Abstract
Autophagy is an intracellular degradation system, by which cytoplasmic contents are degraded in lysosomes. Autophagy is dynamically induced by nutrient depletion to provide necessary amino acids within cells, thus helping them adapt to starvation. Although it has been suggested that mTOR is a major negative regulator of autophagy, how it controls autophagy has not yet been determined. Here, we report a novel mammalian autophagy factor, Atg13, which forms a stable ∼3-MDa protein complex with ULK1 and FIP200. Atg13 localizes on the autophagic isolation membrane and is essential for autophagosome formation. In contrast to yeast counterparts, formation of the ULK1–Atg13–FIP200 complex is not altered by nutrient conditions. Importantly, mTORC1 is incorporated into the ULK1–Atg13–FIP200 complex through ULK1 in a nutrient-dependent manner and mTOR phosphorylates ULK1 and Atg13. ULK1 is dephosphorylated by rapamycin treatment or starvation. These data suggest that mTORC1 suppresses autophagy through direct regulation of the ∼3-MDa ULK1–Atg13–FIP200 complex.
INTRODUCTION
Macroautophagy (simply referred to as autophagy hereafter) is a major degradation system, by which cytoplasmic contents are degraded in the lysosomes (Klionsky, 2007; Mizushima, 2007; Levine and Kroemer, 2008; Mizushima et al., 2008). An isolation membrane, also known as a phagophore, sequesters a portion of the cytoplasm, which results in the formation of an autophagosome. This autophagosome subsequently fuses with a lysosome, where cytoplasm-derived materials are degraded by lysosomal hydrolases. Resultant amino acids are delivered back to the cytoplasm and then reused or further metabolized. Autophagy is basically a nonselective process, although several proteins are selectively degraded by this pathway. Autophagy is highly conserved among eukaryotes and usually activated by nutrient starvation to produce necessary amino acids within cells, thus helping them adapt to starvation conditions. Autophagy is also important for intracellular protein quality control, preimplantation development, degradation of intracellular pathogens, antigen presentation, tumor suppression, and certain types of cell death (Levine and Deretic, 2007; Mizushima et al., 2008; Tsukamoto et al., 2008).
Autophagy is a highly inducible process, but how it is regulated remains to be revealed. Among the many factors that have thus far been reported (Codogno and Meijer, 2005; Meijer and Codogno, 2006; Mizushima, 2007), (mammalian) target of rapamycin [(m)TOR]-mediated suppression appears to be a highly conserved, major regulatory mechanism. (m)TOR forms two distinct complexes, (m)TORC1 and (m)TORC2 (Wullschleger et al., 2006; Guertin and Sabatini, 2007; Yang and Guan, 2007). In mammalian cells, mTORC1 consists of mTOR, mLST8 (GβL), and raptor, and mTORC2 consists of mTOR, mLST8, mSin1, and rictor. mTORC1, not mTORC2, is a nutrient-sensitive complex that can be inhibited by rapamycin. mTORC1 is important for the regulation of protein translation, ribosome biogenesis, and cell growth through phosphorylation of its substrates such as S6-kinese, 4E-BP1, and PRAS40 (Wullschleger et al., 2006; Guertin and Sabatini, 2007; Yang and Guan, 2007). A role of (m)TOR in autophagy has also been established because rapamycin and genetic ablation of the TOR pathway have been shown to trigger autophagy in a wide range of organisms (Noda and Ohsumi, 1998; Ravikumar et al., 2004; Scott et al., 2004). However, how (m)TOR regulates autophagy is still unclear. TAP42, a known TOR substrate in yeast, does not participate in autophagy regulation (Kamada et al., 2000) and involvement of mTOR substrates such as S6-kinase is controversial (Klionsky et al., 2005).
In yeast, the Atg1–Atg13–Atg17 complex is thought to function downstream of Tor (Kamada et al., 2000). Atg13 is hyperphosphorylated by an unidentified kinase under nutrient-rich conditions and is rapidly dephosphorylated after nitrogen depletion or rapamycin treatment, which facilitates interaction with Atg1. Atg13 and Atg17 are important for Atg1 kinase activity, which is required for autophagy (Kamada et al., 2000; Kabeya et al., 2005). Atg1 has two mammalian homologues, UNC-51-like kinase 1 (ULK1 or Unc51.1) and ULK2 (Unc51.2; Yan et al., 1998; Tomoda et al., 1999; Yan et al., 1999). There is no apparent Atg17 homolog in mammals, but we previously proposed that FIP200 (a focal adhesion kinase [FAK] family-interacting protein of 200 kDa) could be a functional counterpart (Hara et al., 2008). Both ULK1 and FIP200 are involved in autophagy (Young et al., 2006; Chan et al., 2007; Hara et al., 2008; Kundu et al., 2008), although the functions of ULK1 and FIP200 are not specific to autophagy (Tomoda et al., 1999; Gan and Guan, 2008). Recently mammalian counterpart of Atg13 was also identified (Meijer et al., 2007) and shown to be required for autophagy (Chan et al., 2009). However, precise nature of the complex including these factors and its relationship with mTOR remains to be revealed.
In this study, we independently identified the mammalian Atg13 by database searching and screening for ULK-interacting proteins. Atg13 is essential for autophagy in mammalian cells and forms a ∼3-MDa protein complex with ULK1 and FIP200. Furthermore, we found that mTOR is incorporated into this complex under nutrient-rich conditions but is excluded during starvation. mTOR was also shown to directly interact with ULK1 and phosphorylate ULK1 and Atg13. These results suggest that mTOR regulates autophagy through the ∼3-MDa ULK1–Atg13–FIP200 complex.
MATERIALS AND METHODS
Plasmids
The cDNA encoding human Atg13 (KIAA0652/AB014552) was obtained from Kazusa DNA Research Institute and was cloned into p3xFLAG CMV10 (Sigma-Aldrich, Tokyo, Japan), pEGFP-C1 (Clontech, Tokyo, Japan), and pCI-neo (Promega, Tokyo, Japan). FLAG-tagged raptor (pcDNA3.1/Zeo(+)-FLAG-raptor) was a gift from Dr. Kenta Hara (Kobe University). Mouse ULK1 and its variants were cloned into p3xFLAG CMV14 (Sigma; Yan et al., 1998). FIP200 plasmids were described previously (Hara et al., 2008).
Cell Culture and Transfection
Wild-type and FIP200−/− mouse embryonic fibroblasts (MEFs) were generated previously (Gan et al., 2006). MEFs, HeLa, and HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), and 50 μg/ml penicillin and streptomycin (complete medium) in a 5% CO2 incubator. Bovine calf serum was used instead of FBS for NIH3T3 cells. For starvation treatment, cells were washed with phosphate-buffered saline (PBS) and incubated in amino acid–free DMEM (Invitrogen, Tokyo, Japan) without FBS (starvation medium). Fugene 6 (Roche Diagnostics, Tokyo, Japan) was used for transfection. For retrovirus-mediated transfection, cDNAs were subcloned into pMXs-IP (kindly provided by T. Kitamura, University of Tokyo). The resulting vectors were used to transfect Plat-E cells and thereby generate recombinant retroviruses. NIH3T3 cells were infected with the recombinant retroviruses and selected as previously described (Hara et al., 2008).
Antibody and Reagents
cDNA encoding human Atg13 was subcloned into pCold I (Takara Bio, Tokyo, Japan) and pGEX6p1 (GE Healthcare, Waukesha, WI). Atg13 fusion proteins tagged with NH2-terminal hexahistidine (His-Atg13) and glutathione S-transferase (GST; GST-Atg13) were expressed in Escherichia coli strain BL21 (DE3) plysS (Novagen, Madison, WI). His-Atg13 was used to immunize rabbits. Antisera were affinity-purified with GST-Atg13. Polyclonal anti-FIP200, anti-ULK1 (Hara et al., 2008), anti-LC3 (Hosokawa et al., 2006), and anti-Atg16L1 antibodies (Mizushima et al., 2003) were described previously. Polyclonal anti-ULK1 antibody (A7481), monoclonal anti-FLAG (M2) antibody, and M2 agarose beads were purchased from Sigma. Polyclonal anti-ULK1 antibodies (sc-10900 and sc-33182) were also purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A monoclonal anti-hemagglutinin (HA) antibody (HA11) was purchased from Covance (Madison, WI). Antibodies to Myc-Tag, p70 S6 kinase (S6K), phospho-S6K (Thr389), mTOR, and raptor were purchased from Cell Signaling Technologies (Tokyo, Japan). Guinea pig polyclonal anti-p62 antibody was purchased from Progen Biotechnik (Heidelberg, Germany). Alexa Fluor 660–conjugated goat anti-rabbit IgG (H + L) antibodies (Invitrogen) were used for immunochemistry. Anti-mTOR mAb (N5D11) was previously described (Nishiuma et al., 1998). Rapamycin were purchased from Sigma. Pepstatin A, chymostatin, leupeptin, E64, and E64d were purchased from Peptide Institute (Osaka, Japan). λ-phosphatase was purchased from New England Biolabs (Tokyo, Japan).
Mass Spectrometry
Liquid chromatography/tandem mass spectrometry (LC–MS/MS) was performed as previously described (Natsume et al., 2002).
Immunoprecipitation and Immunoblotting
Cell lysates were prepared in a regular lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethanesulfonyl fluoride [PMSF], 1 mM Na3VO4, and a protease inhibitor cocktail (Complete EDTA-free protease inhibitor, Roche). The lysates were clarified by centrifugation at 15,000 rpm for 15 min and then subjected to immunoprecipitation using specific antibodies in combination with protein A or G-Sepharose (GE Healthcare). Precipitated immunocomplexes were washed five times in lysis buffer and boiled in sample buffer. For analysis of the mTORC1–ULK1 complex, immunoprecipitation was performed with a second lysis buffer (40 mM HEPES, pH 7.4, 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 0.3% CHAPS, and protease inhibitor cocktail) and a wash buffer (40 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, and 0.3% CHAPS) as described previously to maintain the mTOR complex (Sancak et al., 2007). Samples were subsequently separated by SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore, Tokyo, Japan). Immunoblot analysis was performed with the indicated antibodies and visualized with SuperSignal West Pico Chemiluminescent substrate (Pierce Chemical, Rockford, IL). The signal intensities were analyzed using a LAS-3000mini imaging analyzer and Multi Gauge software version 3.0 (Fujifilm, Tokyo, Japan). Contrast and brightness adjustment was applied to the whole images using Photoshop 7.0.1 (San Jose, CA).
RNA Interference
Stealth RNA interference (RNAi) oligonucleotides were used for small interfering RNA (siRNA) experiments (Invitrogen). The sequences used were as follows: human Atg13_#2-siRNA: sense, 5′-CCAUGUGUGUGGAGAUUUCACUUAA-3′; antisense, 5′-UUAAGUGAAAUCUCCACACACAUGG-3′; human Atg13_#3-siRNA: sense, 5′-GCAUUCAUGUCUACCAGGCAAUUUG-3′; antisense, 5′-CAAAUUGCCUGGUAGACAUGAAUGC-3′; and human ULK1–1-siRNA: sense, 5′-GUGGCCCUGUACGACUUCCAGGAAA-3′; antisense, 5′-UUUCCUGGAAGUCGUACAGGGCCAC-3′. As a negative control, Medium GC duplex of stealth RNAi Negative Control Duplexes (Invitrogen) was used. The stealth RNAi oligonucleotides were transfected into HeLa cells using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's protocols.
Fluorescence Microscopy
NIH3T3 cells stably expressing GFP-Atg13 and HeLa cells stably expressing GFP-LC3 were directly observed with a fluorescence microscope (IX81; Olympus, Tokyo, Japan) equipped with a CCD camera (ORCA ER, HamamatsuPhotonic Systems, Hamamatsu, Japan). A 60× PlanApo oil immersion lens (1.42 NA; Olympus) was used. Immunocytochemistry of Atg16L1 was performed as previously described (Hara et al., 2008).
Electron Microscopy
Conventional electron microscopy was performed as described previously (Hara et al., 2008). For morphometric analysis, 10 cells of each sample were analyzed using MetaMorph image analysis software (Version 6.2; MDS Analytical Technologies, Sunnyvale, CA).
Gel Filtration Analysis
Cells were scrapped from the dish with a rubber policeman and homogenized in hypotonic buffer (40 mM Tris-HCl, pH 7.5, and protease inhibitors [10 μg/ml pepstatin A, chymostatin, leupeptin, E64]) by repeated passage (30 times) through a 1-ml syringe with a 27-gauge needle. The homogenates were centrifuged at 13,000 × g for 15 min, and the supernatants were further centrifuged at 100,000 × g for 60 min. The supernatant (S100) fraction (∼1.6 mg protein in 800 μl) was then applied to a Superose 6 column (GE Healthcare) and eluted at a flow rate of 0.5 ml/min with 40 mM Tris-HCl, pH 7.5, and 150 mM NaCl. Fractions (0.5 ml) were then examined by immunoblotting. The column was calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), albumin (67 kDa), ovalbumin (43 kDa), and cytochrome C (12.5 kDa).
Baculovirus Expression System
A cDNA encoding human Atg13 with an N-terminal GST tag was subcloned into pBacPAK8 (Clontech), and recombinant baculovirus was generated. Recombinant GST-Atg13 was expressed in Sf21 cells and purified as described (Hara et al., 2005).
mTOR and ULK1 Kinase Assays
The mTOR kinase assay was performed as previously described (Hara et al., 2002). ULK1K46N was prepared from HEK293T cells transiently expressing FLAG-ULK1K46N cultured in starvation medium for 3 h with the regular lysis buffer (containing Triton X-100 to break the mTORC1 association) and subjected to immunoprecipitation with M2 agarose beads. The ULK1K46N protein was eluted in elution buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA) containing 0.5 mg/ml 3xFLAG peptide (Sigma). An in vitro protein kinase reaction of ULK1K46N (150 ng per reaction) and GST-Atg13 (1 μg per reaction) was performed for 30 min at 30°C in the presence of [γ-32P]ATP (Perkin Elmer) and mTOR complex obtained from nonstarved HEK293T cells. GST, GST-4E-BP1, and GST-PRAS40 were used as negative and positive controls (Oshiro et al., 2007). The reaction products were separated by SDS-PAGE and visualized with a BAS image analyzer (Fujifilm). The ULK1 kinase assay was performed as previously described using GST-Atg13 as a substrate (Hara et al., 2008). In the ULK1 kinase assay, 10 mM Mg2+ was used instead of Mn2+.
Two-hybrid Assay
Two-hybrid analysis was performed as described previously (James et al., 1996). The strain PJ69–4A was cotransformed with each of the pGBD and pGAD plasmids. Transformants were selected on Trp− Leu− plates and tested for growth on His− Trp− Leu− plates containing 2.5–50 mM 3-amino-triazole (3-AT).
RESULTS
Identification of Mammalian Atg13
We used two different approaches to identify mammalian Atg13. First, we performed a database search using conventional BLAST. Because it was difficult to do so directly with the Saccharomyces cerevisiae Atg13 sequence as a query sequence, we first identified Candida albicans Atg13 (EAK96765) and then used this as the query sequence for the second round BLAST search. As a result, we identified a human cDNA sequence (KIAA0652) encoding a 517-amino acid protein with weak homology to yeast and Candida Atg13. After identifying the sequence, it was assigned as a putative human Atg13 by another group (Meijer et al., 2007). The second approach involved searching for ULK-interacting proteins as described previously (Hara et al., 2008). Mouse ULK1-FLAG and FLAG-ULK2 were expressed in HEK293T cells, and immunoprecipitates were analyzed by highly sensitive direct nano-flow LC-MS/MS (Natsume et al., 2002). In both ULK1-FLAG and FLAG-ULK2 precipitates, we found KIAA0652. On the basis of these findings, together with the results described below, we named KIAA0652 as human Atg13.
Atg13 Interacts with ULK1 and FIP200
Human Atg13 has no particular known domains and has 16% identity to yeast Atg13 (Supplemental Figure S1). The Q-rich region, which is a characteristic feature of yeast Atg13, is not conserved in other species including Drosophila melanogaster and humans.
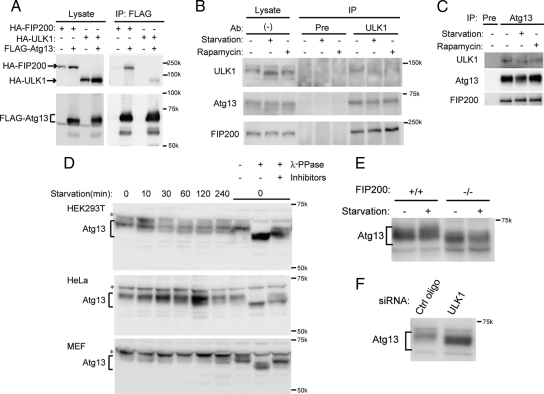
We used transient transfection experiments to first determine whether Atg13 interacts with ULK1 and FIP200. Immunoprecipitation with FLAG-Atg13 revealed that FLAG-Atg13 interacts with both HA-ULK1 and HA-FIP200 (Figure 1A). We also observed interaction between FLAG-Atg13 and HA-ULK2 (Supplemental Figure S2). We then generated an antibody against human Atg13 and confirmed endogenous interaction between Atg13, ULK1, and FIP200 (Figure 1, B and C). Although starvation and rapamycin treatments induce autophagy (Supplemental Figure S3), interaction between these three proteins was not affected by either treatment (Figure 1, B and C), suggesting that endogenous Atg13, ULK1, and FIP200 form a stable protein complex.
Figure 1.
Atg13 forms a complex with ULK1 and FIP200, and is partially dephosphorylated during starvation. (A) HEK293T cells were cotransfected with FLAG-Atg13 and either HA-ULK1 or HA-FIP200. Cell lysates were then subjected to immunoprecipitation (IP) using an anti-FLAG antibody. The resulting precipitates were examined by immunoblot analysis with the indicated antibodies. (B and C) HEK293T cells were subjected to 1-h starvation or rapamycin (100 ng/ml) treatment, and their lysates were immunoprecipitated with anti-ULK1 (B), anti-Atg13 antibody (C) or preimmune rabbit serum (Pre), and analyzed by immunoblot analysis using the indicated antibodies. (D) HEK293T cells, HeLa cells, and MEFs were cultured in complete or starvation medium for the indicated periods. Parts of cell lysates (time 0) were incubated with λ-phosphatase in the presence or absence of phosphatase inhibitors for 15 min and then subjected to immunoblot analysis using anti-Atg13 antibody. Asterisks indicate nonspecific immunoreactive bands. (E) Atg13 is dephosphorylated in FIP200−/− MEFs. FIP200+/+ and FIP200−/− MEFs were cultured in complete or starvation medium for 60 min. Cell lysates were then analyzed by immunoblotting using anti-Atg13 antibody. (F) Atg13 is dephosphorylated in the absence of ULK1. HeLa cells treated with siRNA against ULK1 were analyzed by immunoblotting using anti-Atg13 antibody.
Atg13 Is Hyperphosphorylated and Partially Dephosphorylated during Starvation
Yeast Atg13 is hyperphosphorylated, but rapidly dephosphorylated after nitrogen starvation or rapamycin treatment (Kamada et al., 2000). We tested whether this is also the case in mammalian cells. Endogenous mammalian Atg13 was detected as a smeared band at ∼60–65 kDa (Figure 1D). We found that mammalian Atg13 was also hyperphosphorylated because the position of endogenous Atg13 was downshifted by treatment with λ-phosphatase, and this effect was cancelled by treatment with phosphatase inhibitors (Figure 1D). Atg13 was partially dephosphorylated after starvation in HEK293T and HeLa cells, but it was not clear in MEFs. Thus, dephosphorylation of mammalian Atg13 upon starvation was not as drastic as observed in yeast. We also noticed that Atg13 was downshifted in FIP200−/− MEFs (Figure 1E) and ULK1 siRNA-treated cells (Figure 1F), suggesting that interaction with ULK1 and FIP200 is important for Atg13 phosphorylation.
Atg13 Is Present on Isolation Membranes
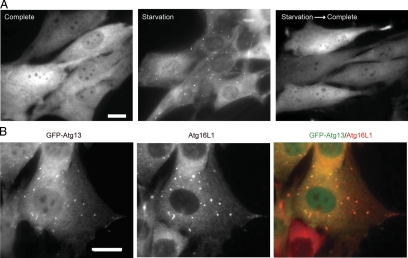
Next we determined the subcellular distribution of Atg13 in NIH3T3 cells stably expressing Atg13 fused with green fluorescent protein (GFP) at the NH2-terminus (GFP-Atg13). Under nutrient-rich conditions, GFP-Atg13 was mostly distributed throughout the cytoplasm (Figure 2A, complete). GFP-Atg13, however, localized to punctuate structures after amino acid and serum starvation (Figure 2A, starvation). These punctuate structures immediately disappeared when the medium was replaced with fresh complete medium (Figure 2A, starvation→complete). These patterns of Atg13 are quite similar to those of ULK1 and FIP200, which are present on isolation membranes (also known as phagophore; Hara et al., 2008). Indeed, GFP-Atg13 almost completely colocalized with Atg16L1, an isolation membrane marker (Mizushima et al., 2003), confirming that Atg13 localizes to isolation membranes (Figure 2B).
Figure 2.
Atg13 localizes to the isolation membrane upon autophagy induction. (A) NIH3T3 cells stably expressing GFP-Atg13 were cultured in regular DMEM (complete) or amino acid-free DMEM without FBS (starvation) for 3 h. They were then cultured in fresh complete medium for an additional 1 h (Starvation→Complete). (B) NIH3T3 cells stably expressing GFP-Atg13 were cultured in starvation medium for 2 h. The cells were then subjected to immunofluorescence microscopy using anti-Atg16L1 antibody and Alexa Fluor 660-conjugated secondary antibody. Scale bars, (A and B) 20 μm.
Atg13 Is Required for Autophagy
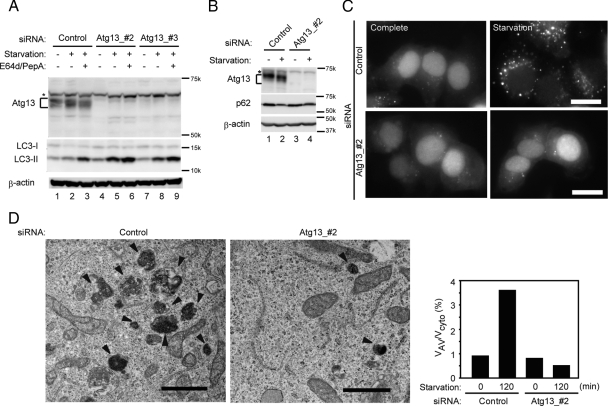
Given that Atg13 interacts with known autophagy proteins and is present on isolation membranes, we next examined the function of Atg13 in mammalian cells. In HeLa cells treated with siRNA against two different regions of Atg13 (Atg13_#2 and Atg13_3#), expression of Atg13 was reduced to almost undetectable levels (Figure 3A). Next, we tested the autophagic activity of these cells. LC3 is one of the mammalian Atg8 homologues and present in two forms: LC3-I (cytosolic form) and LC3-II (membrane-bound form). On induction of autophagy, LC3-I was converted to LC3-II (Figure 3A, lanes 1 and 2). Because LC3-II inside autophagosomes is degraded in autolysosomes, we could see accumulation of LC3-II more clearly if cells were treated with lysosome protease inhibitors (E64d and pepstatin A; Figure 3A, lane 3; Tanida et al., 2005; Mizushima and Yoshimori, 2007). In Atg13_#2 siRNA-treated cells, LC3-II appeared to be generated during starvation, but its amount was not further increased by treatment with lysosome inhibitors (Figure 3A, lanes 4–6), suggesting that the autophagy flux was blocked and LC3-II may be generated in an autophagy-independent manner (see Discussion). The effect of Atg13_#3 siRNA was milder (Figure 3A, lanes 7–9). We also monitored the expression level of p62, a selective autophagy substrate (Bjørkøy et al., 2005; Mizushima and Yoshimori, 2007). The starvation-induced decrease of p62 seen in control cells was almost completely inhibited in Atg13_#2 siRNA-treated cells (Figure 3B).
Figure 3.
Atg13 is essential for autophagy. (A and B) HeLa cells were transfected with two different Atg13 siRNA sets or control siRNA. After 2 d, cells were once again transfected with the same siRNA and cultured for an additional 3 d. Cells were then incubated in regular DMEM or starvation medium in the presence or absence of E64d (50 μM) and pepstatin A (50 μg/ml) for 2 h (A) or in the absence of these inhibitors for 4 h (B). Total lysates were analyzed by immunoblotting using anti-Atg13, anti-LC3, anti-p62, and anti-β-actin antibodies. An asterisk indicates nonspecific immunoreactive bands. (C) HeLa cells stably expressing GFP-LC3 were treated with Atg13 or control siRNA. After 4 d, cells were incubated in complete or starvation medium for 4 h then GFP-LC3 signals were observed by fluorescence microscopy. Scale bar, 20 μm. (D) HeLa cells treated with or without Atg13 siRNA (Atg13_#2) were starved for 2 h with E64d (50 μM) and pepstatinA (50 μg/ml) and then subjected to conventional electron microscopic analysis. Autophagic vacuoles (autophagosomes + autolysosomes) are indicated by arrowheads. Bar, 1 μm. The ratio of the total area of autophagic vacuoles to total cytoplasmic area was determined by morphometric analysis.
Furthermore, we also tested autophagosome formation using HeLa cells stably expressing GFP-LC3. GFP-LC3 was detected diffusely and enriched in the nucleus under normal conditions, although its physiological significance remains unknown (Kuma et al., 2007). GFP-LC3 was translocated to cytoplasmic puncta after autophagy induction in control cells (Figure 3C). However, only a small number of GFP-LC3 puncta were induced by starvation in cells treated with siRNA against Atg13 (Figure 3C). Exclusion of GFP-LC3 from the nucleus was not observed in these cells. We then performed electron microscopy and found that the numbers of autophagic vacuoles were decreased in Atg13 siRNA-treated cells (Figure 3D). Thus, the four independent assays, the LC3 turnover assay, p62 degradation assay, GFP-LC3 dot formation assay and electron microscopy, all suggested that Atg13 is required for autophagosome formation and thereby for the autophagy flux.
Atg13 Forms a 3-MDa Complex Including ULK1 and FIP200
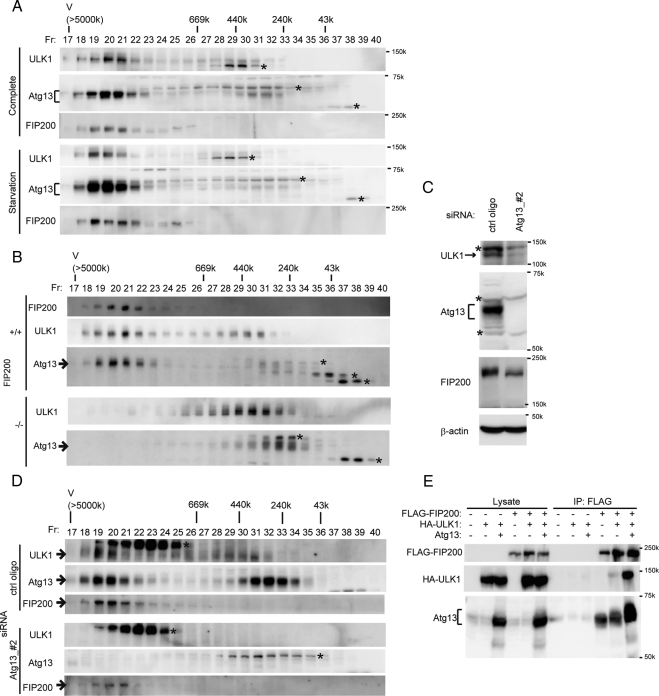
In yeast cells, Atg1, Atg13, Atg17, Atg29, and Atg31 (and probably more factors) form a protein complex (Kamada et al., 2000; Cheong et al., 2005; Kabeya et al., 2005, 2007; Kawamata et al., 2005); however, biochemical information on this complex is limited. We therefore determined how large the ULK1–Atg13–FIP200 complex was in mammalian cells. Cytosol fractions of wild-type MEFs were subjected to gel filtration analysis using a Superose 6 column. ULK1, Atg13, and FIP200 were mainly eluted in a single fraction peak corresponding to ∼3 MDa (Figure 4A). Although we cannot predict its precise molecular weight by gel filtration analysis, these data suggest that ULK1, Atg13, and FIP200 are included in a large protein complex of apparent molecular weight of ∼3 MDa. Much less populations of ULK1 and Atg13 were found in minor peaks of ∼300–500 and ∼200–400 kDa, respectively. Nutrient starvation did not affect formation of the ∼3-MDa ULK1–Atg13–FIP200 complex (Figure 4A), consistent with our immunoprecipitation analysis (Figure 1, B and C). This finding is in marked contrast to previous yeast studies showing that interactions between Atg1, Atg13, and Atg17 are enhanced by starvation treatment (Kamada et al., 2000; Kabeya et al., 2005).
Figure 4.
Atg13 constitutively forms a ∼3-MDa complex including ULK1 and FIP200. (A) MEFs were cultured in complete or starvation medium for 60 min. S100 fractions of the cell lysates (the supernatant obtained after centrifugation at 100,000× g) was separated by size exclusion chromatography on a Superose 6 column. Each fraction was analyzed by immunoblotting with anti-Atg13, anti-ULK1, and anti-FIP200 antibodies. Positions of the molecular mass standards (in kDa) are shown. V, void fraction. Asterisks indicate nonspecific immunoreactive bands. (B) Lysates of wild-type and FIP200−/− MEFs cultured in complete medium were analyzed as described in A. Asterisks indicate nonspecific immunoreactive bands. (C and D) HeLa cells were treated with the control or Atg13 siRNA. Total cell lysates were subjected to immunoblot analysis using the indicated antibodies (C). The S100 fractions were then analyzed by gel filtration as in A (D). Asterisks indicate the nonspecific immunoreactive bands found in human cells. (E) HEK293T cells were cotransfected with FLAG-FIP200, HA-ULK1, and Atg13. Cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG antibody and examined by immunoblot (IB) analysis with anti-Atg13, anti-HA, and anti-FLAG antibodies. Both endogenous and exogenous Atg13 were detected with the anti-Atg13 antibody.
We next determined the role of each mammalian counterpart in formation of the complex. In FIP200−/− MEF cells, no ∼3-MDa complex was generated (Figure 4B). ULK1 and Atg13 were recovered in ∼300–500- and ∼200–400-kDa fractions, respectively. The ULK- and Atg13-positive fractions only partially overlapped. These data suggest that FIP200 is important not only for formation of the ∼3-MDa complex but also for ULK1-Atg13 interaction.
We also analyzed the effect of Atg13 siRNA in HeLa cells because siRNA was the most successful in HeLa cells. In Atg13 siRNA-treated cells, expression levels of not only Atg13 but also ULK1 were markedly reduced, suggesting that Atg13 is important for ULK1 stabilization as is FIP200 (Hara et al., 2008; Figure 4C). The amount of FIP200 was also moderately reduced in Atg13 siRNA-treated cells. In wild-type HeLa cells, Atg13 was found almost equally in both ∼3-MDa and ∼200–400-kDa complexes, whereas most ULK1 and FIP200 were found in the ∼3-MDa complex (Figure 4D). Both Atg13 and ULK1 were not detected in either fraction in Atg13 siRNA-treated cells. Nonetheless, FIP200 was still recovered in the ∼3-MDa (or slightly smaller sized) fractions (Figure 4D). Thus, neither ULK1 nor Atg13 may contribute much to elution volume of this large protein complex. However, we do not know at the moment whether FIP200 is the major component of this complex or whether other proteins are included. We next tested whether Atg13 is important only for stability of ULK1 and FIP200 or also for interaction between ULK1 and FIP200 through an overexpression experiment. Interaction between exogenously introduced FIP200 and ULK1 was markedly enhanced when Atg13 was simultaneously overexpressed (Figure 4E). Yeast two-hybrid analysis also demonstrated interactions between Atg13 and FIP200, and Atg13 and ULK1, suggesting that these are direct interactions (Supplemental Figure S4). Taken together, these data suggest that the ULK1-FIP200 interaction highly depends on Atg13.
mTORC1 Interacts with the ∼3-MDa Complex under Nutrient-rich Conditions
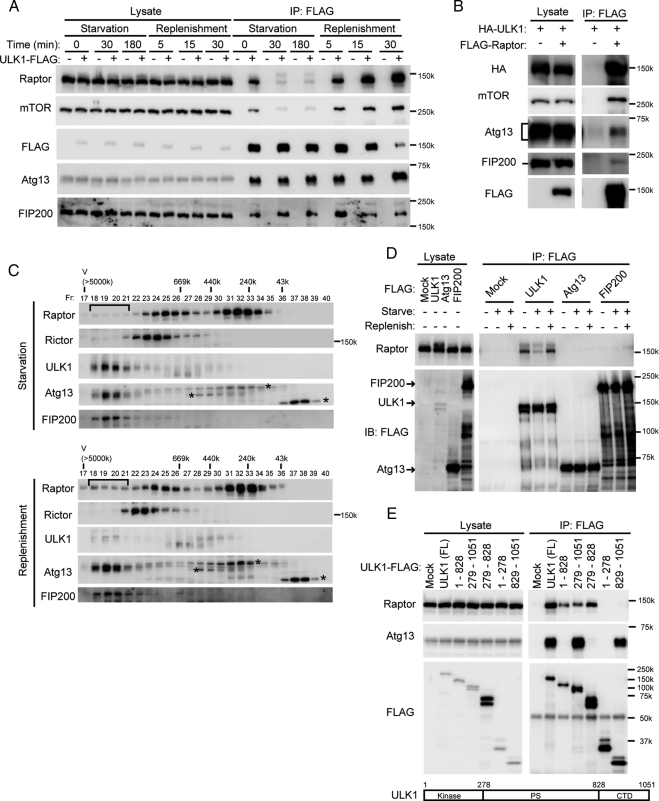
In contrast to the dynamic Atg1–Atg13–Atg17 complex formation in yeast, mammalian ULK1–Atg13–FIP200 formation is rather stable. We sought to further elucidate the regulation mechanism of autophagy through this complex and searched for additional ULK1-interacting molecules by the same method under several nutrient conditions. As a result, we detected raptor in a sample prepared shortly after amino acid restimulation after 3-h starvation, but not in samples derived from starved cells. Raptor is a scaffold that recruits downstream substrates to mTORC1 (Wullschleger et al., 2006; Guertin and Sabatini, 2007; Yang and Guan, 2007). We confirmed that ULK1 interacts with raptor and mTOR under nutrient-rich conditions, but these interactions were markedly diminished during starvation, whereas ULK1 interacted with Atg13 and FIP200 constantly (Figure 5A). When cells were stimulated with the fresh complete medium, ULK1 again interacts strongly with raptor and mTOR within 5 min (Figure 5A). When lysates of replenished cells were precipitated with FLAG-raptor, mTOR, ULK1, Atg13, and FIP200 were detected, suggesting that mTORC1 is associated with the ∼3-MDa complex (Figure 5B). In gel filtration experiments, raptor was broadly distributed but was not included in the ∼3-MDa complex under starvation conditions (Figure 5C). However, after a shift to nutrient-rich medium, a significant population of raptor was recovered in ∼3-MDa fractions. Nutrient replenishment did not cause incorporation of rictor, a specific component of mTORC2, into the ∼3-MDa complex. These data suggest that mTORC1 is incorporated into the large ULK1–Atg13–FIP200 complex in a nutrient-dependent manner.
Figure 5.
mTORC1 interacts with the ∼3-MDa complex under nutrient-rich conditions. (A) HEK293T cells were transfected with an empty vector or ULK1-FLAG. Cells were then cultured in complete or starvation medium as indicated. In the restimulation experiments (replenishment), starved cells were additionally cultured in fresh complete medium for the indicated times. Cell lysates were subjected to immunoprecipitation (IP) using M2 agarose beads. The resulting precipitates were examined by immunoblot analysis with the indicated antibodies. (B) HEK293T cells were cotransfected with FLAG-raptor and HA-ULK1 and analyzed as described in A (30-min replenishment condition). (C) NIH-3T3 cells were cultured in starvation medium for 180 min. In the replenishment experiments, starved cells were cultured in fresh complete medium for additional 30 min. S100 fractions were separated as described in Figure 4A. The shift of the raptor-positive fractions is indicated. Asterisks indicate the nonspecific immunoreactive bands. (D) HEK293T cells transfected with FLAG-tagged ULK1, Atg13, and FIP200 were subjected to 3-h starvation and 30-min replenishment treatments. Cell lysates were immunoprecipitated with anti-FLAG antibody, and coprecipitation of endogenous raptor was analyzed using anti-raptor antibody. (E) HEK293T cells were transfected with FLAG-tagged ULK1 mutants, and interaction with raptor and Atg13 was determined.
We next determined which component of the complex directly interacts with raptor. Endogenous raptor was efficiently coprecipitated with overexpressed ULK1-FLAG, but not with FLAG-Atg13 and FLAG-FIP200, in a nutrient-dependent manner (Figure 5D). Raptor interacts with the PS domain of ULK1 (Figure 5E). As Chan et al. (2009) reported, the C-terminal region of ULK1 is required and sufficient for binding to Atg13 (Figure 5E and Supplemental Figure S4). However, raptor could still interact with a ULK1 mutant lacking the C-terminal region (Figure 5E). Thus, Atg13 does not mediate the raptor-ULK1 interaction. Although we could not entirely exclude a possibility that some unknown factor might mediate the interaction, all these data suggest that raptor interacts with the ∼3-MDa complex through ULK1.
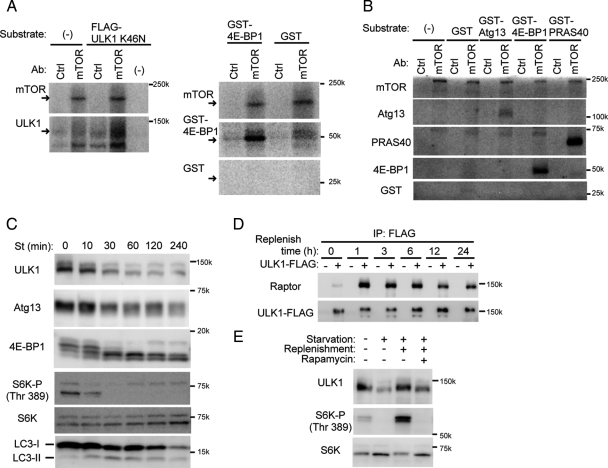
mTOR Phosphorylates ULK1 and Atg13
Because raptor directly interacts with ULK1, we hypothesized that ULK1 could be a substrate of mTOR. We therefore purified mTORC1 from nonstarved cells and FLAG-ULK1K46N (a kinase-dead mutant) from starved cells and subjected them to in vitro kinase assay. We used ULK1K46N in this assay to avoid autophosphorylation. 32P incorporation into ULK1K46N was remarkably enhanced when incubated with mTOR precipitates (Figure 6A). We also detected phosphorylation of Atg13 by the mTOR precipitates (Figure 6B). These data suggest that both ULK1 and Atg13 could be novel mTOR substrates. This is consistent with our observation that Atg13 is partially dephosphorylated under starvation conditions (Figure 1D), during which mTORC1 dissociates from the ∼3-MDa complex. We then tested whether the phosphorylation status of ULK1 also changes during starvation. ULK1 was detected as a slightly smeared band in MEFs and was downsized during starvation, as was Atg13 (Figure 6C). Dephosphorylation of other mTORC1 substrates such as S6-kinase 1 and 4E-BP1, and LC3 conversion were also observed in similar time courses. Conversely, the ULK1 band was shifted up upon restimulation by amino acids and FBS (Figure 6D). This shift correlated well with the interaction with raptor, which was induced by nutrient stimulation and then gradually attenuated. It was determined that the ULK1 band shift was mediated by mTOR because rapamycin treatment under nutrient-rich conditions caused a mobility shift similar to that seen during starvation (Figure 6E). All these data suggest that ULK1 is a bona fide substrate of mTOR, and Atg13 is a second mTOR substrate that binds mTORC1 through ULK1.
Figure 6.
TOR phosphorylates ULK1 and Atg13. (A and B) Endogenous mTOR was immunoprecipitated from HEK293T cells cultured in complete medium with anti-mTOR antibody as described in Materials and Methods. As a control, normal mouse IgG was used for immunoprecipitation. The resulting precipitates were subjected to mTOR kinase assay using FLAG-tagged ULK1K46N (a kinase-dead mutant) (A) or Atg13 (B) as the substrate. GST-PRAS40 and GST-4E-BP1 were used as positive controls, and GST alone was used as a negative control. (C) HEK293T cells were cultured in starvation medium for the indicated times. Total cell lysates were analyzed by immunoblot analysis. (D) HEK293T cells were transfected with an empty vector or ULK1-FLAG. Cells were then starved for 180 min, followed by culturing in fresh complete medium for the indicated times. Cell lysates were analyzed as described in Figure 5A. (E) NIH3T3 cells were cultured in complete or starvation medium for 180 min. In the replenishment experiments, starved cells were pretreated with 100 ng/ml rapamycin or vehicle (DMSO) for 30 min before replenishment and then cultured in fresh complete medium for an additional 30 min with and without rapamycin.
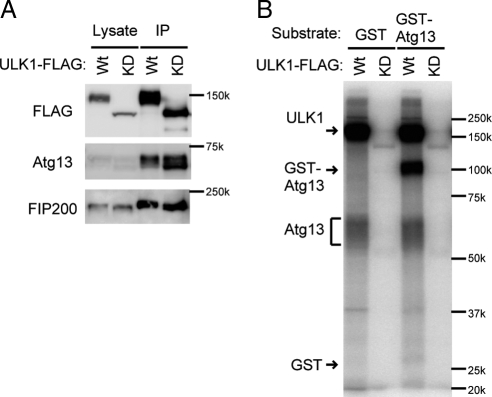
ULK1 Phosphorylates Atg13
Atg13 is dephosphorylated in ULK1 siRNA-treated cells (Figure 1F). A recent report also demonstrated that overexpression of ULK1 or ULK2 caused upward shift of the Atg13 band pattern (Chan et al., 2009). These data suggest that Atg13 could be a direct substrate of ULK1 as well as that of mTOR. We, thus, tested this possibility by in vitro phosphorylation assay. The wild-type and kinase-dead ULK1 complexes were prepared from starved HEK293 cells (Figure 7A) and subjected to the kinase assay using recombinant GST-Atg13 as a substrate. We found that the ULK1 complex directly phosphorylates Atg13 in vitro (Figure 7B). Therefore, Atg13 could be phosphorylated by both mTOR and ULK1, which are activated under nutrient-rich and starvation conditions, respectively (Hara et al., 2008). This complexity might account for the partial dephosphorylation of Atg13 during starvation.
Figure 7.
ULK1 phosphorylates Atg13 in vitro. (A and B) ULK1-FLAG (Wt) and kinase-dead ULK1K46N-FLAG mutant (KD) were immunoprecipitated from HEK293T cells cultured in the starvation medium for 1 h. The resulting precipitates were subjected to immunoblot analysis (A) or ULK1 kinase assay (B) using GST-Atg13 and GST (negative control) as substrates.
DISCUSSION
We identified a mammalian ortholog of yeast Atg13, and found that mammalian Atg13 is incorporated into the ∼3-MDa protein complex together with ULK1 and FIP200. Furthermore, we showed that mTORC1 also interacts with this huge complex in a nutrient dependent manner.
This and previous studies (Young et al., 2006; Chan et al., 2007, 2009; Hara et al., 2008) demonstrated that each component of the ULK1–Atg13–FIP200 complex is essential for autophagy, likely for autophagosome formation. In the present study, we observed accumulation of LC3-II in Atg13 siRNA-treated cells (Figure 3A). We cannot rule out the possibility that the effect of Atg13 siRNA was not sufficient to completely inhibit LC3 conversion. However, we have observed similar phenomenon in FIP200−/− cells (Hara et al., 2008). The amount of LC3-II was increased during starvation in these cells, which was not further increased by lysosome inhibitor treatment. Accumulation of LC3-II was also observed in Atg14 and Vps34 siRNA-treated cells (Itakura et al., 2008) and Beclin 1 siRNA-treated cells (Zeng et al., 2006). Even in yeast, it is known that neither the Atg1 complex nor the Atg14-PI3K complex is essential for Atg8-PE formation (Suzuki et al., 2001; Obara et al., 2008). We speculate that inhibition of these Atg proteins may cause ectopic accumulation of LC3-II on unknown aberrant membranes, not on autophagosome.
Although (m)TOR is known to be a main regulator of autophagy, how (m)TOR regulates the autophagy factors has not yet been determined. We showed that mTORC1 directly interacts with ULK1 and phosphorylates ULK1 and Atg13. These results were also independently found by another group (Jung et al., 2009). Furthermore, Tor-dependent regulation/phosphorylation of Atg1 and Atg13 was also demonstrated in Drosophila (Chang and Neufeld, 2009). We could not find in the ULK1 and Atg13 sequences a classic TOR signaling (TOS) motif, which is thought to be a potential recognition sequence by raptor. However, raptor recognition sequences may not be as strictly conserved as previously predicted (Oshiro et al., 2007). Importantly, mTORC1-ULK1 binding and ULK1 phosphorylation were rapidly accelerated by nutrient replenishment. We propose that mTORC1 suppresses autophagy through direct interaction with the ∼3-MDa ULK1–Atg13–FIP200 complex. We do not think that this is the only regulatory mechanism of autophagy because it has also been reported that there is an mTOR-independent pathway for autophagy induction (Sarkar et al., 2005), and the effect of starvation is larger than rapamycin treatment (Supplemental Figure S3). Determining whether this ∼3-MDa autophagy complex can also be regulated by factors other than mTORC1 is therefore an important future issue.
The mammalian ULK1–Atg13–FIP200 complex shares common features with the yeast Atg1–Atg13–Atg17 complex. Association with each component appears to be interdependent. In yeast, Atg1–Atg17 interaction largely depends on Atg13 (Cheong et al., 2005; Kabeya et al., 2005). In the present article, we showed similar interdependency among the mammalian factors; FIP200 and Atg13 are important for ULK1-Atg13 and ULK1-FIP200 interactions, respectively (Figure 4, B and E). In addition, the C-terminal region of ULK1 is important for binding to Atg13 (Figure 5E and Supplemental Figure S4; Chan et al., 2009) and FIP200 (Hara et al., 2008). In yeast, the C-terminal region of Atg1 is also known to be required for interaction with Atg13 (Cheong et al., 2008). Because of these similarities, we again propose that FIP200 could be a true mammalian counterpart of yeast Atg17, although it is possible that FIP200 has additional functions similar to those of other Atg17-interacting proteins such as Atg11, Atg29, and Atg31 because counterparts of these factors have not yet been identified in mammals.
However, there are several apparent differences between the mammalian and yeast complexes. In yeast, the Atg1–Atg13 interaction is enhanced during starvation, whereas it is stable in mammals. Yeast Atg13 is dramatically dephosphorylated during starvation, whereas this event is only modest in mammalian cells. More importantly, Atg13 is thought to be an upstream factor of Atg1 in yeast for the following reasons: 1) overexpression of Atg1 partially restores the autophagy-defective phenotype of the atg13 mutant (Funakoshi et al., 1997), 2) Atg13 is important for Atg1 kinase activity (Kamada et al., 2000), and 3) PAS targeting of Atg1 is mildly affected in Δatg13, whereas in the Δatg1 mutant PAS targeting of Atg13 is enhanced (Suzuki et al., 2007). However, we demonstrated in this study that mTORC1 directly binds and phosphorylates ULK1 and that raptor-ULK1 interaction is not mediated by Atg13. We therefore postulate that Atg13 might function as a regulator in the complex, rather than a signal mediator between (m)TOR and Atg1/ULK1, although we do not rule out the possibility that the signaling mechanisms of yeast and mammalian cells are different. Further analysis of the function of the ULK1–Atg13–FIP200 complex will provide more insight into the link between autophagy regulation and autophagosome formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Thomas Neufeld and Do-Hyung Kim for discussion and sharing unpublished data, Drs. Masaaki Muramatsu (Tokyo Medical and Dental University) and Noriko Okazaki (Kazusa DNA Research Institute) for providing the ULK1 constructs, Dr. Kenta Hara (Kobe University) for providing the FLAG-raptor plasmid, Dr. Toshio Kitamura (The University of Tokyo) for the retroviral vectors and Plat-E cells, and Dr. Philip James (University of Wisconsin) for providing the yeast strains and vectors necessary for the yeast two-hybrid analysis. We also thank Maki Maeda for technical support during chromatography and Drs. Ushio Kikkawa (Kobe University) and Ei-ichiro Suzuki (Ajinomoto Co.) for continuous support throughout this project. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (N.M.), grants for research fellowships from the Japan Society for the Promotion of Science for Young Scientists (N.H.), and the Toray Science Foundation, and Takeda Science Foundation (N.M.).
Abbreviations used:
- ATG
autophagy-related genes
- FIP200
Focal adhesion kinase family interacting protein of 200 kDa
- GFP
green fluorescent protein
- LC3
microtubule-associated protein light chain 3
- MEF
mouse embryonic fibroblast
- (m)TOR
(mammalian) target of rapamycin
- mTORC
mTOR complex
- ULK
uncoordinated 51-like kinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1248) on February 11, 2009.
REFERENCES
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Øvervatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y.W., Kir S., Tooze S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chang Y.-Y., Neufeld T. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codogno P., Meijer A. J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Gan B., Guan J. L. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787–794. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 2006;175:121–133. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D. A., Sabatini D. M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Hara T., Kamura T., Kotoshiba S., Takahashi H., Fujiwara K., Onoyama I., Shirakawa M., Mizushima N., Nakayama K. I. Role of the UBL-UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol. Cell. Biol. 2005;25:9292–9303. doi: 10.1128/MCB.25.21.9292-9303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara Y., Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Jun C. B., Ro S.-H., Kim Y.-M., Otto N. M., Cao J., Kim D.-H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kawamata T., Suzuki K., Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Suzuki K., Kuboshima N., Akimatsu H., Ota S., Ohsumi M., Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 2005;338:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Meijer A. J., Codogno P., Neufeld T. P., Scott R. C. Autophagy and p70S6 kinase. Autophagy. 2005;1:59–61. doi: 10.4161/auto.1.1.1536. [DOI] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- Kundu M., Lindsten T., Yang C. Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A., Thompson C. B. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood, 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A. J., Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol. Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J.A.K.W. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. How to Interpret LC3 Immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Natsume T., Yamauchi Y., Nakayama H., Shinkawa T., Yanagida M., Takahashi N., Isobe T. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 2002;74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- Nishiuma T., Hara K., Tsujishita Y., Kaneko K., Shii K., Yonezawa K. Characterization of the phosphoproteins and protein kinase activity in mTOR immunoprecipitates. Biochem. Biophys. Res. Commun. 1998;252:440–444. doi: 10.1006/bbrc.1998.9671. [DOI] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Obara K., Noda T., Niimi K., Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–547. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Oshiro N., et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarkar S., Floto R. A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L. J., Rubinsztein D. C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Tomoda T., Bhatt R. S., Kuroyanagi H., Shirasawa T., Hatten M. E. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron. 1999;24:833–846. doi: 10.1016/s0896-6273(00)81031-4. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan J., Kuroyanagi H., Kuroiwa A., Matsuda Y., Tokumitsu H., Tomoda T., Shirasawa T., Muramatsu M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- Yan J., et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- Yang Q., Guan K. L. Expanding mTOR signaling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- Young A. R., Chan E. Y., Hu X. W., Kochl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zeng X., Overmeyer J. H., Maltese W. A. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 2006;119:259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.