Abstract

Positioned at the C-terminus of many eukaryotic proteins, the glycosylphosphatidylinositol (GPI) anchor is a posttranslational modification that anchors the modified protein in the outer leaflet of the cell membrane. The GPI anchor is a complex structure comprising a phosphoethanolamine linker, glycan core, and phospholipid tail. GPI-anchored proteins are structurally and functionally diverse and play vital roles in numerous biological processes. While several GPI-anchored proteins have been characterized, the biological functions of the GPI anchor have yet to be elucidated at a molecular level. This review discusses the structural diversity of the GPI anchor and its putative cellular functions, including involvement in lipid raft partitioning, signal transduction, targeting to the apical membrane, and prion disease pathogenesis. We specifically highlight studies in which chemically synthesized GPI anchors and analogues have been employed to study the roles of this unique posttranslational modification.
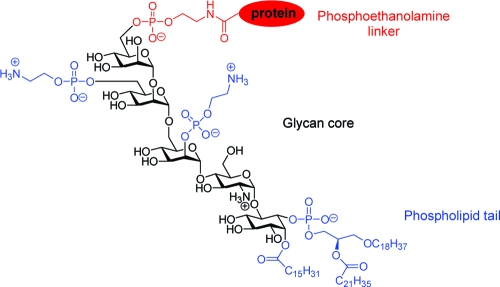
First characterized approximately 20 years ago, the glycosylphosphatidylinositol (GPI1) anchor is a glycolipid structure that is added posttranslationally to the C-terminus of many eukaryotic proteins (1–6). This modification anchors the attached protein in the outer leaflet of the cell membrane (3,7,8). Proteins containing a GPI anchor are functionally diverse and play important roles in signal transduction, prion disease pathogenesis, immune response, and the pathobiology of trypanosomal parasites (1,9). Unlike simple lipid modifications, the GPI anchor has a complex structure that includes a phosphoethanolamine linker, glycan core, and phospholipid tail (Figure 1) (1,2). The phosphoinositol, glucosamine, and mannose residues within the glycan core can be variously modified with phosphoethanolamine groups and other sugars (1,2). Such structural complexity would be expected to encode diverse functional capacity beyond membrane insertion. However, definitive conclusions that relate GPI anchor structure and function have been difficult to draw. While many GPI-anchored proteins have been identified and characterized, the only confirmed biological function of the GPI anchor is to provide the protein with a stable membrane anchoring device (2,10,11). Several excellent reviews have discussed the functional roles of the GPI anchor in protozoan parasites and the biosynthesis of the GPI anchor and its transfer to proteins (3,7,8,12–15). In this review, we will focus on the structure of the GPI anchor and its biological functions in mammalian cells. The putative roles of the GPI anchor in lipid raft partitioning, signal transduction, cellular communication, apical membrane targeting, and prion disease pathogenesis will be discussed. Particular attention will be given to recent studies that attempt to more thoroughly define the functional significance of the GPI anchor using chemically synthesized GPI anchors and GPI anchor analogues.
Figure 1.
Structure of the GPI anchor from human erythrocyte acetylcholinesterase (16). The three domains of the GPI anchor are (i) a phosphoethanolamine linker (red), (ii) the conserved glycan core (black), and (iii) a phospholipid tail (blue). Appendages in blue (including the lipids of the lipid tail) are variable.
Discovery of the GPI Anchor
In 1976, a novel phospholipase that acts upon phosphatidylinositol was purified from Bacillus cereus. This phospholipase, termed phosphatidylinositol phospholipase C (PI-PLC), was found to release alkaline phosphatase (APase) from tissues. Over the next several years, PI-PLCs purified from other types of bacteria (such as Staphylococcus aureus and Clostridium novyi) also were found to contain similar enzymatic activity. Additionally, various other proteins, such as 5′-nucleotidase and erythrocyte acetylcholinesterase (AChE), were released from tissues when treated with PI-PLC. Based on this evidence, these proteins were suggested to be covalently attached to the cell membrane via a site on the protein and a phosphatidylinositol molecule embedded in the lipid bilayer (reviewed in ref (17)).
By 1985, the structural components of the C-termini of two cell surface proteins, variant surface glycoprotein (VSG), found on the parasitic protozoan Trypanosoma brucei, and Thy-1, a glycoprotein expressed on mammalian thymocytes and the brain, had been identified. The C-terminus of VSG contained (i) an ethanolamine amide-linked to the C-terminal amino acid, (ii) a polysaccharide consisting of mannose, glucosamine, and variable amounts of galactose, and (iii) a phospholipid that could be degraded by bacterial PI-PLC to give 1,2-dimyristylglycerol. The C-terminal cysteine of Thy-1 was attached to a structure that contained ethanolamine, glucosamine, galactosamine, mannose, myo-inositol, phosphate, glycerol, and stearic acid. This structural information was combined with data from PI-PLC studies to establish a general structure for the GPI membrane anchor (reviewed in refs (3) and (17)).
Structure of the GPI Anchor
Although the general components of the GPI anchor had been identified, the first detailed structural analysis of a GPI anchor was not completed until 1988. Ferguson and co-workers determined the exact structure of the VSG anchor from T. brucei through a combination of NMR spectroscopy, mass spectrometry, chemical modification, and exoglycosidase digestions (4). As a result of this and other investigations, a general pattern for the GPI anchor structure has emerged (1,2).
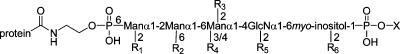
The C-terminus of a GPI-anchored protein is linked through a phosphoethanolamine bridge to the highly conserved core glycan, mannose(α1−2)mannose(α1−6)mannose(α1−4)glucosamine(α1−6)myo-inositol (Figure 1). A phospholipid tail attaches the GPI anchor to the cell membrane. The glycan core can be variously modified with side chains, such as a phosphoethanolamine group, mannose, galactose, sialic acid, or other sugars (blue, Figure 1) (2). Table 1 lists a number of GPI-anchored proteins and the structures of their various side chains and lipids (4,6,13,16,18–24). The phosphoethanolamine side chain, attached to either the second or third mannose of the glycan core, is only found in higher eukaryotes, not in protozoa. The most common side chain attached to the first mannose residue is another mannose. Complex side chains, such as the N-acetylgalactosamine-containing polysaccharides attached to the third mannose of the glycan core, are found in both mammalian and protozoan anchor structures. The core glucosamine is rarely modified, except in the GPI anchor of NETNES, a glycoprotein of unknown function from T. cruzi(23). Depending on the protein and species of origin, the lipid anchor of the phosphoinositol ring is a diacylglycerol, an alkylacylglycerol, or a ceramide (24). The lipid species vary in length, ranging from 14 to 28 carbons, and can be either saturated or unsaturated. Many GPI anchors also contain an additional fatty acid, such as palmitic acid, on the 2-hydroxyl of the inositol ring. This extra fatty acid renders the GPI anchor resistant to cleavage by PI-PLC (24).
Table 1. Representative Structures of Known GPI Anchors (4,6,13,16,18–24)a.
| protein | R1 | R2 | R3 | R4 | R5 | R6 | X |
|---|---|---|---|---|---|---|---|
| rat brain Thy-1 | ±Manα1−2 | OH | PEtN | ±GalNAcβ1−4 | OH | OH | alkylacyl-glycerol |
| human erythrocyte AChE | OH | ±PEtN | PEtN | OH | OH | palmitate | alkylacyl-glycerol |
| hamster brain scrapie prion protein | ±Manα1−2 | OH | PEtN | (±NANA)- (±Gal)-GalNAcβ1−4 | OH | OH | nd |
| human urine CD59 | ±Manα1−2 | OH | PEtN | ±GalNAcβ1−4 | OH | palmitate | nd |
| mouse skeletal muscle NCAM | ±Manα1−2 | nd | PEtN | ±GalNAcβ1−4 | OH | OH | nd |
| bovine liver 5′-nucleotidase | ±Manα1−2 | ±PEtN | PEtN | ±HexNAc | OH | OH | nd |
| human placental APase | OH | ±PEtN | PEtN | OH | OH | OH | alkylacyl-glycerol |
| human CD52 | ±Manα1−2 | ±PEtN | PEtN | OH | OH | palmitate | diacyl-glycerol |
| pig kidney membrane dipeptidase | OH | ±PEtN | PEtN | (±Galβ1−3)GalNAcβ1−4 or (±NANA) - GalNAcβ1−4 | OH | OH | diacyl-glycerol |
| human kidney membrane dipeptidase | ±Manα1−2 | nd | PEtN | (±Galβ1−3)GalNAcβ1−4 | OH | OH | nd |
| T. brucei VSG | OH | OH | OH | ±Galα1−2(Galα1−2Galα1−6)Galα1−3 | OH | OH | dimyristyl-glycerol |
| T. cruzi 1G7 | ±Manα1−2 | OH | OH | OH | OH | OH | alkylacyl-glycerol |
| T. cruzi NETNES | ±Manα1−2 | OH | OH | OH | PEtN | OH | alkylacyl-glycerol |
| L. major gp63 | OH | OH | OH | OH | OH | OH | alkylacyl-glycerol |
| S. cerevisiae gp125 | ±Manα1−2Manα1−2 or Manα1−3Manα1−2 | OH | OH | OH | OH | OH | diacyl-glycerol |
| A. fumigatus PhoAp | ±Manα1−3Manα1−2 | OH | OH | OH | OH | OH | ceramide |
| P. communis arabinogalactan proteins | OH | OH | OH | ±GalNAcβ1−4 | OH | OH | ceramide |
| D. Discoideum PsA | ±Manα1−2 | nd | nd | OH | OH | OH | ceramide |
Various side chain modifications of carbohydrates, phosphoethanolamine, and/or palmitate (R1−R6) are indicated. In some proteins, certain side chains may only be present in a proportion of GPI anchors (indicated by ±). OH indicates that no side chain is known to be present; nd indicates that the side chain or lipid moiety has not been determined. X is the lipid moiety, Man is mannose, Gal is galactose, GalNAc is N-acetylgalactosamine, NANA is sialic acid, HexNAc is N-acetylhexosamine, and PEtN is phosphoethanolamine.
Functions of GPI-Anchored Proteins
The GPI anchor is broadly distributed among eukaryotic organisms, including protozoa, fungi, plants, insects, and mammals (1). Among vertebrates, GPI-anchored proteins have been identified throughout every major cell type and tissue. GPI-anchored proteins vary widely in size, ranging from the 12 amino acid glycopeptide CD52 to the 175 kDa protein CDw109 (1). To date, more than 250 proteins have been found to contain a GPI anchor (3,8). Importantly, GPI anchors are essential for viability. Defects in GPI anchor biosynthesis are embryonic lethal in mammals and conditionally lethal in yeast (25,26).
The connection between GPI anchor structural diversity and function is poorly understood. GPI-anchored proteins display diverse biological functions, some of which are listed in Table 2. Many of these proteins have enzymatic activity, such as APase, which catalyzes the removal of phosphate groups from biomolecules (2). Certain GPI-anchored proteins are involved in cell-cell contact and adhesion, such as an isoform of the neural cell adhesion molecule (NCAM). The GPI-anchored proteins CD55 (decay-accelerating factor or DAF) and CD59 are important in the regulation of the complement cascade, which protects an organism from foreign invaders and pathogens (2). VSG, a GPI-anchored protein from T. brucei, forms a protective coat around the trypanosome parasite (3). Although many GPI-anchored proteins have been characterized, some GPI-anchored proteins, like the prion protein, do not yet have an assigned function (27).
Table 2. Representative Functions of GPI-Anchored Proteins (2,3,17,27).
| biological role | protein | source |
|---|---|---|
| enzymes | alkaline phosphatase | mammalian tissues, Schistosoma |
| 5′-nucleotidase | mammalian tissues | |
| acetylcholinesterase | Torpedo electric organ, insect brain, mammalian blood cells | |
| dipeptidase | pig and human kidney, sheep lung | |
| cell−cell interaction | LFA-3 | human blood cells |
| NCAM | mammalian and chicken brain and muscle | |
| PH-20 | guinea pig sperm | |
| complement regulation | CD55 (DAF) | human blood cells |
| CD59 | human blood cells | |
| mammalian antigens | Thy-1 | mammalian brain and lymphocytes |
| Qa-2 | mouse lymphocytes | |
| CD14 | human monocytes | |
| carcinoembryonic antigen (CEA) | human tumor cells | |
| CD52 | human lymphocytes | |
| protozoan antigens | VSG | T. brucei |
| 1G7 | T. cruzi | |
| procyclin | T. brucei | |
| miscellaneous | scrapie prion protein | hamster brain |
| CD16b | human neutrophils | |
| folate-binding protein | human epithelial cells |
Unique Properties of GPI-Anchored Proteins Mediated by the GPI Anchor
GPI-Anchored Proteins May Associate with Lipid Raft Domains GPI-anchored proteins are believed to associate with lipid rafts, membrane microdomains enriched in glycosphingolipids, cholesterol, and certain types of lipidated proteins (Figure 2) (28,29). Lipid rafts organize the plasma membrane into a series of discrete smaller domains that can serve as platforms for a variety of cellular functions, such as vesicular trafficking and signal transduction (28,29). Lipid rafts are hypothesized to form by the self-association of sphingolipids, favored by their long and mostly saturated hydrocarbons that allow them to pack tightly in a bilayer. Cholesterol molecules are believed to fill the voids between the associating sphingolipids (28). The presence of cholesterol may be necessary for the function and formation of lipid rafts, as depletion of cellular cholesterol has been shown to disrupt these rafts (28). Due to the tight packing of sphingolipids, lipid rafts are believed to be less fluid than the surrounding phospholipid bilayer (28). The highly ordered environment of the lipid rafts may also allow for the close packing of GPI-anchored proteins. These lipid rafts were first characterized by their insolubility at 4 °C in the nonionic detergent Triton X-100, which has become the most widely used assay for raft existence (28,29). GPI-anchored proteins also are detergent insoluble under these conditions, presumably due to their association with lipid rafts (28). Common signaling proteins are also found in these complexes, which has led to the hypothesis that the GPI anchor may be important in signal transduction (30).
Figure 2.
Membrane-associated proteins in a lipid bilayer containing lipid raft domains. GPI-anchored proteins and other lipidated proteins are believed to associate with lipid rafts.
Cellular lipid rafts have been difficult to characterize due to their proposed small size and dynamic nature (29,31,32). Common assays used to probe for the presence of rafts include cholesterol depletion and detergent extraction, but these assays are indirect and plagued by artifacts (29,31). Methods used to determine the size of lipid rafts have given conflicting results, and both fluorescence and electron microscopy have consistently failed to prove the existence of lipid rafts enriched in GPI-anchored proteins in living cells (29,32). Using imaging fluorescence resonance energy transfer (FRET) microscopy, Kenworthy and Edidin visualized antibody-labeled 5′-nucleotidase in MDCK cells (33). Their data were in agreement with a model that suggested that most 5′-nucleotidase molecules were randomly distributed on the plasma membrane of these cells. Glebov and Nichols found that the FRET signal from GPI-anchored fluorescent proteins in COS-7 and Jurkat cells was similar to the signal measured for nonraft proteins (34). Cholesterol depletion using β-methyl cyclodextrin also did not affect the FRET signal, suggesting that the GPI-anchored fluorescent proteins were not clustered in cholesterol-dependent lipid rafts (34). However, these conclusions can be questioned if the rafts are very small (5 nm or less) or if the GPI-anchored proteins are not present at high enough concentrations in the plasma membrane (32,34).
Other studies, using specialized microscopy and additional techniques, have given support for the existence of GPI-anchored proteins in lipid rafts. Using depolarization FRET microscopy, Varma and Mayor demonstrated that GPI-anchored proteins were organized in cholesterol-dependent microdomains with diameters less than 70 nm in living cells (35). Friedrichson and Kurzchalia also investigated the existence of GPI-anchored proteins in lipid rafts by chemically cross-linking GPI-anchored growth hormone (GH-GPI) with short (1.1 nm) cross-linkers and analyzing the cross-linking efficiency (36). The extent of cross-linking was found to be independent of the amount of GH-GPI expressed by the cells, suggesting that GPI-anchored proteins clustered in lipid rafts. Recently, Sharma and co-workers employed a technique known as homo-FRET to look at GPI-anchored fluorescent proteins and determined that a small fraction of GPI-anchored proteins were organized into nanometer size (∼4−5 nm) raft domains (32). The authors concluded that 20−40% of GPI-anchored proteins were present in rafts and that each cluster consisted of four or fewer GPI-anchored proteins (32). Although the existence of lipid rafts and the enrichment of GPI-anchored proteins in these domains is a highly controversial subject (29), a variety of new tools and techniques have recently been developed that can be used to further investigate the association of GPI-anchored proteins with lipid rafts.
GPI-Anchored Proteins Can Be Exogenously Incorporated onto Cell Surfaces.
Since the lipid tail of the GPI anchor does not completely extend through the lipid bilayer, GPI-anchored proteins are associated more loosely with the plasma membrane than transmembrane proteins. In fact, many GPI-anchored proteins can transfer spontaneously to cell membranes both in vitro and in vivo, a process that has been termed “cell surface painting” (2,37). Before the structure of the GPI anchor was known, purified human DAF was shown to insert onto sheep erythrocytes (37). Exogenously added DAF was freely mobile on the sheep cell surface and was able to function normally as shown by its inhibition of convertase complexes. Since then, numerous GPI-anchored proteins have been incorporated onto a variety of different cell types (2,37). Generally, these exogenously added GPI-anchored proteins retained the same characteristics and functions as endogenously expressed GPI-anchored proteins (2,37). While the mechanism by which this transfer process occurs is unknown, the lipid moieties of the GPI anchor must be intact for cell membrane insertion (37).
Intermembrane transfer of GPI-anchored proteins also can occur in vivo. Kooyman et al. engineered transgenic mice to express the human GPI-anchored proteins DAF and CD59 solely on the surface of their red blood cells (38). Immunohistology studies on the tissues from these mice detected both proteins on vascular endothelial cells from several organs, in addition to erythrocytes. Erythrocyte studies on human patients with African trypanosomiasis found that their cells contained membrane-bound VSG trypanosomal coat proteins (37). The results from these and other studies indicate that GPI-anchored proteins can spontaneously transfer from one cell to another in vivo.
The ability of GPI-anchored proteins to be inserted into cell membranes has been exploited to modulate host immune responses. Huang and colleagues generated the purified GPI-anchored MHC class I molecule HLA-A2.1 complexed to an antigenic peptide from hepatitis B virus (39). This GPI-anchored protein was transferred to MHC-class-I-negative cells, which were then able to activate specific T-cells. In another study, McHugh and co-workers immunized mice with EG7 tumors expressing GPI-anchored B7-1 via cell surface painting, which induced tumor-specific T-cell proliferation and cytolytic T lymphocytes (40). These mice were protected when challenged with live wild-type tumor cells. These studies demonstrate that exogenously added GPI-anchored proteins are functional in vivo and can potentially be used as therapeutic agents.
Significance and Functions of the GPI Anchor
Despite continued attempts to characterize the functions of GPI-anchored proteins, the significance of the GPI anchor structure has yet to be deduced (1,10,11). The GPI anchor could have a genuine functional role in some or all anchored proteins, or it could merely be a vestigial relic. The only confirmed role of the GPI anchor is to provide the attached protein with a stable membrane anchoring device that is resistant to most extracellular proteases and lipases (10,11). Given that there are many ways in which a protein can be attached to the cell membrane, the GPI anchor is a fairly complicated structure when compared to a simple lipid or transmembrane domain. It is possible that the GPI anchor serves other biological functions besides a membrane anchor.
The GPI Anchor May Affect the Structure of Its Associated Protein.
The GPI anchor may influence the conformation and structure of the protein to which it is attached. For example, an antibody that binds the GPI-anchored protein procyclin from T. brucei shows greatly reduced affinity toward the same protein lacking the lipid tail (41). The OX7 antibody that recognizes the GPI-anchored Thy-1 protein also fails to bind Thy-1 after treatment with PI-PLC (42). In addition, the circular dichroism spectra of GPI-anchored human Thy-1 differ from that of soluble human Thy-1 (42). Taken together, these results suggest that the GPI anchor may affect the overall conformation of its attached protein.
The GPI anchor may also influence protein structure by interacting directly with or causing the protein to interact with the cell membrane. Based on a combination of two-dimensional NMR analysis and molecular modeling, Homans and co-workers proposed that the glycan core of the VSG GPI anchor exists in an extended conformation that lies along the plane of the plasma membrane (43). Computer modeling has also been used to suggest that the glycan portion of the Thy-1 GPI anchor occupies a carbohydrate-binding site of the protein domain (44). In this model, the protein portion of Thy-1 sits directly on the cell membrane with most of its GPI anchor buried within the protein. FRET studies of fluorescently labeled, GPI-anchored human placental alkaline phosphatase (PLAP) in artificial lipid bilayers found that the protein portion sits less than 10−14 Å away from the bilayer (45). Contact between the PLAP protein moiety and the lipid bilayer might allow for transmission of structural changes or signals between the cell membrane and the GPI-anchored protein (45).
The GPI Anchor May Be Involved in Signal Transduction.
The GPI anchor may also act as an intermediary between the exterior of a cell and internal signaling molecules (46,47). As mentioned earlier, the GPI anchor may allow for signal transduction by GPI-anchored proteins (2,30). Antibody cross-linking of some GPI-anchored proteins can effect the transduction of cellular activation or inhibition signals, resulting in Ca2+ fluxes, protein tyrosine phosphorylation, or cytokine secretion (2,46,47). These effects are not generally observed with genetically engineered forms of GPI proteins, where the GPI anchor has been replaced with a transmembrane domain, indicating that the GPI anchor is crucial for these signaling events (2,46,47). Although the GPI anchor does not completely cross the cell membrane, it has been postulated that the transduction of cellular signals occurs through the physical association of the GPI anchor with other transmembrane proteins involved in intracellular signaling (2). In support of this hypothesis, certain GPI-anchored proteins have been found to associate with transmembrane signal transduction partners, such as protein tyrosine kinases, integrins, and heterotrimeric GTP-binding proteins (2,30,46,47).
The GPI Anchor May Facilitate Cellular Communication.
Many GPI-anchored proteins involved in signaling and cell−cell communication, such as DAF and Thy-1, diffuse freely on the cell surface, allowing these proteins to move rapidly in response to external stimuli (11). This high mobility has been postulated to facilitate cell−cell interactions and communication (11). For instance, the high lateral mobility of DAF may allow it to interact with and inhibit membrane-associated complement fragments. Other GPI-anchored proteins, such as NCAM, are involved in cellular adhesion and communication and might benefit from the ability to move rapidly on the cell surface in response to external stimuli.
The GPI Anchor May Act as an Apical Targeting Signal.
Another possible function of the GPI anchor is to act as a targeting device. In polarized cells, different domains of the plasma membrane display different protein and lipid compositions, allowing for a variety of specialized functions in each domain (48). Known as the apical and basolateral domains, these domains are separated by tight junctions and are important to the cell for maintaining asymmetric growth, directional migration, or transport and delivery of signals and nutrients. Since many GPI-anchored proteins are delivered to the apical membrane, the GPI anchor has been proposed to act as an apical targeting signal (reviewed in ref (48)). In 1992, Brown and Rose used detergent extraction to determine that human PLAP expressed in MDCK cells, a polarized epithelial cell line, was associated with lipid rafts during transport through the Golgi and subsequently to the apical surface (49). Based on this and other data, it was postulated that lipid rafts may act as platforms for the formation of apical targeting vesicles (48,49). Due to their presumed inclusion in lipid rafts, the GPI anchor was believed to be an apical targeting domain by mediating association of a protein with these lipid raft domains (48,49). However, epithelial Fisher rat thyroid cells trafficked most of their GPI-anchored proteins to the basolateral surface, while some apical proteins in polarized MDCK cells did not even associate with lipid rafts (48,50). Further investigations have implicated N-glycosylation and oligomerization in the apical sorting of GPI-anchored proteins (51,52). Taken together, these studies suggest that a number of mechanisms may be responsible for the sorting of many GPI-anchored proteins to the apical surface in polarized epithelial cells.
The GPI Anchor May Allow for Regulation of Its Associated Protein via Phospholipase Cleavage.
The susceptibility of the GPI anchor to cleavage from its associated protein by phospholipases, such as PI-PLC and phopholipase D, has been suggested as a mechanism for the selective regulation of GPI-anchored proteins (10,11). Phospholipase-mediated release is rapid and may be used by the cell to secrete certain GPI-anchored proteins at a specific time. GPI anchors with an extra fatty acid attached to the inositol moiety are phospholipase-resistant, which may allow for cell- or protein-specific control over the release of GPI-anchored proteins. The cleavage of the GPI anchor from a protein may also be used to disrupt the adhesion between cells. Alternatively, the products released from phospholipase cleavage of a GPI-anchored protein, such as inositol phospholipids, may be involved in signal transduction pathways or cellular communication (10).
The GPI Anchor Binds to Bacterial Toxins.
Aerolysin is a bacterial toxin secreted by Aeromonas hydrophilia implicated in the virulence of this human pathogen (53). This toxin is a hydrophilic protein that binds to certain sensitive cells and forms oligomers that insert into the cell membrane (53). The aerolysin oligomers form channels in the plasma membrane that kill the cell (53). Known aerolysin receptors, such as Thy-1 and contactin, seem to be unrelated in function; however, they all contain GPI anchors (53). In 1998, Diep and colleagues demonstrated that the GPI anchor of these target proteins was an important binding determinant for aerolysin (53). In addition, Fukushima et al. determined that β-N-acetylglucosamine, a side chain on the GPI anchor of human PLAP, was necessary for aerolysin binding (54). However, certain GPI-anchored proteins bound strongly to aerolysin, while others did not, suggesting that the variable regions of the GPI anchor, such as the sugar or phosphoethanolamine side chains, were responsible for this specificity (53). Recently, another pore-forming toxin, alpha toxin from Clostridium septicum, has been found to bind to the GPI anchor (55). Since GPI-anchored proteins appear to be clustered on the cell surface, it has been speculated that binding of these bacterial toxins to the GPI anchor facilitates their concentration and oligomerization, thus allowing for toxin insertion into host cell membranes (53).
The GPI Anchor May Be Involved in Prion Disease Pathogenesis.
Prion disease is characterized by the formation of insoluble protein plaques within neurons and related cells in the brain, which are associated with neurodegeneration (27). Plaque formation involves a conformational change of the normal cellular prion protein PrPC, a GPI-anchored protein, to the pathogenic scrapie form, PrPSc(27). Although the normal function of PrPC is still unknown, it has been suggested to be a signaling molecule (27). Plaque formation might interfere with the normal prion signaling function, leading to neuronal death.
PrPC, like many GPI-anchored proteins, is able to migrate from one cell membrane to another (56). This transfer requires cellular activation by phorbol 12-myristate 13-acetate, direct cell−cell contact, and an intact GPI anchor (56). It is not known whether PrPSc also undergoes intercellular transfer. If so, the process may permit PrPSc to infect healthy, PrPC-containing cells. Alternatively, intercellular transfer of PrPC may allow PrPSc-infected cells to recruit PrPC from healthy cells, thus providing the infected cells with more PrPC substrate for propagation of PrPSc.
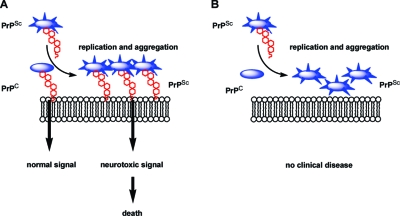
Recently, the GPI anchor of PrPC was discovered to play a potential role in prion disease pathogenesis (9,57). When infected with PrPSc, transgenic mice that expressed an anchorless, secreted version of PrPC never developed clinical prion disease (9). However, the brains of these mice still contained PrPSc plaques that are normally associated with clinical disease progression. Thus, removal of the GPI anchor from PrPC did not prevent the generation of PrPSc plaques but somehow undermined the genesis of prion disease. It is possible that the lack of a GPI anchor on PrPC prevents the delivery of neurotoxic signals after PrPSc plaque formation (Figure 3).
Figure 3.
A proposed model for the role of the GPI anchor in the conversion of PrPC to PrPSc and progression to clinical disease (9). (A) When exposed to PrPSc, GPI-anchored PrPC is converted into aggregates of PrPSc. The aggregates may interfere with the normal signaling events involving PrPC, leading to neuron death. (B) Transgenic mice expressing PrPC lacking a GPI anchor still form PrPSc aggregates upon infection with exogenous PrPSc. However, these aggregates may be unable to disrupt signal transduction pathways due to the lack of a GPI anchor. Figure adapted from ref (57). Copyright 2005 AAAS.
Structural Significance of the GPI Anchor
The relationship between the structures and functions of the GPI anchor is difficult to study due to the lack of sufficient quantities of pure anchors and anchored proteins. When produced in cells, GPI-anchored proteins exist as heterogeneous mixtures with considerable variation in their glycan core modifications and lipid moieties, a complicating feature with respect to functional analysis (1,13,18,58). Furthermore, well-defined modifications to the GPI anchor structure cannot be imposed using conventional biological methods; the biosynthetic enzymes are not well characterized, and their disruption in cells simply leads to loss of the entire GPI structure (7,8,14,25,59).
Chemical synthesis can provide access to both native and novel GPI-anchored protein structures, providing valuable material for functional studies. Several total syntheses of native GPI anchors have been reported; however, these routes are complicated and not amenable to structural modification (reviewed in ref (60)). More importantly, most synthetic routes do not provide an avenue for coupling the anchor structure to a protein, the state in which they function naturally (60). Recently, Shao et al. attached a synthetic 12-amino acid glycopeptide from CD52, a GPI-anchored peptide, to a synthetically produced GPI anchor (61). However, almost all known GPI-anchored proteins are considerably larger than 12 amino acids and are not readily accessible by routine peptide synthesis.
An additional motivation for the synthesis of GPI anchors derives from their potential clinical utility. Certain eukaryotic parasites, such as T. brucei, Leishmania, and Plasmodium falciparum have an abundance of GPI-anchored proteins on the plasma membrane. Their GPI anchor structures differ from those found in mammals with respect to decorations of the core pentasaccharide and/or lack of an associated protein. Because of these differences, the parasite’s GPI anchor is often an immunodominant epitope and, accordingly, synthetic variants have been explored as vaccine candidates and for the characterization of malaria-induced antibody responses (62,63).
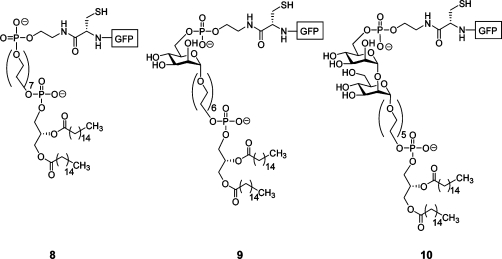
To circumvent the difficulty in native GPI anchor synthesis, a number of research groups have generated peptides or proteins attached to GPI anchor substitutes (Figure 4) (64–70). These GPI anchor replacements were designed to act solely as membrane-anchoring devices rather than emulating the complex structure of a native GPI anchor. Since none of these GPI anchor substitutes contained sugars, the contributions of the various monosaccharides within the glycan core to the biological functions of the GPI anchor could not be assessed. Nevertheless, these substitutes did allow for some interesting structural and functional studies of lipid-modified prion proteins (PrPs). For example, both the circular dichroism spectra of 2, when incorporated into liposomes, and the infrared spectrum of liposome-incorporated 4 were similar to the respective spectra of soluble PrP (65,68). These results suggest that structures determined from the soluble protein may represent the conformations adopted by the cell surface-bound, GPI-anchored PrPC65,68. In another study, lipidated PrP 7 was able to incorporate into cellular membranes and was found to float at a different concentration of sucrose than natively anchored PrPC in a sucrose gradient floatation assay (70). This discrepancy is most likely the result of structural differences between the native PrPC GPI anchor and the GPI anchor substitution found on 7.
Figure 4.
Structures of peptides/proteins attached to GPI anchor substitutes (64–70). The GPI anchor replacement structures were chemically synthesized and coupled to either expressed proteins or chemically synthesized peptides or proteins. Single letter abbreviations are used for amino acids (2 and 7). PrP106 is the 106-amino acid truncated mouse prion protein (64), PrP(23−231) is the truncated mouse (2) or hamster (4) prion protein (65,68), PrP(214−231) is a fragment of the human prion protein (66), PrP(S230C) is the truncated mouse prion protein with a serine-to-cysteine mutation at residue 230 (69), GFP is enhanced green fluorescent protein (67), and PrP(90−232) is the truncated mouse prion protein (70).
In an effort to define the functional significance of the GPI glycan core, our laboratory has recently synthesized a series of GPI anchor analogues bearing systematic modifications to the core structure (Figure 5) (71). The analogues were similar in length to the native GPI anchor, contained no (8), one (9), or two (10) mannose units, and replaced the phosphoinositol and glucosamine units with a simple hydrophilic poly(ethylene glycol) (PEG) linker. These analogues were coupled to the green fluorescent protein (GFP) using native chemical ligation (71). The GPI-protein analogues all incorporated into cellular membranes and trafficked to recycling endosomes similarly to GFP bearing a native GPI anchor (GFP-GPI) (72). This result suggests that the glycan core of the GPI anchor is not a major determinant of the intracellular fate of GPI-anchored proteins. However, deletions in the GPI anchor glycan core significantly altered the diffusion kinetics of these proteins in the cell membrane. Fluorescence correlation spectroscopy revealed that all three GPI-protein analogues diffused more slowly on the cell membrane than natively anchored GFP-GPI, suggesting that the sugars of the glycan core affect the lateral mobility of GPI-anchored proteins (72). The GPI anchor analogues we designed contained flexible PEG linkers, which may permit greater movement of the attached protein, thus allowing the protein to engage in contacts with both the lipid bilayer and other cell surface proteins. Such transient interactions would be expected to retard diffusion. The additional sugar moieties in the native GPI structure might sufficiently rigidify the anchor so as to avoid nonspecific membrane interactions. These studies demonstrate that the GPI anchor may be more than a membrane anchor and that the sugars of the GPI anchor may play an important role in regulating the behavior of the attached protein. Furthermore, this cellular system provides a basic platform for dissecting the contributions of various GPI anchor components to their biological function.
Figure 5.
Structures of GPI-protein analogues bearing systematic deletions in the glycan core (71). The GPI anchor analogues possess no monosaccharides (8), one mannosyl unit (9), or two mannosyl units (10) and were coupled to GFP using native chemical ligation. These GPI-protein analogues were used to investigate the functional significance of the GPI anchor glycan core (72).
Conclusions and Perspectives
The GPI anchor is a structurally complex posttranslational modification that remains a mystery with respect to its biological activities. A major obstacle to functional investigations is the difficulty in producing structurally defined GPI moieties and GPI-anchored proteins. Cells generate heterogeneous mixtures of both anchor and modified protein structures, and chemical synthesis requires numerous difficult synthetic transformations. Nonetheless, there have been many advances in the production of these structures, including a recent report detailing a method for the expression of GPI-anchored proteins in insect cells (73). The GPI anchor biosynthetic pathway has been elucidated, and many of the enzymes have been characterized. Application to the large-scale production of GPI anchors and modified proteins may be on the horizon. Furthermore, recent progress in the chemical synthesis of GPI anchors and GPI-anchored proteins allows for the generation of native and modified structures that can be used for the systematic investigation of the biological function. In particular, the approach pursued by our laboratory can be further expanded upon with GPI analogues bearing more subtly modified monosaccharide units, various side chain modifications, or lipid modifications. Comparison of the proteins bearing modified and natural anchor structures might reveal differences in membrane protein organization or association with other proteins. Additionally, GPI anchor analogues might be coupled to PrPC for studies of its conversion to PrPSc in cell-based functional assays.
Detailed structural information would be extremely useful in forming hypotheses regarding the functions of the GPI anchor. Unfortunately, the flexibility of glycosidic linkages and the solubility issues inherent to lipids have rendered GPI-modified proteins difficult to crystallize or to study by NMR. To date, none of the GPI-anchored protein structures that have been determined by X-ray crystallography contained their GPI anchor. No NMR solution structures of GPI-anchored proteins are available either. Emerging techniques that limit the flexibility of the GPI anchor or assemble GPI-anchored proteins into ordered arrays (e.g., bicelles (74)) should facilitate structural studies in the future.
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: AChE, acetylcholinesterase; APase, alkaline phosphatase; DAF, decay-accelerating factor; FRET, fluorescence resonance energy transfer; Gal, galactose; GalNAc, N-acetylgalactosamine; GFP, green fluorescent protein; GFP-GPI, GPI-anchored green fluorescent protein, GH-GPI, GPI-anchored growth hormone; GPI, glycosylphosphatidylinositol; HexNAc, N-acetylhexosamine; PI-PLC, phosphatidylinositol phospholipase C; Man, mannose; NANA, sialic acid; NCAM, neural cell adhesion molecule; PEG, poly(ethylene glycol); PEtN, phosphoethanolamine; PLAP, placental alkaline phosphatase; PrP, prion protein; PrPC, normal cellular prion protein; PrPSc, scrapie prion protein; VSG, variant surface glycoprotein.
References
- Nosjean O.; Briolay A.; Roux B. (1997) Mammalian GPI proteins: sorting, membrane residence and functions. Biochim. Biophys. Acta 1331, 153–186. [DOI] [PubMed] [Google Scholar]
- Low M. (1999) GPI-anchored biomolecules—an overview, in GPI-anchored membrane proteins and carbohydrates (Hoessli D. C., and Ilangumaran S., Eds.) pp. 1−14, Landes Company, Austin, TX. [Google Scholar]
- Ferguson M. A. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A.; Homans S. W.; Dwek R. A.; Rademacher T. W. (1988) Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science 239, 753–759. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A.; Low M. G.; Cross G. A. (1985) Glycosyl-sn-1,2-dimyristylphosphatidylinositol is covalently linked to Trypanosoma brucei variant surface glycoprotein. J. Biol. Chem. 260, 14547–14555. [PubMed] [Google Scholar]
- Homans S. W.; Ferguson M. A.; Dwek R. A.; Rademacher T. W.; Anand R.; Williams A. F. (1988) Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature 333, 269–272. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B.; Maurer-Stroh S.; Novatchkova M.; Schneider G.; Eisenhaber F. (2003) Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. BioEssays 25, 367–385. [DOI] [PubMed] [Google Scholar]
- Tiede A.; Bastisch I.; Schubert J.; Orlean P.; Schmidt R. E. (1999) Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol. Chem. 380, 503–523. [DOI] [PubMed] [Google Scholar]
- Chesebro B.; Trifilo M.; Race R.; Meade-White K.; Teng C.; LaCasse R.; Raymond L.; Favara C.; Baron G.; Priola S.; Caughey B.; Masliah E.; Oldstone M. (2005) Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Low M. G. (1989) Glycosyl-phosphatidylinositol: a versatile anchor for cell surface proteins. FASEB J. 3, 1600–1608. [DOI] [PubMed] [Google Scholar]
- Low M. G.; Saltiel A. R. (1988) Structural and functional roles of glycosyl-phosphatidylinositol in membranes. Science 239, 268–275. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A.; Brimacombe J. S.; Brown J. R.; Crossman A.; Dix A.; Field R. A.; Guther M. L.; Milne K. G.; Sharma D. K.; Smith T. K. (1999) The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim. Biophys. Acta 1455, 327–340. [DOI] [PubMed] [Google Scholar]
- Ikezawa H. (2002) Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25, 409–417. [DOI] [PubMed] [Google Scholar]
- Kinoshita T.; Inoue N. (2000) Dissecting and manipulating the pathway for glycosylphos-phatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4, 632–638. [DOI] [PubMed] [Google Scholar]
- Orlean P.; Menon A. K. (2007) Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011. [DOI] [PubMed] [Google Scholar]
- Deeg M. A.; Humphrey D. R.; Yang S. H.; Ferguson T. R.; Reinhold V. N.; Rosenberry T. L. (1992) Glycan components in the glycoinositol phospholipid anchor of human erythrocyte acetylcholinesterase. Novel fragments produced by trifluoroacetic acid. J. Biol. Chem. 267, 18573–18580. [PubMed] [Google Scholar]
- Low M. G. (1989) The glycosyl-phosphatidylinositol anchor of membrane proteins. Biochim. Biophys. Acta 988, 427–454. [DOI] [PubMed] [Google Scholar]
- Brewis I. A.; Ferguson M. A. J.; Mehlert A.; Turner A. J.; Hooper N. M. (1995) Structures of the glycosyl-phosphatidylinositol anchors of porcine and human renal membrane dipeptidase. Comprehensive structural studies on the porcine anchor and interspecies comparison of the glycan core structures. J. Biol. Chem. 270, 22946–22956. [DOI] [PubMed] [Google Scholar]
- Nakano Y.; Noda K.; Endo T.; Kobata A.; Tomita M. (1994) Structural study on the glycosyl-phosphatidylinositol anchor and the asparagine-linked sugar chain of a soluble form of CD59 in human urine. Arch. Biochem. Biophys. 311, 117–126. [DOI] [PubMed] [Google Scholar]
- Mukasa R.; Umeda M.; Endo T.; Kobata A.; Inoue K. (1995) Characterization of glycosylphosphatidylinositol (GPI)-anchored NCAM on mouse skeletal muscle cell line C2C12: the structure of the GPI glycan and release during myogenesis. Arch. Biochem. Biophys. 318, 182–190. [DOI] [PubMed] [Google Scholar]
- Fontaine T.; Magnin T.; Melhert A.; Lamont D.; Latge J. P.; Ferguson M. A. (2003) Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 13, 169–177. [DOI] [PubMed] [Google Scholar]
- Oxley D.; Bacic A. (1999) Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc. Natl. Acad. Sci. U.S.A. 96, 14246–14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae J. I.; Acosta-Serrano A.; Morrice N. A.; Mehlert A.; Ferguson M. A. (2005) Structural characterization of NETNES, a novel glycoconjugate in Trypanosoma cruzi epimastigotes. J. Biol. Chem. 280, 12201–12211. [DOI] [PubMed] [Google Scholar]
- McConville M. J.; Ferguson M. A. (1993) The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 294, 305–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe K.; Kitamura D.; Okabe M.; Taniuchi I.; Ikawa M.; Watanabe T.; Kinoshita T.; Takeda J. (1996) Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood 87, 3600–3606. [PubMed] [Google Scholar]
- Leidich S. D.; Drapp D. A.; Orlean P. (1994) A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem. 269, 10193–10196. [PubMed] [Google Scholar]
- Taylor D. R.; Hooper N. M. (2006) The prion protein and lipid rafts. Mol. Membr. Biol. 23, 89–99. [DOI] [PubMed] [Google Scholar]
- Rajendran L.; Simons K. (2005) Lipid rafts and membrane dynamics. J. Cell Sci. 118, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Munro S. (2003) Lipid rafts: elusive or illusive?. Cell 115, 377–388. [DOI] [PubMed] [Google Scholar]
- Simons K.; Toomre D. (2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- Pierce S. K. (2004) To cluster or not to cluster: FRETting over rafts. Nat. Cell Biol. 6, 180–181. [DOI] [PubMed] [Google Scholar]
- Sharma P.; Varma R.,.; Sarasij R. C.; Ira, Gousset K.; Krishnamoorthy G.; Rao M.; Mayor S. (2004) Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589. [DOI] [PubMed] [Google Scholar]
- Kenworthy A. K.; Edidin M. (1998) Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J. Cell Biol. 142, 69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov O. O.; Nichols B. J. (2004) Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat. Cell Biol. 6, 238–243. [DOI] [PubMed] [Google Scholar]
- Varma R.; Mayor S. (1998) GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801. [DOI] [PubMed] [Google Scholar]
- Friedrichson T.; Kurzchalia T. V. (1998) Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394, 802–805. [DOI] [PubMed] [Google Scholar]
- Medof M. E.; Nagarajan S.; Tykocinski M. L. (1996) Cell-surface engineering with GPI-anchored proteins. FASEB J. 10, 574–586. [DOI] [PubMed] [Google Scholar]
- Kooyman D. L.; Byrne G. W.; McClellan S.; Nielsen D.; Tone M.; Waldmann H.; Coffman T. M.; McCurry K. R.; Platt J. L.; Logan J. S. (1995) In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium. Science 269, 89–92. [DOI] [PubMed] [Google Scholar]
- Huang J. H.; Getty R. R.; Chisari F. V.; Fowler P.; Greenspan N. S.; Tykocinski M. L. (1994) Protein transfer of preformed MHC-peptide complexes sensitizes target cells to T cell cytolysis. Immunity 1, 607–613. [DOI] [PubMed] [Google Scholar]
- McHugh R. S.; Nagarajan S.; Wang Y. C.; Sell K. W.; Selvaraj P. (1999) Protein transfer of glycosyl-phosphatidylinositol-B7−1 into tumor cell membranes: a novel approach to tumor immunotherapy. Cancer Res. 59, 2433–2437. [PubMed] [Google Scholar]
- Butikofer P.; Malherbe T.; Boschung M.; Roditi I. (2001) GPI-anchored proteins: now you see ’em, now you don’t. FASEB J. 15, 545–548. [DOI] [PubMed] [Google Scholar]
- Barboni E.; Rivero B. P.; George A. J.; Martin S. R.; Renoup D. V.; Hounsell E. F.; Barber P. C.; Morris R. J. (1995) The glycophosphatidylinositol anchor affects the conformation of Thy-1 protein. J. Cell Sci. 108, 487–497. [DOI] [PubMed] [Google Scholar]
- Homans S. W.; Edge C. J.; Ferguson M. A.; Dwek R. A.; Rademacher T. W. (1989) Solution structure of the glycosylphosphatidylinositol membrane anchor glycan of Trypanosoma brucei variant surface glycoprotein. Biochemistry 28, 2881–2887. [DOI] [PubMed] [Google Scholar]
- Rademacher T. W.; Edge C. J.; Dwek R. A. (1991) Dropping anchor with the lipophosphoglycans. Curr. Biol. 1, 41–42. [DOI] [PubMed] [Google Scholar]
- Lehto M. T.; Sharom F. J. (2002) Proximity of the protein moiety of a GPI-anchored protein to the membrane surface: a FRET study. Biochemistry 41, 8368–8376. [DOI] [PubMed] [Google Scholar]
- Jones D. R.; Varela-Nieto I. (1998) The role of glycosyl-phosphatidylinositol in signal transduction. Int. J. Biochem. Cell Biol. 30, 313–326. [DOI] [PubMed] [Google Scholar]
- Robinson P. J. (1997) Signal transduction via GPI-anchored membrane proteins. Adv. Exp. Med. Biol. 419, 365–370. [DOI] [PubMed] [Google Scholar]
- Schuck S.; Simons K. (2006) Controversy fuels trafficking of GPI-anchored proteins. J. Cell Biol. 172, 963–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A.; Rose J. K. (1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- Zurzolo C.; Lisanti M. P.; Caras I. W.; Nitsch L.; Rodriguez-Boulan E. (1993) Glycosylphosphatidylinositol-anchored proteins are preferentially targeted to the basolateral surface in Fischer rat thyroid epithelial cells. J. Cell Biol. 121, 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benting J. H.; Rietveld A. G.; Simons K. (1999) N-Glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby canine kidney cells. J. Cell Biol. 146, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino S.; Sarnataro D.; Pillich R.; Tivodar S.; Nitsch L.; Zurzolo C. (2004) Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 167, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep D. B.; Nelson K. L.; Raja S. M.; Pleshak E. N.; Buckley J. T. (1998) Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming toxin aerolysin. J. Biol. Chem. 273, 2355–2360. [DOI] [PubMed] [Google Scholar]
- Fukushima K.; Ikehara Y.; Kanai M.; Kochibe N.; Kuroki M.; Yamashita K. (2003) A beta-N-acetylglucosaminyl phosphate diester residue is attached to the glycosylphosphatidylinositol anchor of human placental alkaline phosphatase: a target of the channel-forming toxin aerolysin. J. Biol. Chem. 278, 36296–36303. [DOI] [PubMed] [Google Scholar]
- Gordon V. M.; Nelson K. L.; Buckley J. T.; Stevens V. L.; Tweten R. K.; Elwood P. C.; Leppla S. H. (1999) Clostridium septicum alpha toxin uses glycosylphosphatidylinositol-anchored protein receptors. J. Biol. Chem. 274, 27274–27280. [DOI] [PubMed] [Google Scholar]
- Liu T.; Li R.; Pan T.; Liu D.; Petersen R. B.; Wong B. S.; Gambetti P.; Sy M. S. (2002) Intercellular transfer of the cellular prion protein. J. Biol. Chem. 277, 47671–47678. [DOI] [PubMed] [Google Scholar]
- Aguzzi A. (2005) Cell biology. Prion toxicity: all sail and no anchor. Science 308, 1420–1421. [DOI] [PubMed] [Google Scholar]
- Thomas J. R.; Dwek R. A.; Rademacher T. W. (1990) Structure, biosynthesis, and function of glycosylphosphatidylinositols. Biochemistry 29, 5413–5422. [DOI] [PubMed] [Google Scholar]
- Bastisch I.; Tiede A.; Deckert M.; Ziolek A.; Schmidt R. E.; Schubert J. (2000) Glycosylphosphatidylinositol (GPI)-deficient Jurkat T cells as a model to study functions of GPI-anchored proteins. Clin. Exp. Immunol. 122, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.; Bishop L. (2004) Chemical synthesis of GPIs and GPI-anchored glycopeptides. Eur. J. Org. Chem. 2004, 3585–3596. [Google Scholar]
- Shao N.; Xue J.; Guo Z. (2004) Chemical synthesis of a skeleton structure of sperm CD52-A GPI-anchored glycopeptide. Angew. Chem., Int. Ed. 43, 1569–1573. [DOI] [PubMed] [Google Scholar]
- Kamena F.; Tamborrini M.; Liu X.; Kwon Y. U.; Thompson F.; Pluschke G.; Seeberger P. H. (2008) Synthetic GPI array to study antitoxic malaria response. Nat. Chem. Biol. 4, 238–240. [DOI] [PubMed] [Google Scholar]
- Schofield L.; Hewitt M. C.; Evans K.; Siomos M. A.; Seeberger P. H. (2002) Synthetic GPI as a candidate anti-toxic vaccine in a model of malaria. Nature 418, 785–789. [DOI] [PubMed] [Google Scholar]
- Ball H. L.; King D. S.; Cohen F. E.; Prusiner S. B.; Baldwin M. A. (2001) Engineering the prion protein using chemical synthesis. J. Pept. Res. 58, 357–374. [DOI] [PubMed] [Google Scholar]
- Eberl H.; Tittmann P.; Glockshuber R. (2004) Characterization of recombinant, membrane-attached full-length prion protein. J. Biol. Chem. 279, 25058–25065. [DOI] [PubMed] [Google Scholar]
- Musiol H. J.; Dong S.; Kaiser M.; Bausinger R.; Zumbusch A.; Bertsch U.; Moroder L. (2005) Toward semisynthetic lipoproteins by convergent strategies based on click and ligation chemistry. ChemBioChem 6, 625–628. [DOI] [PubMed] [Google Scholar]
- Grogan M. J.; Kaizuka Y.; Conrad R. M.; Groves J. T.; Bertozzi C. R. (2005) Synthesis of lipidated green fluorescent protein and its incorporation in supported lipid bilayers. J. Am. Chem. Soc. 127, 14383–14387. [DOI] [PubMed] [Google Scholar]
- Hicks M. R.; Gill A. C.; Bath I. K.; Rullay A. K.; Sylvester I. D.; Crout D. H.; Pinheiro T. J. (2006) Synthesis and structural characterization of a mimetic membrane-anchored prion protein. FEBS J. 273, 1285–1299. [DOI] [PubMed] [Google Scholar]
- Breydo L.; Sun Y.; Makarava N.; Lee C. I.; Novitskaia V.; Bocharova O.; Kao J. P.; Baskakov I. V. (2007) Nonpolar substitution at the C-terminus of the prion protein, a mimic of the glycosylphosphatidylinositol anchor, partially impairs amyloid fibril formation. Biochemistry 46, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschewski D.; Seidel R.; Miesbauer M.; Rambold A. S.; Oesterhelt D.; Winklhofer K. F.; Tatzelt J.; Engelhard M.; Becker C. F. (2007) Semisynthetic murine prion protein equipped with a GPI anchor mimic incorporates into cellular membranes. Chem. Biol. 14, 994–1006. [DOI] [PubMed] [Google Scholar]
- Paulick M. G.; Wise A. R.; Forstner M. B.; Groves J. T.; Bertozzi C. R. (2007) Synthetic analogues of glycosylphosphatidylinositol-anchored proteins and their behavior in supported lipid bilayers. J. Am. Chem. Soc. 129, 11543–11550. [DOI] [PubMed] [Google Scholar]
- Paulick M. G.; Forstner M. B.; Groves J. T.; Bertozzi C. R. (2007) A chemical approach to unraveling the biological function of the glycosylphosphatidylinositol anchor. Proc. Natl. Acad. Sci. U.S.A. 104, 20332–20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shams-Eldin H.; Azzouz N.; Niehus S.; Smith T. K.; Schwarz R. T. (2008) An efficient method to express GPI-anchor proteins in insect cells. Biochem. Biophys. Res. Commun. 365, 657–663. [DOI] [PubMed] [Google Scholar]
- Wang G. (2008) NMR of membrane-associated peptides and proteins. Curr. Protein Pept. Sci. 9, 50–69. [DOI] [PubMed] [Google Scholar]