGGC | R21: ACATGCCGCGCACCTGGAAA | |||
Chromosome location.
Figure 1.
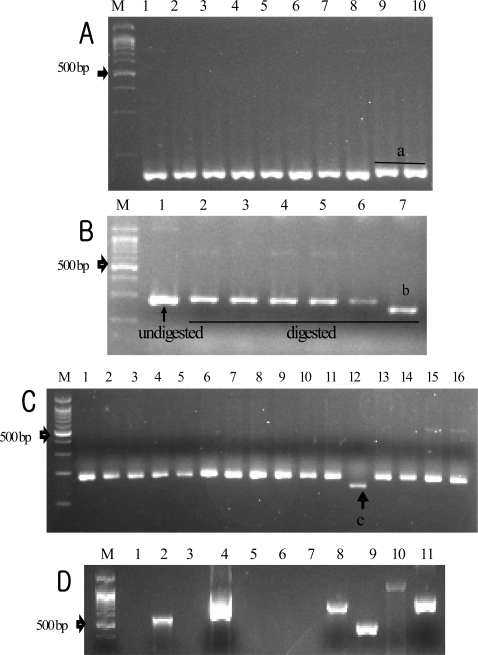
Examples of PCR amplicons of several types of markers. (A) Polymorphic indel in Tre (M, 100 bp DNA marker; 1, Koshihikari; 2, Samgwang; 3, Ilpum; 4, Sinkeumo; 5, Dobong; 6, Samnam; 7, Palkong; 8, Hitomebore; 9, Hexi41; 10, Samdeok); a, insertion of CTTT. (B) Polymorphic SNP in S3 (1, Koshihikari; 2, Samgwang; 3, Ilpum; 4, Sinkeumo; 5, Dobong; 6, Samnam; 7, Hexi41); b, point mutation T to G allele. (C) Variation in (CTT)n repeats in CBG (1, Koshihikari; 2, Gopum; 3, Samgwang; 4, Ilpum; 5, Chucheong; 6, Dongjin; 7, Sinkeumo; 8, Hwaseong; 9, Hwacheong; 10, Dobong; 11, Samnam; 12, Palkong; 13, Hitomebore; 14, Baekjinju1; 15, Seonong 4; 16, Manmi); c, (CTT)8 alleles, others are (CTT)19 alleles. (D) DNA banding patterns produced from 11 STS markers developed by Ohtsubo et al. (13, 17) and Ohtsubo and Nakamura (12) observed on Koshihikari (marker; 1, B7; 2, A7; 3, G81; 4, B43; 5, E30; 6, F6; 7, G4; 8, G22; 9, G28; 10, J6; 11, M11).
Development of New Markers
On the basis of selected regions either close to or within the genes linked to interesting QTLs (11, 18, 19) for rice eating quality traits (Table 2), analysis of nucleotide polymorphism of the sequences among japonica varieties was performed to develop primers for eating quality. To design primers, Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3) (20) was used. The purified PCR products from various japonica varieties were TA-cloned into pGEM-T Easy vectors and transformed into Eschericia coliDH5α-competent cells prepared according to the protocol of Sambrook and Russell (21). Plasmids were isolated using the DNA-spin Plasmid DNA Purification Kit (Intron Biotechnology, Korea) and sequenced with an ABI-3700 DNA sequencer following the manufacturer’s instructions (Applied Biosystems, Inc.). To identify SNPs, insertions and deletions (Indels), and/or microsatellite repeats in the rice varieties, sequence results were aligned using the CLUSTAL W program (22) from EMBL-European Bioinformatics Institute (http://www.ebi.ac.uk/tools), with assistance from Codoncode Aligner 2.0.6 (CodonCode Corporation, Dedham, MA) as well as BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). To detect a 1 bp substitution in a specific fragment, a dCAPS (derived cleaved amplified polymorphic sequences) primer was designed, facilitated by dCAPS Finder 2.0 (23) (http://helix.wustl.edu/dcaps). Finally, nine molecular markers were successfully developed on the basis of Indels, SNPs, and microsatellite repeats identified in this study (Table 3).
Table 2. Candidate Genes for QTLs Related to Rice Eating Quality and a Search of the Genomics DBa.
| candidate gene | clone | markers developed | chrb | QTL | source |
|---|---|---|---|---|---|
| granule−bound starch synthase1 (GBSS1) | AP002542 | GBSS1 | 6 | OSR19−RM587 | Kwon et al. (18) |
| sucrose synthase 3 (S3) | AP004988 | S3cI, S3cII | 7 | RM234−RM47 | Kwon et al. (18) |
| trehalose phosphatase(Tre) | AP004341 | TreB | 7 | RM234−RM47 | Kwon et al. (18) |
| UDP−N−acetylglucosamine pyrophosphorylase (AcPh) | AP003875 | AcPh | 8 | RM547−RM72 | Kwon et al. (18) |
| glucosamine−fructose−6−phosphate aminotransferase (GPA) | AC138454 | GPA | 11 | RM20b−RM332 | Kwon et al. (18) |
| aspartate aminotransferase (AM) | AP003991 | Ams | 2 | OSR8−OSR9 | Suh et al. (19) |
| noncyanogenic β−glucosidase (CBG) | AC074354 | CBG | 10 | RM2887 | Wada et al. (12) |
| ADP−glucose pyrophosphorylase/shrunken gene (SH) | AP004317 | SH51 | 1 | Genomics DB |
http://www.ncbi.nlm.nih.gov/, http://rgp.dna.affrc.go.jp/E/IRGSP/index.html, http://rice.plantbiology.msu.edu/, and http://www.gramene.org/.
Chromosome location.
Table 3. Markers Developed in This Study for Evaluation of Rice Eating Quality.
| sequence |
|||||
|---|---|---|---|---|---|
| marker name | marker type | chra | forward (5′−3′) | reverse (5′−3′) | amplicon size (bp) |
| S3cI | Indel | 7 | CCACTCTCATGTCCTTGAAC | GCCATGACATTTGGACAT | 153/150 |
| S3cII | dCAPS/TaqI | 7 | TTCCATGATGTGCCACTCTC | GGACAAATGTTTTCAGTGAATAAAT | 277/251 + 26 |
| TreB | Indel | 7 | CACTCCAGTTCCTGCTCAAA | CACTCCAGTTCCTGCTCAAA | 167/171 |
| AMs | SSR | 2 | CTTCCAAGGACCCCATCCT | CCCAACATCTCCGTCAGAAT | 187/179 |
| GPA | SSR | 11 | AATACGCGGCCTTCTCCTAT | TTGATCCGAATGGGTCAAAT | 178/149 |
| GBSS1 | SSR | 6 | CAAATAGCCACCCACACCAC | CTTGCAGATGTTCTTCCTGATG | 172/170 |
| AcPh | dCAPS/MseI | 8 | AGTTGTGGTTTAAGCATAGG | ATTGTCCTTTTCTTTAAAGTTTATTA | 14 + 10 + 125 + 12/14 + 135 + 12 |
| CBG | SSR | 10 | AGCTTCCCTAATGGCTTCGT | ATTTGCCAACTTTTGGATGG | 184/151 |
| SH51 | dCAPS/SpeI | 1 | ATTCTTGATGAAAATAATTAACTAG | GGTTAACCATCTTATAAAATTTGTC | 475/454 + 21 |
Chromosome location.
PCR Protocols
PCR amplification of markers was carried out in a PTC-200 Peltier Thermal Cycler (MJ Research, Inc.) in a total volume of 20 μL with the following genotyping PCR reagents: 2 μL of DNA at 20 ng/μL, 2 μL of 10× buffer containing 25 mM MgCl2, 1 μL of 2.5 mM dNTPs, 1 unit of Taq Polymerase (Intron Biotechnology, Korea), and 1 μL each of forward and reverse primers (10 μM). The PCR reaction for further sequencing analysis consisted of 1 unit of ExTaq polymerase (TaKaRa) in a total reaction of 50 μL. All amplifications were performed for a total of 35 cycles of 1 min at 95 °C, 30 s at 55 °C, and 1 min at 72 °C. For STS markers developed by Ohtsubo et al. (13, 17) and Ohtsubo and Nakamura (12), the initial denaturation was at 95 °C for 5 min, followed by 40 cycles of 96 °C for 1 min, 62 °C for 1 min, and 72 °C for 2 min. All amplifications using primers reported by Bao et al. (2, 3) were performed under the following conditions: 5 min at 94 °C followed by 35 cycles of 45 s at 94 °C, 1 min at 55 °C, and 1 min at 72 °C, with a final extension of 7 min at 72 °C. Amplified PCR products were analyzed by electrophoresis on 3% agarose gels stained with ethidium bromide and/or by nondenaturing electrophoresis on 8% polyacrylamide gels stained with ethidium bromide (model MGV, CBS Scientific Co.).
Evaluation of Eating Quality Traits of Rice Varieties
For palatability and physicochemical analyses, the rice grains were hulled and milled to 91% yield. Palatability was measured using a rice taste measuring system (Toyo taste meter, model MA-90) in accordance with the operation manual (TRCM Co.) (Toyo Rice Polishing Machine Factory, Japan). PC was calculated using total nitrogen multiplied by 5.95 after determination of the nitrogen content of rice material by the micro-Kjeldahl method (24). The AC of milled rice was determined by the relative absorbency of starch−iodine color in a digested solution of 100-mesh rice flour according to the method of Perez and Juliano (25). RVA pasting properties were determined on a rapid visco analyzer (RVA) in accordance with the operation manual (NewPort Sci. Co., Australia). Rice starch paste profile characteristics were described by six parameters: peak viscosity (PV), hot paste viscosity (HPV), cool paste viscosity (CPV), breakdown viscosity (BDV = PV − HPV), setback viscosity (SBV = CPV − PV), and consistency viscosity (CTV = CPV − HPV) in accordance with the procedure of Bao and Xia (26).
Sensory Evaluation by a Taste Panel
The milled rice was cooked according to the protocol of NICS, the Rural Development Administration (RDA), Korea. Dry-milled head rice (300 g) was rinsed four times and soaked for 30 min with distilled water, and the water was then strained for 10 min. Rice was cooked using an electric rice cooker with the ratio of rice/water = 1:1.25 w/w. After completion of the automatic cooking cycle, the rice was allowed to remain in the cooker for 30 min. Samples were transferred to plates and kept at room temperature for about 10 min until cooled to 35−37 °C. Sensory evaluation of cooked rice samples by 11 well-trained panel members was performed with five replications. The overall palatability was assessed according to appearance (glossiness), fragrance, taste, stickiness, texture, and palatability score (overall score; overall eating quality) and scored from +3 to −3 compared to a cooked rice reference sample, Chucheong (score = 0). The palatability score of each variety by the sensory test was the average value scored by 11 panel members.
Statistical Analysis
The collected data were analyzed and subjected to analysis of variance using SAS software version 8.2 (27). The least significant difference (Duncan) method was used to evaluate differences between trait means. Regression and correlation analyses were also performed to determine the relationships between rice eating quality traits. Multiple regression analysis was also conducted to determine the relationship between palatability scored both by taste analyzer and by sensory test and also palatability evaluated by molecular markers. STS marker data were scored as 1 (present) and 0 (absent). Similarly, the two different alleles resulting from each SSR and SNP marker were also converted into binary values of 1 or 0. By using the palatability scores as dependent variables and the binary data from molecular markers as independent variables or regressors, the best model equation to predict rice palatability was obtained. The most accurate prediction gave the lowest standard error and significantly highest coefficient of determination (R2), which consisted of the highly significant regression coefficient for each marker in the model. Using the binary matrix, a cluster analysis was performed with the Unweighted Pair Group Method (UPGMA) in NTSYS (Exeter Software, Setauket, NY) (28).
Results
Evaluation and Development of Markers
DNA sequences of PCR amplicons produced by 20 previously reported STS primer pairs (12, 13, 17) were compared against all sequences in the nonredundant databases using the BLASTX and BLASTN programs. The amplicon sizes ranged from about 450 to 1800 bp in length. However, no candidate gene of any known function possibly related to quality traits was found among the sequences derived from STS primers. Therefore, a total of 21 markers, including SSIIa (3), were used directly in this study (Table 1) as 5 of the markers listed in Table 1 from Bao et al. (2) were not polymorphic among the 22 varieties.
On the basis of QTL analyses for rice eating quality (11, 18, 19), we were able to select seven candidate genes underlying the QTL regions: sucrose synthase 3 (S3, clone AP004988), trehalose phosphatase (Tre, clone AP004341), granule bound starch synthase 1 (GBSS1/Waxy gene, clone AP002542), UDP-N-acetylglucosamine pyrophosphorylase (AcPh, clone AP003875), glucosamine-fructose-6-phosphate aminotransferase (GPA, clone AC138454), aspartate aminotransferase (AM, clone AP003991), and noncyanogenic β-glucosidase (CBG, clone AC074354) (Table 2). In addition, by searching the Genomics Date Base (DB), ADP-glucose pyrophosphorylase (shrunken gene, SH51, AP004317), a gene involved in starch biosynthesis and located on chromosome 1, was also chosen for marker development. By comparing the sequence of each gene (Table 2) among japonica varieties, we developed a total of nine DNA markers (Table 3). Examples of PCR amplicons of different marker types among japonica rice varieties are shown in Figure 1. Some unique alleles existed in only some varieties. The insertion of CTTT alleles in the Tre locus (Figure 1A) and the G allele in the S3 locus (Figure 1B) were rarely found in japonica varieties. In the CBG locus were distinguished two (CTT)n repeats of (CTT)8 and (CTT)19, but only a rice of low palatability (Palkong) possessed the (CTT)8 allele (Figure 1C). Moreover, the DNA banding pattern from the STS primers for palatability on Koshihikari was similar to those of previous studies (12, 13, 17) (Figure 1D; Table 4).
Table 4. Genotyping of 22 Rice Varieties Using 32 Markers.
| (A) Using Previously Reported Markers (12, 13, 17) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| primer |
|||||||||||||||||||||
| variety | A6 | A7 | B1 | B43 | E30 | F6 | G4 | G22 | G28 | J6 | M2CG | M11 | P5 | S13 | T16 | WK9 | B7 | G49A | G81 | P3 | digitized value |
| Koshihikari | + | + | + | + | − | − | − | + | + | + | − | + | + | − | − | − | − | − | − | + | 11,110,001,110,110,000,001 |
| Gopum | + | + | + | + | + | − | − | − | + | − | + | − | − | − | − | − | − | − | + | + | 11,111,000,101,000,000,011 |
| Samgwang | + | + | + | + | + | − | − | − | + | + | + | − | − | + | − | − | + | + | − | + | 11,111,000,111,001,001,101 |
| Ilpum | + | + | + | + | + | − | − | − | + | − | − | − | − | − | − | + | − | + | − | + | 11,111,000,100,000,010,101 |
| Chucheong | + | + | + | + | + | + | − | − | + | − | + | − | − | − | − | − | − | + | − | + | 11,111,100,101,000,000,101 |
| Dongjin | + | + | + | + | − | + | − | − | − | + | − | − | − | + | − | − | − | − | + | + | 11,110,100,010,001,000,011 |
| Sinkeumo | + | + | + | + | − | − | − | − | + | + | − | − | − | − | − | + | − | + | − | + | 11,110,000,110,000,010,101 |
| Hwaseong | + | + | + | + | − | + | − | − | − | + | − | − | − | − | − | − | − | + | − | − | 11,110,100,010,000,000,100 |
| Hwacheong | + | + | + | + | + | − | − | − | + | − | + | − | − | − | − | − | − | + | − | + | 11,111,000,101,000,000,101 |
| Dobong | + | + | + | + | − | − | − | − | + | − | + | + | − | − | − | − | − | − | − | + | 11,110,000,101,100,000,001 |
| Samnam | + | + | + | + | + | − | − | − | − | − | + | − | − | − | − | − | − | + | − | + | 11,111,000,001,000,000,101 |
| Palkong | + | − | + | + | − | − | − | − | + | + | − | + | − | − | − | − | − | + | − | + | 10,110,000,110,100,000,101 |
| Hitomebore | + | + | + | + | − | − | − | + | + | + | + | + | + | + | + | + | − | − | − | + | 11,110,001,111,111,110,001 |
| Baekjinju1 | + | + | + | − | + | + | − | − | + | + | + | + | − | − | − | + | − | + | + | − | 11,101,100,111,100,010,110 |
| Seonong4 | + | + | + | − | + | + | − | + | − | + | + | − | − | + | + | + | − | + | + | + | 11,101,101,011,001,110,111 |
| Onnuri | + | + | + | − | + | + | − | − | − | + | + | + | − | + | − | + | − | − | − | − | 11,101,100,011,101,010,000 |
| Manmi | + | + | + | + | + | − | − | + | + | + | − | + | − | + | − | − | − | + | − | + | 11,111,001,110,101,000,101 |
| Giho | + | + | + | − | + | − | − | + | − | + | + | − | − | + | − | − | − | + | + | + | 11,101,001,011,001,000,111 |
| Geuman | + | + | + | + | + | − | − | + | + | + | + | + | − | − | − | − | − | − | + | + | 11,111,001,111,100,000,011 |
| Nakdong | + | + | + | + | − | − | − | + | + | + | + | − | − | + | − | − | − | − | − | − | 11,110,001,111,001,000,000 |
| Hexi41 | + | + | + | + | + | + | + | + | + | + | + | − | − | − | + | − | − | − | − | + | 11,111,111,111,000,100,001 |
| Samdeok | + | + | + | − | + | − | − | + | + | + | + | − | − | + | − | + | − | − | − | + | 11,101,001,111,001,010,001 |
| (B) Using SSIIa (3) and Nine Newly Developed Markers | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| binary data of each primer |
||||||||||
| variety | SSIIaa | TreBb | S3cIIc | S3cId | AMse | GPAf | GBSSIg | AcPhh | SH51i | CBGj |
| Koshihikari | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| Gopum | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
| Samgwang | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| Ilpum | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Chucheong | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Dongjin | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Sinkeumo | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Hwaseong | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Hwacheong | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Dobong | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Samnam | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Palkong | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| Hitomebore | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Baekjinju1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Seonong4 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| Onnuri | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| Manmi | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Giho | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Geuman | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 |
| Nakdong | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Hexi41 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| Samdeok | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 |
TT:GGTTTC (0) and GC:GGGCTC (1) at nt 4329−4330 (exon).
Insertion of CTTT (1) and no insertion (0) at nt 79−82 of consensus region (intron).
Point mutation from T (1) to G allele (0) at nt 1454 of consensus region (intron).
No deletion (1) and deletion (CTC) (0) at nt 1255−1257 of consensus region (intron).
(CT)31 (1) and (CT)27 (0).
(CT)26 (0) and (CT)11 (1).
(CT)18 (1) and (CT)17 (0).
Point mutation from T (1) to G allele (0) at nt 397 of consensus region (intron).
Point mutation from A (0) to T allele (1) at nt 51 (intron).
(CTT)19 (1) and (CTT)8 (0).
Genotyping of Rice Varieties and Cluster Analysis
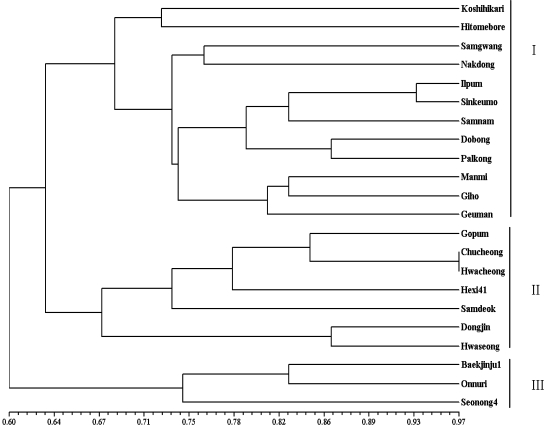
We genotyped 22 japonica rice varieties with diverse palatability values using a total of 30 markers comprising 21 markers developed previously and 9 markers developed in this study (Table 4). Cluster analysis was performed on similarity coefficient matrices calculated from molecular markers to generate a dendrogram (Figure 2). The varieties Chucheong and Hwacheong showed maximum genetic similarity (0.96), whereas Koshihikari and Seonong4 showed the least genetic similarity (0.41). When a cutoff value of 0.64 was used for genetic similarity among all varieties, three clusters were formed, I, II, and III (Figure 2), which contained 12, 7, and 3 varieties, respectively. The two Japanese cultivars, Koshihikari and Hitomebore, formed an independent subcluster, supporting the fact that Hitomebore was bred using Koshihikari as a parent. Similarly, other varieties sharing parentage grouped in the same subclusters.
Figure 2.
Dendrogram of 22 japonica varieties constructed on the basis of similarity coefficients in UPGMA analysis.
Eating Quality Traits
The palatability value according to the Toyo taste meter (P), the palatability score according to the sensory test (ST), and the physicochemical properties of 22 japonica rice varieties are summarized in Tables 5 and 6. All traits except SBV showed significant differences among varieties. P and ST exhibited a wide range of variation as expected. Four varieties (Koshihikari, Ilpum, Samgwang, and Geuman) showed good palatability in both P and ST. Eight varieties exhibited better ST than Chucheong, which was used as a check variety in the sensory test. AC was similar among varieties except for two, Baekjinju1 and Seonong4, which were developed as extremely low AC varieties.
Table 5. Means and Ranges of Eating Quality Parameters Detected for the Japonica Rice Varieties under Study.
| parametera | mean ± SDb | range | CV (%) | skewness | kurtosis |
|---|---|---|---|---|---|
| P | 74.46 ± 8.11 ** | 49.83−84.00 | 10.90 | −1.56 | 2.09 |
| ST | −0.22 ± 0.48 * | −1.24 to 0.39 | 46.61 | −0.87 | −0.27 |
| M | 74.44 ± 8.09 ** | 49.99−83.98 | 10.8 6 | −1.55 | 2.07 |
| AC (%) | 17.33 ± 3.10 ** | 8.75−19.93 | 17.87 | −1.80 | 2.23 |
| PC (%) | 6.83 ± 0.61 ** | 5.91−7.89 | 8.89 | 0.30 | −0.94 |
| CPVRVU | 232.62 ± 42.08 ** | 93.53−284.39 | 18.09 | −1.91 | 3.88 |
| BDVRVU | 89.19 ± 21.16 ** | 39.83−123.08 | 23.73 | −0.49 | −0.23 |
| PVRVU | 242.37 ± 28.52 ** | 181.89−298.95 | 11.77 | 0.05 | 0.27 |
| SBVRVU | −9.75 ± 31.08 ns | −94.75 to 25.00 | 36.56 | −1.39 | 1.40 |
| HPVRVU | 153.17 ± 30.99 ** | 65.20−208.80 | 20.23 | −0.72 | 1.28 |
| CTVRVU | 79.44 ± 18.03 ** | 28.33−98.92 | 22.70 | −1.37 | 1.49 |
P, palatability value; ST, palatability score from sensory test; AC, amylose content; PC, protein content; CPV, cold paste viscosity; BDV, breakdown viscosity; PV, peak viscosity; HPV, hot paste viscosity; SBV, setback viscosity; CTV, consistency viscosity; M, palatability value estimated from the equation based on marker data; RVU, Rapid Visco Unit.
ns, nonsignificant at 5% level; ** and *, significant at 1 and 5% level, respectively.
Table 6. Characteristics Associated with Eating Quality of Rice Varieties in This Study.
| cultivar | P | ST | M | AC (%) | PC (%) | PV (RVU)a | HPV (RVU) | BDV (RVU) | CPV (RVU) | SBV (RVU) | CTV (RVU) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Koshihikari | 84.00 | 0.25 | 83.98 | 18.38 | 5.91 | 285.39 | 208.80 | 76.58 | 284.39 | −1.00 | 75.59 |
| Gopum | 78.00 | 0.18 | 76.86 | 19.93 | 6.40 | 255.11 | 172.95 | 82.17 | 251.72 | −3.39 | 78.77 |
| Samgwang | 82.60 | 0.28 | 82.35 | 18.67 | 6.30 | 248.86 | 168.47 | 80.39 | 258.39 | 9.53 | 89.92 |
| Ilpum | 83.10 | 0.39 | 83.50 | 18.87 | 6.28 | 235.92 | 182.97 | 52.95 | 252.86 | 16.94 | 69.89 |
| Chucheong | 75.40 | 0.00 | 76.86 | 19.44 | 6.31 | 223.83 | 146.08 | 77.75 | 236.67 | 12.83 | 90.59 |
| Dongjin | 78.40 | −0.05 | 77.72 | 19.36 | 6.11 | 239.61 | 131.86 | 107.75 | 223.39 | −16.22 | 91.53 |
| Sinkeumo | 61.00 | −0.40 | 60.82 | 16.62 | 7.32 | 231.61 | 157.69 | 73.92 | 256.61 | 25.00 | 98.92 |
| Hwaseong | 77.10 | −0.20 | 77.72 | 18.73 | 6.19 | 245.78 | 141.31 | 104.47 | 238.72 | −7.05 | 97.41 |
| Hwacheong | 77.77 | −0.32 | 76.86 | 18.85 | 7.00 | 222.28 | 150.94 | 71.33 | 237.86 | 15.58 | 86.92 |
| Dobong | 49.83 | −1.24 | 49.99 | 15.11 | 7.89 | 298.95 | 192.47 | 106.47 | 257.14 | −41.81 | 64.67 |
| Samnam | 65.03 | −0.95 | 65.03 | 16.51 | 7.10 | 246.00 | 181.64 | 64.36 | 263.19 | 17.20 | 81.55 |
| Palkong | 68.60 | −0.83 | 68.59 | 17.33 | 6.93 | 275.2 | 181.08 | 94.11 | 276.47 | 1.28 | 95.39 |
| Hitomebore | 75.50 | 0.15 | 75.48 | 18.56 | 6.90 | 292.33 | 186.22 | 106.11 | 264.42 | −27.91 | 78.20 |
| Baekjinju1 | 76.63 | 0.10 | 76.62 | 8.75 | 7.82 | 231.28 | 110.11 | 121.17 | 151.72 | −79.56 | 41.61 |
| Seonong4 | 79.03 | 0.12 | 78.56 | 9.51 | 6.60 | 188.28 | 65.20 | 123.08 | 93.53 | −94.75 | 28.33 |
| Onnuri | 77.00 | −0.05 | 76.62 | 18.82 | 7.51 | 229.36 | 144.75 | 84.61 | 231.14 | 1.78 | 86.39 |
| Manmi | 76.60 | −0.10 | 76.40 | 13.33 | 7.88 | 242 | 136.50 | 105.50 | 204.83 | −37.17 | 68.33 |
| Giho | 75.50 | −0.40 | 75.94 | 18.64 | 6.81 | 234.28 | 144.53 | 89.75 | 237.25 | 2.97 | 92.72 |
| Geuman | 80.00 | 0.20 | 80.14 | 19.83 | 7.22 | 255.75 | 149.75 | 106.00 | 246.69 | −9.06 | 96.94 |
| Nakdong | 76.90 | −0.25 | 76.94 | 19.83 | 6.70 | 228.55 | 134.94 | 93.61 | 226.83 | −1.72 | 91.89 |
| Hexi41 | 63.77 | −1.20 | 63.76 | 17.60 | 6.02 | 181.89 | 142.05 | 39.83 | 205.56 | 23.67 | 63.51 |
| Samdeok | 76.33 | −0.54 | 76.94 | 18.56 | 7.01 | 239.83 | 139.53 | 100.30 | 218.17 | −21.67 | 78.64 |
The value is presented as a Rapid Visco Unit (RVU).
Correlation analysis among quality traits revealed that P and ST were significantly correlated (r = 0.85**) as expected (Table 7). However, P and ST were not significantly correlated with other traits, indicating that palatability is a complex trait in which a number of factors are involved. AC was significantly correlated with RVA pasting properties.
Table 7. Correlation Matrix of Eating Quality Parametersa.
| parameter | AC | P | ST | PC | CPV | BDV | PV | HPV | SBV | CTV |
|---|---|---|---|---|---|---|---|---|---|---|
| P | 0.18 ns | |||||||||
| ST | 0.06 ns | 0.85 ** | ||||||||
| PC | −0.48 * | −0.43 * | −0.24 ns | |||||||
| CPV | 0.74 ** | −0.14 ns | −0.12 ns | −0.17 ns | ||||||
| BDV | −0.46 * | 0.12 ns | 0.23 ns | 0.40 ns | −0.44 * | |||||
| PV | 0.22 ns | −0.14 ns | 0.02 ns | 0.18 ns | 0.67 ** | 0.25 ns | ||||
| HPV | 0.52 * | −0.21 ns | −0.14 ns | −0.11 ns | 0.92 ** | −0.45 * | 0.75 ** | |||
| SBV | 0.79 ** | −0.07 ns | −0.18 ns | −0.39 ns | 0.74 ** | −0.83 ** | −0.01 ns | 0.56 ** | ||
| CTV | 0.82 ** | 0.02 ns | −0.04 ns | −0.20 ns | 0.75 ** | 0.25 ns | 0.28 ns | 0.43 * | 0.75 ** | |
| M | 0.19 ns | 1.00 ** | 0.85 ** | −0.43 * | −0.14 ns | 0.11 ns | −0.14 ns | −0.21 ns | −0.06 ns | 0.03 ns |
ns, nonsignificant at 5% level; * and **, significant at 5 and 1% level, respectively.
Regression Analysis between Marker Genotypes and Eating Quality Traits
Association analyses between marker genotypes based on 30 primer sets (Table 4) and palatability values according to the Toyo taste meter (P) (Table 6) and sensory test (ST) were performed (Table 8). General linear model regression analysis indicated that individual molecular markers could not differentiate the eating qualities. Thus, multiple regression analysis of the entire set of markers was performed. By using only significant markers as independent variables, model equations were generated that could predict the eating quality with highly significant resolution (R2 = 0.99) (Table 8). This indicates that palatability values of 22 varieties estimated by the Toyo taste meter and/or by the sensory test can be estimated with high resolution by these regression equations based on the marker genotypes. The marker sets for the equations consisted of 13 markers for palatability by the Toyo taste meter and 14 markers for palatability by the sensory test, with 9 markers shared for both equations. Three newly developed markers (GPA, S3cI, and CBG) were included among these 9 markers. The other 3 markers (AMs, SSIIa, and AcPh) were included among either 13 markers for palatability by the Toyo taste meter or 14 markers for palatability by the sensory test.
Table 8. Model Equations for Evaluating Rice Eating Quality Containing the Significant Coefficient of Each Marker t Value Aided by Multiple Regression Analysisa.
| palatability by Toyo taste meter (P) |
palatability by sensory test (ST) |
|||||
|---|---|---|---|---|---|---|
| PCR primer | parameter estimate | t value | R2 | parameter estimate | t value | R2 |
| G4 | −16.97 ± 1.19 | −14.22 ** | 0.087 | −1.20 ± 0.06 | −21.77 ** | 0.212 |
| M11 | −1.94 ± 0.60 | −3.25 ** | 0.096 | −0.14 ± 0.03 | −5.03 ** | 0.010 |
| E30 | 26.55 ± 0.83 | 32.12 ** | 0.104 | 0.86 ± 0.04 | 19.62 ** | 0.059 |
| M2CG | −2.40 ± 0.56 | −4.33 ** | 0.060 | −0.38 ± 0.03 | −15.21 ** | 0.041 |
| GPA | −21.14 ± 1.11 | −19.12 ** | 0.129 | −0.82 ± 0.05 | −17.28 ** | 0.021 |
| S3cI | −1.62 ± 0.62 | −2.60 * | 0.017 | −0.38 ± 0.03 | −13.41 ** | 0.005 |
| P5 | 19.01 ± 1.32 | 14.44 ** | 0.307 | 1.09 ± 0.04 | 26.81 ** | 0.288 |
| B1 | 6.42 ± 0.77 | 8.30 ** | 0.047 | 0.41 ± 0.03 | 12.90 ** | 0.015 |
| CBG | 13.45 ± 1.11 | 12.12 ** | 0.087 | 0.68 ± 0.07 | 10.27 ** | 0.228 |
| J6 | 3.87 ± 0.74 | 5.21 ** | 0.083 | |||
| WK9 | 2.62 ± 0.59 | 4.42 ** | 0.003 | |||
| A7 | −12.33 ± 1.27 | −9.72 ** | 0.031 | |||
| AMs | −8.72 ± 1.56 | −5.58 ** | 0.021 | |||
| G81 | 0.27 ± 0.03 | 10.41 ** | 0.021 | |||
| F6 | 0.32 ± 0.03 | 10.36 ** | 0.033 | |||
| SSIIa | −0.27 ± 0.03 | −8.06 ** | 0.001 | |||
| G28 | 0.33 ± 0.04 | 8.81 ** | 0.005 | |||
| AcPh | −0.48 ± 0.04 | −11.44 ** | 0.060 | |||
| intercept | 76.66 + 2.71 | 28.29 ** | −0.54 ± 0.11 | −4.74 ** | ||
| total | 0.990 | 0.990 | ||||
| eq | Y = 76.66 − 16.97(G4) − 1.94(M11) + 26.55(E30) − 2.40(M2CG) − 21.14(GPA) − 1.62(S3cI) + 19.01(P5) + 6.42(B1) + 13.45(CBG) + 3.87(J6) + 2.62(WK9) −12.33(A7) − 8.72(Ams) | Y = − 0.54 − 1.20(G4) − 0.14(M11) + 0.86(E30) − 0.38(M2CG) − 0.82(GPA) − 0.38(S3cI) + 1.09(P5) + 0.41(B1) + 0.68(CBG) + 0.27(G81) + 0.32(F6) − 0.27(SSIIa) + 0.33(G28) − 0.48(AcPh) | ||||
** and *, significant at 1% and 5% level, respectively.
Validation of the Regression Equation Using Breeding Lines
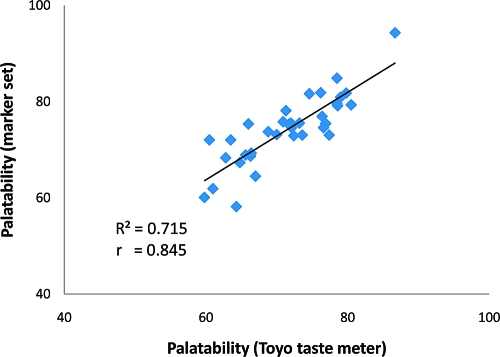
To evaluate the validity of the regression equation, 32 breeding lines along with 3 japonica check varieties were genotyped with the set of 13 markers mentioned above and their palatability values were evaluated by the Toyo taste meter (Table 9). Palatability estimates were calculated by the regression equation based on marker genotypes. There was a highly significant correlation (R2 = 0.715**) between palatability estimates by the regression equation and palatability values according to the Toyo taste meter (Figure 3), indicating that the model equation we developed reliably predicts the palatability of random breeding lines of japonica rice.
Table 9. Palatability Values of 32 Breeding Lines and 3 Check Varieties Measured by the Toyo Taste Meter and Estimated by the Regression Equation.
| palatability |
||
|---|---|---|
| breeding line/variety | Toyo taste meter (P) | regression eq (M) |
| Suweon503 | 72.40 | 72.93 |
| Suweon507 | 80.51 | 79.34 |
| Suweon508 | 76.40 | 76.94 |
| Suweon509 | 73.21 | 75.55 |
| Suweon510 | 72.02 | 75.55 |
| Suweon511 | 78.51 | 79.56 |
| Suweon513 | 66.32 | 68.69 |
| Suweon514 | 68.79 | 73.76 |
| Suweon515 | 66.02 | 75.38 |
| Suweon516 | 79.81 | 81.79 |
| Suweon518 | 64.80 | 67.35 |
| Nampyeonga | 76.18 | 81.87 |
| Iksan486 | 86.71 | 94.32 |
| Iksan488 | 76.89 | 75.47 |
| Iksan490 | 78.51 | 84.89 |
| Iksan493 | 76.60 | 74.61 |
| Iksan494 | 73.62 | 73.07 |
| Iksan495 | 79.02 | 80.92 |
| Iksan496 | 71.67 | 75.32 |
| Iksan497 | 72.11 | 74.61 |
| Iksan502 | 71.32 | 78.13 |
| Iksan504 | 65.58 | 68.91 |
| Keumoa | 63.53 | 72.06 |
| Juana | 77.41 | 73.07 |
| Milyang211 | 70.90 | 75.83 |
| Milyang215 | 59.81 | 60.10 |
| Milyang218 | 70.02 | 73.15 |
| Milyang219 | 74.58 | 81.65 |
| Milyang220 | 64.31 | 58.20 |
| Milyang222 | 60.52 | 72.06 |
| Milyang223 | 62.80 | 68.34 |
| Milyang224 | 61.00 | 61.93 |
| Milyang230 | 78.60 | 79.16 |
| Milyang231 | 66.41 | 69.31 |
| Milyang232 | 67.03 | 64.52 |
Three check varieties.
Figure 3.
Correlation between palatability values of 32 breeding lines and 3 check varieties measured by the Toyo taste meter and estimated by the regression equation.
Discussion
Evaluation of Rice Eating Quality Traits
The highly significant correlation between palatability values according to the Toyo taste meter and palatability values according to the sensory test is in good agreement with previous studies (9). Japanese researchers have also reported that palatability values from the Toyo taste meter and palatability scores from the sensory test exhibit a high positive correlation (7, 8). This indicates that the palatability value according to the Toyo taste meter can be used as a good measure of the eating quality of rice. For breeding better eating quality rice, the Toyo taste meter is the measurement method of choice because of its relative simplicity and reproducibility compared to the sensory test (7−9). This is why the Toyo taste meter has been widely used in breeding programs for eating quality in Korea, Japan, and northeastern Asia. No correlation between palatability and any pasting property was found in this study, suggesting that pasting properties could not be a good indicator for palatability value due to its complex nature. PC was the only trait to affect the palatability value as reported previously (29).
Feasibility of PCR Marker-Based Evaluation of Eating Quality of Cooked Rice
As nucleotide differences among genotypes are a major source of heritable variation, molecular markers derived from them should provide an effective measure of genotypic variation and hence phenotypic differences among varieties. When 22 varieties were classified on the basis of the cluster analysis of 30 markers, they formed three major clusters at a genetic similarity level of 0.64−0.67. However, the clusters did not appear to be related to eating quality, but to the parental origin of the strains, although the 30 markers were derived from quality-associated traits or QTLs for eating quality. This may be explained by the fact that the cluster diagram was constructed on the basis of genetic similarities simply calculated on the basis of the assumption that each of the markers had the same effect on the genotype determination. This demonstrates that simple genotyping and similarity analysis might not be a good measure to evaluate the eating quality of rice, and thus there should be differential weighting of markers. Therefore, multiple regression analysis was performed. Multiple regression equations developed in this study (Table 8) proved to be effective in predicting the palatability of rice varieties and breeding lines (Figure 3). Ohtsubo et al. (13, 17) and Ohtsubo and Nakamura (12) were able to estimate rice palatability (sensory test of Japanese people) by using a combination of STS markers and estimation equations with significant multiple regression coefficients. Similarly, other marker sets derived from starch synthesizing genes (GBSS, SS, SBE, and SDBE) (2, 3) and storage protein genes (glutelin and prolamin) were also tested to evaluate AC, PC, and the adhesiveness of cooked rice (30).
Of the significant markers in the equations (Table 8), nine were important for palatability as measured by both the Toyo taste meter (P) and the sensory test (ST), and the R2 values of the nine markers explained most of the variation in both equations. This indicates that palatability by the Toyo taste meter and palatability by the sensory test are in good accordance as revealed by the significant correlation (r = 0.85 **) (Table 7). Of the nine significant markers, the STS marker, P5, showed the highest partial R2 in both equations, implying that it might represent a major QTL for palatability.
Because we used most of the reported markers developed for taste evaluation in japonica rice, the regression equations (Table 8) are of sufficiently high resolution to estimate the palatability of japonica rice in Korea. However, because palatability varies among countries and varietal groups even within japonica rice, the equations may need to be modified to be applicable in other countries. Particularly in countries where indica rice is preferred, the markers should be re-evaluated. We expect that the set of 13 markers used in this study will be useful for the selection of early breeding or even individual F2 plants to accelerate breeding for improvement of rice eating quality in Korea or other japonica rice-consuming countries.
This study was supported by a grant from the Technology Development Program for Agriculture and Forestry, Ministry of Agriculture, Forestry, and Fisheries, Republic of Korea.
References
- Juliano B. O.Criteria and tests for rice grain qualities. In Chemistry and Technology of Rice, 2nd ed.; Juliano B. O., Ed.; American Association of Cereal Chemists: St. Paul, MN, 1985; pp443−513. [Google Scholar]
- Bao J. S.; Corke H.; Sun M. Microsatellites, single nucleotide polymorphisms and a sequence tagged site in starch-synthesizing genes in relation to starch physicochemical properties in non-waxy rice (Oryza sativa L.). Theor. Appl. Genet. 2006, 113, 1185–1196. [DOI] [PubMed] [Google Scholar]
- Bao J. S.; Corke H.; Sun M. Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 2006, 113, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Ramesh M.; Bhattacharya K. R.; Mitchell J. R. Developments in understanding the basis of cooked-rice texture. Crit. Rev. Food Sci. Nutr. 2000, 40, 449–460. [DOI] [PubMed] [Google Scholar]
- Izumi O. E.; Yuji M.; Kuniyuki S.; Toshiro K. Effects of rising temperature on grain quality, palatability and physicochemical properties of rice. Sci. Rep. Fac. Agric., Okayama Univ. 2007, 96, 13–18. [Google Scholar]
- Hong M. C.; Kuo B. J. Studies on investigating the eating quality of cooked rice using small sample size (II). Taichung District Agric., Res. Rep. 2001, 70, 9–19. [Google Scholar]
- Azuma S.; Sasaki Y.; Ishizaki K.; Kondou T.; Hoshi T. Improvement of grain and eating quality in rice breeding of Niigata Prefecture: VII. Comparison of some measuring methods for effective selection about eating quality. Hokuriku Crop Sci. 1994, 29, 35–36(in Japanese). [Google Scholar]
- Tanaka R.; Ino K.; Kanagawa M. Cultivation method and eating quality of paddy rice 1. Evaluation with mechanical taster of boiled rice so called “MIDO meter”. Tohuku J. Crop Sci. 1992, 35, 45–46(in Japanese). [Google Scholar]
- Takeuchi Y.; Nonoue Y.; Ebitani T.; Suzuki K.; Aoki N.; Sato H.; Ideta O.; Hirabayashi H.; Hirayama M.; Ohta H.; Nemoto H.; Kato H.; Ando I.; Ohtsubo K.; Yano Y.; Imbe T. QTL detection for eating quality including glossiness, stickiness, taste and hardness of cooked rice. Breed. Sci. 2007, 57, 231–242. [Google Scholar]
- Wada T.; Uchimura Y.; Ogata T.; Tsubone M.; Matsue Y. Mapping of QTLs for physicochemical properties in japonica rice. Breed. Sci. 2006, 56, 253–260. [Google Scholar]
- Wada T.; Ogata T.; Tsubone M.; Matsue Y.. Identification of QTLs for eating quality of japonica rice “Koshihikari”. The 6th Asian Crop Association Conference and The 2nd International Conference on Rice for the Future, Bangkok, Thailand, Nov 5−9, 2007.
- Ohtsubo K.; Nakamura S. Variety identification of rice (Oryza sativa L.) by Polymerase Chain Reaction method and its application to processed rice products. J. Agric. Food Chem. 2007, 55, 1501–1509. [DOI] [PubMed] [Google Scholar]
- Ohstubo K.; Nakamura S.; Okadome H. Investigation on estimation of rice palatability by DNA analysis (studies on estimation of rice palatability by DNA analysis part I). Nippon Nogeikagaku Kaishi 2003, 50, 122–132. [Google Scholar]
- Larkin P. D.; McClung A. M.; Ayres N. M.; Park W. D. The effect of the waxy locus (granule bound starch synthase) on pasting curve characteristics in specially rice (Oryza sativa L.). Euphytica 2003, 131, 243–253. [Google Scholar]
- Han Y. P.; Xu M. L.; Liu X. Y.; Yan C. J.; Korban S. S.; Chen X. L.; Gu M. H. Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci. 2004, 166, 357–364. [Google Scholar]
- Murray M. G.; Thompson W. F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohstubo K.; Nakamura S.; Imamura T. Development of the primer sets for identification of a rice variety, Koshihikari, by PCR. Nippon Nogeikagaku Kaishi 2002, 76, 388–397. [Google Scholar]
- Kwon S. W.; Cho Y. C.; Lee J. H.; Suh J. P.; Choi I. S.; Oh M. K.; Kim Y. G.; Kim M. K.; Koh H. J.; Yang S. J. Linkage map construction and QTL analysis in the cross between two japonica cultivars. Korean J. Breed. Sci 2007, 39, 94. [Google Scholar]
- Suh J. P.; Choi Y. H.; Kim K. J.; Cho Y. C.; Kwon S. J.; Jeong Y. P.; Jeung J. U.; Choi I. S.; Kim Y. G.; Choi H. C.; Hwang H. G. Genetic diversity and QTLs for grain quality in japonica rice. Korean J. Breed. Sci. 2004, 36, 31–37. [Google Scholar]
- Rozen S.; Skaletsky H. J. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Sambrook J.; Russell D. W.. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2001. [Google Scholar]
- Thompson J. D.; Higgins D. G.; Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. M.; Neff J. D.; Chory J.; Pepper A. E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphism: experimental application in Arabidopsis thaliana genetics. Plant J. 1998, 14, 387–392. [DOI] [PubMed] [Google Scholar]
- AOAC.AOAC Official Methods of Analysis, 16th ed.; AOAC: Gaithersburg, MD, 1995; 30 pp. [Google Scholar]
- Perez C. M.; Juliano B. O. Modification of the simplified amylose test for milled rice. Starch/Staerke 1978, 30, 424–426. [Google Scholar]
- Bao J. S.; Xia Y. W. Genetic control of the paste viscosity characteristics in indica rice (Oryza sativa L.). Theor. Appl. Genet. 1999, 98, 1120–1124. [Google Scholar]
- Statistical Analysis Systems (SAS).SAS User’s Guide, release 8.2; Statistical Analysis Systems Institute: Cary, NC, 2001. [Google Scholar]
- Rohlf F. J.NTSYS-PC Numerical Taxonomy and Multivariate Analysis System, bersion 1.8; Exeter Publishers; : Setauket, NY, 1993. [Google Scholar]
- Yu T. Q.; Jiang W.; Ham T. H.; Chu S. H.; Lestari P.; Lee J. H.; Kim M. K.; Xu F. R.; Han L.; Dai L.; Koh H. J. Comparison of grain quality traits between japonica rice cultivars from Korea and Yunnan province of China. J. Crop Sci. Biotechnol. 2008, 11, 135–140. [Google Scholar]
- Nakamura S.; Okadome H.; Yoza K.; Haraguchi K.; Okunishi T.; Suzuki K.; Satoh H.; Ohtsubo K. Differentiation and search for palatability-factors of world-wide rice grains by PCR method. Nippon Nogeikagaku Kaishi 2004, 78, 764–779. [Google Scholar]