Abstract
Autophagy, the starvation-induced degradation of bulky cytosolic components, is up-regulated in mammalian cells when nutrient supplies are limited. Although mammalian target of rapamycin (mTOR) is known as the key regulator of autophagy induction, the mechanism by which mTOR regulates autophagy has remained elusive. Here, we identify that mTOR phosphorylates a mammalian homologue of Atg13 and the mammalian Atg1 homologues ULK1 and ULK2. The mammalian Atg13 binds both ULK1 and ULK2 and mediates the interaction of the ULK proteins with FIP200. The binding of Atg13 stabilizes and activates ULK and facilitates the phosphorylation of FIP200 by ULK, whereas knockdown of Atg13 inhibits autophagosome formation. Inhibition of mTOR by rapamycin or leucine deprivation, the conditions that induce autophagy, leads to dephosphorylation of ULK1, ULK2, and Atg13 and activates ULK to phosphorylate FIP200. These findings demonstrate that the ULK-Atg13-FIP200 complexes are direct targets of mTOR and important regulators of autophagy in response to mTOR signaling.
INTRODUCTION
When nutrient supplies are limited, eukaryotic cells undergo autophagy, an evolutionarily conserved process through which cytoplasm, organelles, or long-lived proteins or protein aggregates are sequestered in a double-membrane vesicle and subsequently degraded in lysosomes (Klionsky and Ohsumi, 1999). Through destruction of cellular organelles and proteins, autophagy provides energy for starved cells or allows for the balanced regulation between biogenesis and degradation of cellular structures, thereby playing essential roles for growth, survival, differentiation, and development (Neufeld and Baehrecke, 2008; Tsukamoto et al., 2008; Vellai et al., 2008). Dys-regulation of autophagy is associated with many human diseases, including cancer, myopathies, and Parkinson's and Huntington's diseases, and also is involved in degradation of invasive microbes (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003; Gutierrez et al., 2004; Ravikumar et al., 2004; Ogawa et al., 2005; Sugimoto, 2007; Levine and Kroemer, 2008; Mizushima et al., 2008). Despite the widespread importance of autophagy, the molecular mechanism underlying autophagy induction by starvation remains unclear.
Genetic studies in Saccharomyces cerevisiae have revealed the important functions of two protein kinases, TOR and Atg1, in autophagy induction (Noda and Ohsumi, 1998; Kamada et al., 2000). TOR is a serine/threonine protein kinase whose activity is inhibited by nutrient starvation, and its inhibition induces autophagy (Noda and Ohsumi, 1998; Schmelzle and Hall, 2000; Scott et al., 2004). Atg1 is also a serine/threonine kinase, and its activity is enhanced under starvation in yeast (Kamada et al., 2000). The significance of Atg1 in regulating autophagy has been shown in several eukaryotes including S. cerevisiae, Dictyostelium discoideum, Drosophila melanogaster, and Caenorhabditis elegans (Meléndez et al., 2003; Otto et al., 2004; Scott et al., 2004, 2007). In S. cerevisiae, Atg1 null or kinase-dead mutant strains have shown defects in autophagy even under nutrient starvation or when TOR is inhibited, supporting that Atg1 plays downstream of TOR (Matsuura et al., 1997; Kamada et al., 2000).
Because TOR was identified as an upstream kinase of Atg1, the mechanism by which TOR regulates Atg1 has been an important question regarding the molecular cascade of autophagy induction. In yeast, Atg1 interacts with the autophagy regulatory proteins Atg13 and Atg17 (Funakoshi et al., 1997; Kamada et al., 2000; Cheong et al., 2005). Atg13 is phosphorylated in a TOR-dependent manner and is required for Atg1 activity, autophagy induction, and the cytoplasm-to-vacuole targeting (Cvt; Kamada et al., 2000). Atg17, which is also required for Atg1 activity, is specifically involved in autophagy but not in the Cvt pathway and is proposed to play a scaffold role (Suzuki et al., 2007). In mammals, two Atg1 homologues ULK1 and ULK2 have been identified and investigated (Chan et al., 2007; Hara et al., 2008; Kundu et al., 2008). Although overexpression of the kinase-dead mutants of ULK1 or ULK2 led to inhibition of autophagy, their roles in the regulation of autophagy have not been clearly defined. Until recently, the lack of knowledge on Atg13 and Atg17 homologues in higher eukaryotes has limited our understanding of the mechanism underlying autophagy induction. With this regard, a putative mammalian homologue of Atg13 that was recently predicted by a sequence homology search has revealed important values (Meijer et al., 2007). Also, a focal adhesion kinase family–interacting protein of 200 kDa (FIP200) was identified as a binding protein of ULK1 and ULK2 and was proposed as having the functional and structural features of Atg17 (Hara et al., 2008). These putative homologues of Atg13 and Atg17 have led us to investigate the roles of mTOR in the regulation of ULK more closely.
In this study, we have determined that the putative human homologue of Atg13 binds both ULK1 and ULK2 and plays an important role for autophagosome formation. We identified that Atg13 as well as ULK1 and ULK2 are phosphorylated by mTOR and the mTOR-mediated phosphorylations act inhibitory to ULK activity. Our study has revealed that Atg13 mediates the interaction between FIP200 and ULK and is required for the phosphorylation of FIP200 by ULK in response to starvation. From this study, we propose that ULK, Atg13, and FIP200 constitute the protein complexes that are direct effectors of mTOR and mediate nutrient signaling to the autophagy machinery.
MATERIALS AND METHODS
Chemicals and Antibodies
Anti-mTOR (sc-1549), ULK1 (sc-10900 and sc-33182), epidermal growth factor receptor (sc-03), Fas (sc-20140), p21 (sc-397), tubulin (sc-12462), GAPDH (sc-25778) and 14-3-3 (sc-732) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against ULK2, FIP200, and p62 were obtained from Abcam (ab56736; Cambridge, MA), the Bethyl Laboratories (no. A301-536A; Montgomery, TX), and BD Biosciences (no. 610832; San Jose, CA), respectively. Antibodies against S6K1 (9202) and phospho-S6K1 Thr389 (p-S6K1; 9205) were from Cell Signaling Technology (Danvers, MA). Anti-hemagglutinin (HA) antibody (HA.11) was from Covance (Berkeley, CA). Anti-Myc 9E10 mAb and rapamycin were from EMD Biosciences (San Diego, CA). Glutathione 4B beads were from GE Healthcare (Piscataway, NJ). Anti-Atg13, ULK1, and ULK2 antibodies were generated by the Yenzym custom antibody service (South San Francisco, CA) from rabbits using glutathione S-transferase (GST)-tagged full-length proteins or epitope peptides CETDLNSQDRKDLDK-amide and CDLGTFYREFQNPPQ-amide (Atg13) and MEPGRGGTETVGKFEFSC-amide (ULK1) as antigens.
DNA Constructions
Mouse and human ULK1 cDNA clones were obtained from Dr. Takahiko Shimizu (Tokyo Metropolitan Institute of Gerontology) and the Katzusa Institute, respectively. Human ATG13 homolog cDNAs for three isoforms were obtained from the Katzusa Institute (KIAA0652; isoform 1) and the Open Biosystems (Huntsville, AL; Image no. 2961127 and 3936851; isoforms 2 and 3). ULK1 and Atg13 fragments were obtained by PCR amplification and subcloned into HA- and myc-prk5 vector. The kinase-dead mutant of ULK1, M92A, was made by using the site-directed mutagenesis kit (Stratagene, La Jolla, CA) and oligonucleotides listed in Supplemental Table S1. The cDNAs for human FIP200 (Image no. 3908134) and ULK2 (KIAA0623) constructs were obtained from the Open Biosystems and the Katzusa Institute, respectively.
Cell Culture and Transfection
HEK293T, HeLa, and mouse embryonic fibroblast (MEF) cells were cultured in DMEM containing 10% fetal bovine serum and penicillin/streptomycin at 37°C in 5% CO2. For transient expression of proteins, HEK293T cells were transfected with recombinant DNAs or short hairpin RNA (shRNA) plasmids using FuGENE 6 (Roche Applied Science, Indianapolis, IN) following the manufacturer's protocol. Cells were harvested 2 d after transfection for coimmunoprecipitation assay or other biochemical or Western blot analysis.
Lentiviral Preparation and Viral Infection
Lentiviral shRNA transduction was performed as described previously (Vander Haar et al., 2007). Briefly, the pLKO shRNA vectors encoding shRNAs were transfected into HEK293T cells with lentiviral packaging vectors pHR'8.2ΔR and pCMV-VSV-G using FuGENE 6. Viruses were collected 60 h after transfection, and HeLa or 293T cells were infected with the collected viruses for 4 h in the presence of polybrene. The target sequences for Atg13, ULK1, and ULK2 are listed in Supplemental Table S1.
Microscopic Analysis of Autophagosome Formation
Viral-transduced cells were seeded onto glass coverslips. Next day, medium was refreshed with DMEM containing 10% fetal bovine serum, and after an additional 24 h, autophagy was induced by 100 nM rapamycin or vehicle (methanol or DMSO) in the presence of pepstatin (10 μg/ml), an inhibitor of the lysosomal protein degradation (Sigma). After 2, 5, or 24 h of induction, cells were fixed with formaldehyde, permeabilized using 1% Triton X-100, and stained with 4, 6-diamidinophenylindole (DAPI) and anti-LC3 antibody from either Nanotools (France) or Abcam. Stained cells were visualized under a confocal microscope attached to an IX70 inverted microscope (Olympus, Melville, NY). Multiple fields at random positions were collected for each condition. Quantification of LC3-labeled structures in the images was performed by counting the number of LC3-positive cells.
Coimmunoprecipitation and Western Blotting
Whole cell extracts were prepared in a lysis buffer containing 0.3% Chaps as described in Kim et al. (2002) and immunoprecipitated with antibodies described for each experiment. Precipitated proteins were washed four times with the lysis buffer, loaded onto 8% Tris-glycine gels, transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Richmond, CA), and detected with enhanced chemiluminescence (ECL) Western blotting detection reagents (Perkin Elmer-Cetus, Norwalk, CT).
GST Pulldown Assay
The DNA constructs for GST-tagged Atg13, ULK1 (651-end), ULK2 (651-end), and FIP200 (860-end) were cloned in the Escherichia coli expression plasmid pGEX-6P-2 (Amersham Biosciences, Piscataway, NJ) and introduced into BL21(DE3) cells (EMD Biosciences, San Diego, CA). The GST fusion proteins were expressed by induction with 0.1 mM isopropyl-1-thio-b-galatopyranoside for 16 h and purified with glutathione-Sepharose 4B beads according to the manufacturer's protocol.
Western Blot Assay of Autophagy
The lentiviral shRNA-transduced cells or MEF cells were treated with rapamycin or vehicle for 4 h in the presence or absence of pepstatin A (10 μg/ml) and E-64 (10 μg/ml). Cell lysates were run on SDS-PAGE, and proteins were transferred to PVDF membranes and probed with anti-LC3 mouse polyclonal antibody (Nanotools) and anti-p62 antibody (Bethyl Laboratories).
In Vivo Labeling
293T cells on 6-cm plates were transduced with plasmids and washed with phosphate-free medium (Invitrogen) twice and incubated with the phosphate-free medium containing 10% dialyzed fetal bovine serum for 4 h before 0.1 mCi [32P]orthophosphate was added. Cells were then treated with rapamycin at 50 nM for 1 h in the presence of the isotope. Myc-ULK1 or Atg13 immunoprecipitates were obtained by immunoprecipitation using anti-myc antibody, run on SDS-PAGE, and transferred to PVDF membrane, and an autoradiogram was obtained.
In Vitro Kinase Assay
For ULK kinase assay, endogenous ULK1 or recombinant ULK1 or ULK2 were isolated by immunoprecipitation using anti-ULK1 antibody (Santa Cruz Biotechnology, sc-10900) or anti-myc antibody (9E10, EMD Biosciences) from 293T cells. The reaction buffer contained 25 mM MOPS, pH 7.5, 1 mM EGTA, 0.1 mM Na3VO4, 15 mM MgCl2, and hot and cold ATP at 100 μM final concentration. As substrates, myelin basic protein (MBP; Sigma) or the recombinant proteins Atg13 and FIP200 (860-end) purified from E. coli expressing GST-tagged Atg13 or FIP200 were used at 1 μg for each reaction. As for mTOR kinase assay, mTOR or myc-tagged immunoprecipitates were obtained from 293T cells using anti-mTOR antibody (Santa Cruz Biotechnology, sc-1549) or anti-myc antibody (9E10) or a purified, active form of mTOR (Millipore, Bedford, MA) was used. As substrates, Atg13, ULK1 (651-end), and ULK2 (651-end) were purified from E. coli. The kinase reaction was performed in a buffer containing 50 mM HEPES, pH 7.5, 1 mM EGTA, 0.01% Tween 20, 10 mM MnCl2, 2.5 mM DTT, 100 μM ATP, 1 μg substrate, and the trace [γ-32P]ATP for 30 and 60 min at 30°C. All the kinase reactions were stopped by adding 5× SDS-sample loading buffer.
RESULTS
Atg13 Binds ULK1, ULK2, and FIP200
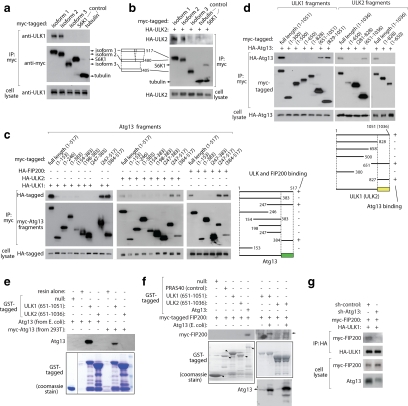
From the genome database, the human gene KIAA0652 was predicted to encode a homologue of yeast ATG13 (Meijer et al., 2007). We cloned its cDNA encoding 517 amino acids and tested if the protein encoded by the gene can interact with ULK1 and ULK2. Using recombinant constructs, we determined that Atg13 is coimmunoprecipitated with ULK1 or ULK2 but not with control proteins (Figure 1, a–c). We generated polyclonal antibodies specific to the Atg13 N-terminus or its full length. Using the antibodies, we confirmed that endogenous ULK1 and Atg13 interact with each other (Figure 1d and Supplemental Figure S1). We also observed that ULK2 is coimmunoprecipitated with Atg13, although the affinity of the interaction between Atg13 and ULK2 is weaker than the interaction between Atg13 and ULK1 (Figure 1b).
Figure 1.
Atg13 interacts with ULK1, ULK2, and FIP200. (a) ULK1 and ULK2 are coimmunoprecipitated with Atg13. HA-tagged ULK1 was expressed with myc-tagged Atg13 or control proteins (S6K1 and tubulin) in 293T cells. Anti-myc immune complexes were assessed for the presence of HA-ULK1 and HA-ULK2 by Western blotting. (b) Atg13 binds ULK1 and ULK2. Myc-tagged Atg13 was expressed with HA-tagged ULK1, ULK2, or control proteins (S6K1, tubulin) in 293T cells. Anti-HA immune complexes were isolated and the amount of Atg13 was assessed by Western blotting. (c) Recombinant Atg13 is coimmunoprecipitated with ULK1. Myc-tagged Atg13 or control proteins was expressed in 293T cells. Anti-myc immune complexes were isolated, and the presence of endogenous ULK1 was assessed by Western blotting. (d) Endogenous Atg13 is coimmunoprecipitated with ULK1. ULK1 immunoprecipitate was obtained from 293T cells in the presence (+pep) or absence (−pep) of ULK1 antibody epitope peptides and the amounts of Atg13 in the immune complexes were assayed by Western blotting. (e) Atg13 interacts with FIP200. Myc-tagged Atg13 or tubulin (control) was coexpressed with HA-FIP200 in 293T cells. Anti-myc immune complexes were analyzed for the presence of FIP200 on Western blots. (f) FIP200 interacts with Atg13, ULK1, and ULK2. HA-tagged Atg13, ULK1, and ULK2 were coexpressed with myc-tagged FIP200 in 293T cells. HA immunoprecipitates were analyzed for the amount of myc-FIP200 on Western blots.
Knowing that Atg13 interacts with ULK1 and ULK2, we wondered if Atg13 interacts with FIP200, which has been shown to interact with ULK1 and ULK2 (Hara et al., 2008). FIP200 was detected in immune complexes containing Atg13 but not in control immune complexes (Figure 1e). Furthermore, the amount of FIP200 in Atg13 immune complexes was higher than those in ULK1 and ULK2 immune complexes, supporting that Atg13 might bind FIP200 more tightly than ULK1 and ULK2 (Figure 1f). It was noteworthy that coexpression with either ULK1 or ULK2 induced upward shifts of Atg13 and FIP200 bands on SDS-PAGE. This supports that ULK1 and ULK2 likely induce phosphorylation of Atg13 and FIP200 (Figure 1, b and f).
Atg13 Directly Interacts with ULK1 and Mediates the Interaction between ULKs and FIP200
A polyclonal antibody generated against the full length of Atg13 allowed us to detect several isoforms in human cell lines (Supplemental Figure S1). We searched for Atg13 cDNA sequences that are available in the NCBI database and found several splicing variants. We cloned three of the isoforms and tested their interaction with ULKs. We found that deletion of the C-terminal 75 amino acids disabled the ability of Atg13 to bind ULK1 and ULK2 (Figure 2, a and b, and Supplemental Figure S1). Consistent with an important role of the C-terminal residues of Atg13 for ULK binding, ULK1-, ULK2-, and FIP200-binding regions on Atg13 were all mapped to C-terminal residues 384-517 (Figure 2c). Atg13-binding sites on ULK1 and ULK2 were mapped to C-terminal regions containing residues 829-1051 and 651-1036, respectively (Figure 2d).
Figure 2.
Atg13 interacts with ULK C-terminus and mediates the interaction between ULKs and FIP200. (a and b) Atg13 isoforms 1 and 2, but not isoform 3, bind ULK1 and ULK2. Myc-tagged Atg13 isoforms were expressed alone (a) or with HA-ULK2 (b) in 293T cells, and the amounts of endogenous ULK1 or HA-ULK2 in myc immunoprecipitaes was analyzed by Western blotting. (c) ULK1, ULK2, and FIP200 interact with Atg13 C-terminus. Myc-tagged fragments of Atg13 were coexpressed with HA-tagged ULK1, ULK2, or FIP200 in 293T cells. The levels of HA-constructs coimmunoprecipitated with myc-tagged Atg13 fragments were analyzed on Western blots. (d) Atg13 interacts with ULK C-terminus. Myc-tagged fragments of ULK1 or ULK2 were coexpressed with HA-Atg13 in 293T cells. The amount of HA-Atg13 recovered with myc-tagged constructs was analyzed on Western blots. (e) Atg13 directly interacts with ULK1. GST alone or GST-tagged ULK1 or ULK2 fragments were incubated with either E. coli–purified Atg13 or myc-Atg13 expressed in 293T cells, and the amounts of Atg13 recovered with GST-tagged proteins were analyzed by Western blotting. (f) Atg13 interacts with FIP200 directly and mediates the interaction between FIP200 and ULKs. GST-tagged ULK1 and ULK2 C-terminal fragments purified from E. coli were incubated with myc-tagged FIP200 expressed in 293T cells in the presence or absence of Atg13 purified from E. coli. The amounts of myc-FIP200 and Atg13 recovered with GST-tagged proteins were analyzed by Western blotting. (g) Atg13 is necessary for the interaction between ULK and FIP200. Myc-tagged FIP200 was coexpressed with HA-tagged ULK1 in 293T cells transduced with either scrambled shRNA or Atg13 shRNA. The amount of myc-tagged FIP200 coimmunoprecipitated with HA-tagged ULK1 in HA immunoprecipitate was analyzed by Western blotting.
We next questioned if the interactions are direct. To determine if Atg13 can directly bind ULK1 or ULK2, we purified the full-length Atg13 and incubated it with a fragment of ULK1 containing residues 651-1051 or a fragment of ULK2 containing residues 651-1036. The C-terminal fragment of ULK1, but neither the control protein GST nor the ULK2 fragment, were bound to Atg13 (Figure 2e), validating that the interaction between ULK1 and Atg13 is direct but the interaction between Atg13 and ULK2 might require other cellular factors.
To determine if Atg13, ULK1, and ULK2 directly bind FIP200, we prepared cell lysate from 293T cells expressing myc-tagged FIP200 and incubated it with GST-tagged Atg13, ULK1 (651-1051), and ULK2 (651-1036). Myc-FIP200 was pulled down only with GST-Atg13 but neither with ULK1 nor ULK2 constructs (Figure 2f). However, coincubation with purified Atg13 allowed GST-ULK1 (651-1051) and GST-ULK2 (651-1036) to be pulled down with FIP200, supporting that Atg13 is required for the interaction between FIP200 and ULKs (Figure 2f). Consistent with the important role of Atg13 for the interaction between FIP200 and ULK1, silencing of Atg13 in 293T cells reduced the amount of FIP200 bound to ULK1 (Figure 2g). These results confirm that Atg13 directly binds FIP200 and mediates the interaction between FIP200 and ULKs. We also found that Atg13 is pulled down with GST-ULK2 (651-1036) when myc-tagged FIP200 is added to the incubation with Atg13 (Figure 2f), indicating that the stable interaction between Atg13 and ULK2 requires FIP200.
Atg13, ULK1, and ULK2 Are Important for Autophagosome Formation
Knowing that Atg13 interacts with ULK1 and ULK2, we questioned if Atg13 is involved in the regulation of autophagy induction. First, we knocked down Atg13, ULK1, or ULK2 in HEK293T cells through transduction using a lentiviral shRNA and conducted the autophagy flux assay by comparing the levels of the lipid modified form of LC3 (microtubule-associated protein 1 light chain 3), LC3-II, in the absence and presence of lysosomal inhibitors such as pepstatin A and E-64 (Mizushima and Yoshimori, 2007). Rapamycin, in combination with the lysosomal inhibitors, induced an increase of LC3-II levels in scrambled control shRNA cells, indicating an accumulation of LC3-II due to the inhibition of LC3-II degradation in lysosomes (Figure 3, a and b). On the other hand, the accumulation was significantly suppressed in Atg13- or ULK2-silenced cells or ULK1-deficient MEF cells. This result suggests that disruption of the ULK-Atg13 complex suppresses the autophagic flux of LC3-II, thereby inhibiting LC3-II accumulation in autophagosomes. To further clarify the effects of Atg13, ULK1, or ULK2 silencing on autophagy induction, we monitored the amounts of p62, a protein that is degraded by autophagy (Mizushima and Yoshimori, 2007). Supporting the important roles of Atg13, ULK1, and ULK2 in autophagic degradation of p62, rapamycin induced p62 degradation in scrambled control cells, whereas the degradation was suppressed in Atg13-, ULK1-, and ULK2-silenced cells (Figure 3c).
Figure 3.
Atg13, ULK1, and ULK2 are important for autophagy. (a) Knockdown of Atg13 or ULK2 or knockout of ULK1 inhibits rapamycin-induced autophagic flux of LC3-II. HEK293T cells stably transduced by lentiviral shRNAs or MEF cells were treated with rapamycin (100 nM) or vehicle for 4 h in the presence or absence of lysosomal inhibitors, pepstatin A (10 μg/ml) and E-64 (10 μg/ml), and the indicated proteins in cell lysates were analyzed by Western blotting. (b) Quantitative analysis of relative amounts of LC3-II from two independent measurements (mean ± SD). (c) Knockdown of Atg13, ULK1, or ULK2 or knockout of ULK1 inhibits rapamycin-induced degradation of p62 and enhances S6K1 phosphorylation. The shRNA-transduced HEK293T cells or MEF cells were treated with rapamycin or vehicle for 4 h, and the indicated proteins were analyzed by Western blotting. (d) Knockdown of Atg13, ULK1, or ULK2 inhibits the formation of LC3-positive autophagosomes. HeLa cells stably transduced by lentiviral shRNAs were treated with rapamycin (100 nM) or vehicle for 24 h and stained with anti-LC3 antibody (green) and DAPI (violet). (e) Quantitative analysis of the number of cells with LC3-positive autophagosome structures relative to the total number of cells (mean ± SD). Three or four areas of microscopic images were captured and analyzed by counting the stained cells (Fig S2). (f) Knockdown of ULK2 inhibits the formation of LC3-positive autophagosomes. HeLa cells stably transduced by lentiviral shRNA were treated with rapamycin (100 nM) or vehicle for 2 h or 5 h in the presence of pepstatin (10 μg/ml) and stained with anti-LC3 antibody (green) and DAPI (violet). (g) Quantitative analysis of the number of cells with LC3-positive autophagosome structures relative to the total number of cells (mean ± SD).
Lastly, to confirm that Atg13, ULK1, and ULK2 are involved in the regulation of autophagy, we analyzed the rapamycin-induced autophagosome formation. HeLa cells transduced by Atg13 or ULK1 shRNA exhibited a reduction in the formation of autophagosomes decorated with LC3 after rapamycin treatment relative to control cells transduced with scrambled shRNA (Figure 3d and Supplemental Figure S2). About 19% of Atg13-silenced cells and 18% of ULK1-silenced cells exhibited the LC3-positive autophagosome structures compared with 44% of control cells (Figure 3e). Rapamycin increased the number of LC3-positive cells by 2.0-fold for Atg13- and 2.7-fold for ULK1-silenced cells relative to 7.1-fold for scrambled cells. Similarly, ULK2 silencing reduced the number of cells having LC3-positive autophagosome structures from 73 to 37% at 2 h and from 42 to 28% at 5 h of rapamycin treatment (Figure 3, f and g). Rapamycin increased the number of LC3-positive cells by only 2.3- and 1.3-fold in ULK2-silenced cells relative to 17.5- and 3.2-fold in scrambled cells for 2 and 5 h, respectively.
Another noticeable change was an increase in S6K1 phosphorylation at Thr389 in Atg13-, ULK1-, or ULK2-silenced cells (Figure 3c and Supplemental Figure S2). This result agrees with the previous findings that overexpression of Atg1 in Drosophila fat body has a negative effect on S6K1 phosphorylation and knockdown of ULK1 or ULK2 in mammalian cells increases S6K1 phosphorylation (Lee et al., 2007; Scott et al., 2007). The up-regulation of S6K1 phosphorylation by Atg13, ULK1, or ULK2 knockdown occurred in both 293T and HeLa cells (Figure 3c and Supplemental Figure S2). This result suggests that the ULK complexes may have functions for the regulation of mTORC1 activity as well as autophagy.
ULK1 and ULK2 Phosphorylate Atg13 and FIP200
Knowing that Atg13 is an important element for autophagosome formation, we wondered about the mechanism involving the Atg13-ULK complexes in the regulation of autophagy. From Figure 1B, we knew that ULK induces phosphorylation of Atg13. Treatment of immunoprecipitated Atg13 with lambda phosphatase reversed the mobility shift of Atg13 and coexpression of ULK1 with Atg13 led to incorporation of 32P into Atg13 in 293T cells, thus confirming that ULK1 induces Atg13 phosphorylation (Figure 4, a and b). Furthermore, the band of Atg13 on SDS-PAGE was shifted up when Atg13 was isolated from cells expressing wild-type ULK1 or ULK2 but not a kinase-dead mutant of ULK1 or a control protein (Figure 4c). To determine if ULKs directly phosphorylate Atg13, we isolated ULK1 from 293T cells expressing myc-tagged ULK1 by immunoprecipitation using anti-myc antibody and incubated it with E. coli–purified Atg13 in vitro in the presence of 32P-ATP. ULK1 wild type, but not its kinase-dead mutant, induced incorporation of 32P into Atg13 (Figure 4d). We also observed a drastic increase of 32P incorporation into ULK1 wild type but not mutant, implying that ULK1 undergoes autophosphorylation. We also determined that ULK2 phosphorylates Atg13 in vitro and undergoes autophosphorylation (Figure 4d).
Figure 4.
ULK1 and ULK2 phosphorylate Atg13 and FIP200. (a) ULK1 induces phosphorylation of Atg13. Myc-tagged Atg13 was coexpressed with HA-tagged ULK1 in 293T cells. Myc immunoprecipitate was obtained and treated with lambda phosphatase for 30 min. The migration patterns of Atg13 and ULK1 isolated from myc immunoprecipitation were analyzed on SDS-PAGE. (b) ULK1 induces 32P labeling of Atg13. Myc-tagged Atg13 was coexpressed with HA-tagged ULK1 or control proteins in 293T cells. Transduced cells were incubated with 32P in phosphate-free DMEM for 4 h. Myc-Atg13 immunoprecipitate was isolated and analyzed for its phosphorylation state by autoradiography. (c) The kinase activity of ULK1 and ULK2 is required for phosphorylation of Atg13. HA-tagged Atg13 was expressed together with ULK1 wild type, its kinase-dead mutant M92A, or ULK2 wild type in 293T cells. The migration patterns of HA-Atg13 and myc-ULK on SDS-PAGE were analyzed by Western blotting. (d) ULK1 and ULK2 directly phosphorylate Atg13. Myc-tagged ULK1 or ULK2 was isolated by myc immunoprecipitation from 293T cells and incubated with Atg13 purified from E. coli in the presence of 32P-ATP in vitro. The incorporation of 32P into ULK1, ULK2, and Atg13 was analyzed by autoradiography. (e) The kinase activity of ULK1 and ULK2 is required for phosphorylation of FIP200. Myc-tagged FIP200 was coexpressed with ULK1 wild type or M92A mutant (kinase dead) or ULK2 wild type in 293T cells, and the migration patterns of myc-FIP200 in cell lysate was analyzed on SDS-PAGE. (f) ULK1 and ULK2 directly phosphorylate FIP200. Myc-tagged ULK1 wild type or M92A mutant or ULK2 was isolated from 293T cells by immunoprecipitation using anti-myc antibody and incubated in the presence of 32P-ATP with the FIP200 fragment containing residues 860-end that was purified from E. coli. The incorporation of 32P into ULK and FIP200 was analyzed by autoradiography. (g) ULK1 and ULK2 are important for Atg13 and FIP200 phosphorylation in living cells. ULK1 MEF cells or shRNA-transduced 293T cells were harvested and the phosphorylation states of FIP200, ULK1, ULK2, and Atg13 were analyzed on SDS-PAGE by Western blotting.
Figure 1 also showed that FIP200 band on SDS-PAGE is shifted up when ULK1 or ULK2 is coexpressed (Figures 1f and 4e). To investigate if ULKs phosphorylate FIP200, we purified a fragment of FIP200 containing C-terminal residues 860-end from E. coli and incubated it with ULK1 or ULK2 immunoprecipitate in the presence of 32P-ATP. Both ULK1 and ULK2, but not the ULK1 kinase-dead mutant, showed a high kinase activity toward phosphorylation of the FIP200 fragment (Figure 4f). Knowing that ULK1 and ULK2 phosphorylate Atg13 and FIP200 in vitro, we analyzed if the phosphorylations occur in living cells. Knockdown of ULK1 or ULK2 in 293T cells or knockout of ULK1 in MEF cells induced drastic downshifts of Atg13 and FIP200 bands, supporting that ULK1 and ULK2 determine the phosphorylation status of Atg13 and FIP200 in cells (Figure 4g). Collectively, these results suggest that ULK1 and ULK2 are the kinase phosphorylating their binding proteins Atg13 and FIP200.
mTOR Phosphorylates Atg13, ULK1, and ULK2
The above results suggest that ULK1 and ULK2 form the protein complexes with Atg13 and FIP200 and phosphorylate the proteins. Now, we question how the complex machinery is regulated by mTOR. We first investigated if rapamycin or leucine deprivation could cause any change in the phosphorylational status of the proteins. Rapamycin or leucine deprivation for 1 h induced downshifts of ULK1, ULK2, and Atg13 bands on SDS-PAGE (Figure 5a). Rapamycin almost completely inhibited incorporation of 32P into ULK1 in 293T cells but only marginally for Atg13 (Figure 5, b and c). We reasoned that the marginal inhibition of Atg13 phosphorylation by rapamycin might be due to a low basal level of Atg13 phosphorylation. Consistent with this reasoning, overexpression of Ras homolog enriched in brain (Rheb), the GTPase that stimulates the kinase activity of mTOR (Saucedo et al., 2003; Stocker et al., 2003; Tee et al., 2003; Zhang et al., 2003), caused a drastic upward shift of Atg13 band and incorporation of 32P into Atg13 and rapamycin suppressed the changes (Figure 5c). Rheb overexpression also caused an upward shift of the ULK1 band, but rapamycin inhibited it. Further support shows that Atg13 bands were shifted up when Atg13 was isolated from cells expressing mTOR wild type but not its kinase dead mutant (Supplemental Figure S3). The mTOR-mediated phosphorylation of Atg13 occurred even in ULK1- or ULK2-silenced cells, indicating that the Atg13 phosphorylation may occur independently of ULK1 and ULK2 (Supplemental Figure S3). Taken together, these results suggest that mTOR induces phosphorylations of Atg13, ULK1, and ULK2.
Figure 5.
mTOR phosphorylates Atg13, ULK1, and ULK2. (a) Rapamycin or leucine deprivation induces dephosphorylation of ULK1, ULK2, and Atg13. HEK293T cells were treated with rapamycin or vehicle for 1 h or deprived of leucine for 2 h (−leu) or deprived of leucine for 1 h and supplemented with leucine for 1 h (−leu → +leu). The migration patterns of endogenous Atg13, ULK1, and ULK2 on SDS-PAGE were analyzed by Western blotting. (b) Rapamycin blocks phosphorylation of ULK1. Myc-tagged ULK1 was expressed in 293T cells and incubated with 32P in phosphate-free DMEM for 4 h. Cells were treated with rapamycin (100 nM) or vehicle for 1 h. Myc-ULK1 immunoprecipitate was isolated, and the phosphorylation state of myc-ULK1 was analyzed by autoradiography. (c) Rheb induces phosphorylation of Atg13 and ULK1, whereas rapamycin suppresses the phosphorylation. Myc-tagged Atg13 was expressed with or without HA-tagged Rheb in 293T cells and incubated with 32P in phosphate-free DMEM for 4 h. Cells were treated with rapamycin (100 nM) or vehicle for 1 h. The migration patterns and 32P incorporation of myc-Atg13 and endogenous ULK1 isolated from myc immunoprecipitate were analyzed by Western blotting and autoradiography. (d) mTOR immunoprecipitate has the capacity to phosphorylate Atg13. Endogenous mTOR was isolated from 293T cells by mTOR immunoprecipitation and incubated with Atg13 purified from E. coli in the presence of 32P-ATP. The reaction was analyzed by autoradiography. (e) mTOR phosphorylates Atg13 in vitro. The active form of mTOR containing residues 1362-end (Millipore) and myc-mTOR wild type (WT) and its kinase dead (KD) mutant, D2357E, were tested for their activity to phosphorylate Atg13 in vitro. The incorporation of 32P into Atg13 and mTOR was analyzed by autoradiography. (f) mTOR phosphorylates ULK1 in vitro. Myc-tagged ULK1 M92A kinase dead (KD) mutant or ULK1 fragment containing residues 281-end was incubated with the active form of mTOR (Millipore) in the presence of 32P-ATP. The levels of 32P-labeled ULK1 KD, ULK1 fragment, and mTOR fragment were analyzed by autoradiography. (g) mTOR immunoprecipitate has the capacity to phosphorylate ULK1. Endogenous mTOR or myc-tagged wild type or the kinase dead mutant of mTOR was isolated from 293T cells by immunoprecipitation and incubated with E. coli—purified ULK1 fragment containing residues 651-end. The levels of 32P-labeled mTOR and the ULK1 fragment were analyzed by autoradiography. (h) mTOR immunoprecipitate has the capacity to phosphorylate ULK2. mTOR immunoprecipitates were prepared as described in panel g and incubated with E. coli–purified ULK2 fragment containing residues 651-end. The autoradiogram was obtained to analyze the phosphorylation status of mTOR and ULK2.
Next, we questioned if mTOR can directly phosphorylate the proteins. We isolated mTOR immunoprecipitate from 293T cells and incubated it with E. coli–purified Atg13 in the presence of 32P-ATP. The mTOR immunoprecipitate induced high incorporation of 32P into Atg13 (Figure 5d). Supporting that the Atg13 phosphorylation is due to the kinase activity of mTOR, only wild-type mTOR but not a kinase-dead mutant was able to phosphorylate Atg13 in vitro (Figure 5e). We also tested an active fragment of mTOR that contains residues 1362-end as an enzyme source and confirmed that the active form of mTOR phosphorylates Atg13 (Figure 5e). On the other hand, Atg13 was barely phosphorylated by the protein kinase S6K1 in vitro, indicating that S6K1 may not be the kinase phosphorylating Atg13 (unpublished data).
To investigate if mTOR can directly phosphorylate ULK1, we prepared two ULK1 constructs as substrates: 1) the kinase-dead mutant M92A and 2) a fragment containing 281-end that lacks the N-terminal kinase domain, from 293T cells by immunoprecipitation using anti-myc antibody. Both substrates were highly phosphorylated by the active fragment of mTOR (Figure 5f). To further confirm the direct phosphorylation of ULK1 by mTOR, we purified a soluble fragment of ULK1 containing residues 651-end from E. coli. Endogenous or recombinant mTOR wild type, but not the kinase-dead mutant, induced the phosphorylation of the ULK1 fragment (Figure 5g). In a similar manner, we confirmed that mTOR wild type, but not its kinase-dead mutant, phosphorylates a fragment of ULK2 containing C-terminal residues 651-end in vitro (Figure 5h). On the basis of all these results from in vitro kinase assay and analysis of the phosphorylation in living cells, we conclude that mTOR is the kinase phosphorylating Atg13, ULK1, and ULK2.
mTOR Inhibits ULK1 and ULK2 Kinase Activity
Now that we have identified that mTOR phosphorylates the Atg13-ULK complexes, we questioned the functional consequence of the phosphorylations. Because rapamycin induces autophagy, we reasoned that rapamycin might activate ULK. We isolated endogenous and recombinant ULK1 from 293T cells treated with either rapamycin or vehicle for 1 h and analyzed the kinase activity of ULK1. ULK1 isolated from rapamycin-treated cells, relative to the vehicle-treated cells, exhibited a higher kinase activity toward phosphorylation of MBP (Figure 6, a and b). Like rapamycin, leucine deprivation also led to a high kinase activity of ULK1 (Figure 6, c and d). Consistent with the negative effects of mTOR on the ULK activity, Rheb overexpression reduced the kinase activity of ULK1 (Figure 6, e and f). We also confirmed that ULK2, like ULK1, is stimulated by rapamycin using two different substrates, MBP and the FIP200 fragment containing C-terminal residues 860-1591 (Figure 6, g and h).
Figure 6.
mTOR negatively regulates the kinase activity of ULK. (a) Rapamycin increases the kinase activity of ULK1. 293T cells were treated with rapamycin or vehicle for 1 h. ULK1 immunoprecipitate was isolated using anti-ULK1 antibody and incubated with MBP (1 μg) in the presence of 32P-ATP. The levels of 32P-labeled MBP were analyzed by autoradiography. (b) Quantitative analysis of ULK1 kinase activity from two independent experiments (mean ± SD). (c) Leucine starvation enhances the kinase activity of ULK1. 293T cells were incubated in the presence or absence of leucine (52 μg/ml) for 1 h. ULK1 immunoprecipitate was isolated and analyzed for its kinase activity toward phosphorylation of MBP in the presence of 32P-ATP. (d) Quantitative analysis of leucine-dependent ULK1 kinase activity. (e) Rheb overexpression inhibits ULK1 kinase activity. Myc-tagged ULK1 was expressed alone or with HA-tagged Rheb in 293T cells. Myc-ULK1 was isolated by immunoprecipitation using anti-myc antibody and analyzed for the kinase activity as described in panel a. (f) Quantitative analysis of the ULK1 kinase activity from two independent measurements (mean ± SD). (g) Rapamycin enhances the kinase activity of ULK2. Myc-tagged ULK2 was expressed in 293T cells and treated with rapamycin or vehicle for 1 h. Myc-ULK2 was isolated by immunoprecipitation using anti-myc antibody and incubated with either MBP or the FIP200 fragment (aa 860-end) purified from E. coli in the presence of 32P-ATP. The levels of 32P-labeled MBP and FIP200 were analyzed by autoradiography. (h) Quantitative analysis of ULK2 kinase activity toward MBP and FIP200. (i) Rapamycin or starvation does not have any significant effect on the interaction between ULK1, Atg13, and FIP200 but induces phosphorylation of ULK1 during a prolonged rapamycin treatment. HEK293T cells were treated with rapamycin or vehicle for 1 or 24 h or incubated in the presence or absence of leucine for 1, 2, or 24 h, and ULK1 immunoprecipitate was isolated using anti-ULK1 antibody. The amounts of Atg13 and FIP200 coimmunoprecipitated with ULK1 and in cell lysate were analyzed by Western blotting. (j) ULK1 and ULK2 are important for rapamycin- and starvation-induced phosphorylation of FIP200. ULK1 MEF cells or shRNA-transduced 293T cells were treated with rapamycin or vehicle or incubated in the presence or absence of leucine for 1 h, and the phosphorylation states of FIP200, ULK, and Atg13 were analyzed on SDS-PAGE by Western blotting.
The second line of evidence in support of mTOR inhibition of ULK1 activity came from the observation of the mobility shift of ULK1 bands on SDS-PAGE. When cells were treated with rapamycin for a long period of time, ULK1 bands were converted from fast-migrating forms to slow-migrating ones (Figure 6i and Supplemental Figure S4). This supports that the dephosphorylated form of ULK1 generated during rapamycin treatment might have a kinase activity and undergo autophosphorylation. The third line of evidence was that rapamycin or leucine deprivation induced a drastic upward shift of the FIP200 band on SDS-PAGE (Figure 6, i and j). We interpreted this as a result of activation of ULKs that are the kinases phosphorylating FIP200. Using ULK1 null MEF cells and ULK2-silenced 293T cells, we confirmed that the rapamycin- and starvation-induced phosphorylation of FIP200 depends on ULK1 and ULK2 (Figure 6j). We could not determine contribution of each ULK to FIP200 and Atg13 phosphorylation, which may require double knockout or knockdown cells. Given that ULK1 null mice is viable (Kundu et al., 2008), it may be possible that ULK1 and ULK2 have redundant functions. However, based on the mobility shift assay in Figure 6j, it is likely that ULK1 is the major kinase phosphorylating FIP200 and Atg13 in response to rapamycin and nutrient starvation at least in MEF and 293T cells.
In S. cerevisiae, nutrient deprivation or rapamycin alters the affinity of the interaction between Atg1 and Atg13 (Kamada et al., 2000). We investigated whether rapamycin has similar effects on the interaction between Atg13 and ULK. Although we observed that the affinity of the interaction increases at 24 h of rapamycin treatment, the expression levels of ULK1 and Atg13 were also increased (Figure 6i). Furthermore, the affinity of the interaction was not drastically altered by leucine deprivation for 1, 2, or 24 h. We also found that the interaction between ULK2 and Atg13 is not significantly altered by rapamycin or leucine deprivation (data not shown). Given that rapamycin or leucine deprivation for 1 h is sufficient to have the stimulatory effect on ULK activity, we conclude that the activity of ULKs is not altered by any drastic change in the affinity of the interaction with Atg13.
Atg13 Is Important for the ULK1 Kinase Activity and FIP200 Phosphorylation by ULKs
Knowing that rapamycin increases the kinase activities of ULK1 and ULK2, we wondered whether Atg13 plays any role in the ULK activation. We isolated myc-tagged ULK1 from 293T cells and incubated it with different amounts of Atg13 isolated from E. coli and analyzed the kinase activity of ULK1. The levels of the substrate MBP incorporated with 32P increased when higher amounts of Atg13 were added to the reaction (Figure 7, a and b). The 32P-labeled MBP level was no longer increased when the amount of Atg13 was over a certain concentration (lane 5), which might be because the amount of Atg13 has exceeded the binding capacity of ULK1. Agreeing with the positive role of Atg13 for ULK1 activity, the kinase activity of ULK1 was increased when ULK1 was isolated from cells expressing Atg13 (Figure 7, c and d, and Supplemental Figure S4). Similar to the results in Figure 7, a and b, when the expression levels of Atg13 were higher than a certain concentration, the kinase activity and the isolated amount of ULK1 were reduced. This might be due to sequestration of a cellular component crucial for the stability and the activity of ULK1 by Atg13 at a high level of expression. Another possibility is that Atg13 might be a competitive inhibitor for the phosphorylation of MBP because Atg13 itself is a substrate of ULK.
Figure 7.
Atg13 positively regulates ULK activity and is required for starvation-induced phosphorylation of FIP200. (a) Atg13 increases the kinase activity of ULK1 in vitro. Myc-tagged ULK1 was isolated from 293T cells and incubated with purified Atg13 at various amounts (0.5, 1, 2, 5 μg) and 1 μg of MBP in the presence of 32P-ATP. (b) Quantitative analysis of ULK1 activity from panel a. (c) Coexpression of Atg13 with ULK1 increases the kinase activity of ULK1. Myc-tagged ULK1 was coexpressed with HA-Atg13 at different amounts (0, 0.05, 0.5, and 1.5 μg of DNA) in 293T cells. Myc-ULK1 immunoprecipitate was isolated and incubated with 1 μg of MBP in the presence of 32P-ATP and the levels of 32P-labeled MBP were analyzed by autoradiography. (d) Quantitative analysis of ULK1 activity from panel c. (e) Atg13 is important for the stability of ULK1 and the rapamycin- and starvation-induced phosphorylation of FIP200. HeLa or HEK293T cells stably transduced by lentiviral shRNAs were treated with rapamycin (100 nM) or vehicle for 1 h or incubated in the presence or absence of leucine (52 μg/ml) for 1 h. The expression levels and the mobility shifts of endogenous FIP200, ULK1, ULK2, and Atg13 were analyzed by Western blotting. (f) Model: The ULK-Atg13-FIP200 complexes mediate mTOR signaling downstream to the autophagy machinery. Starvation suppresses mTOR-mediated phosphorylation of ULK and Atg13 that have inhibitory effects on the kinase activity of ULK, resulting in ULK-mediated phosphorylations of Atg13, FIP200, and ULK itself.
Consistent with the dependence of ULK stability on Atg13, we observed that knockdown of Atg13 induces a drastic reduction in the expression level of ULK1 in HeLa cells (Figure 7e). The stability of ULK1 was only marginally affected by Atg13 knockdown in 293T cells (Figures 3, a and c, and 7e), indicating that the regulation of ULK stability by Atg13 may depend on other cellular factors or cellular contexts. Another noticeable change was observed in Atg13-silenced cells with regard to the phosphorylation of FIP200. Knockdown of Atg13 suppressed phosphorylation of FIP200 and disabled rapamycin or leucine deprivation to induce FIP200 phosphorylation (Figure 7e). This result suggests that Atg13 is required for the mTOR-mediated activation of ULK or at least for the phosphorylation of FIP200 by ULKs. Starvation-induced phosphorylation of ULK1 still occurred in Atg13-silenced cells, suggesting that mTOR can phosphorylate ULK1 independently of Atg13 (Figure 7e). Taken together, these results suggest that Atg13 plays a positive role in mTOR-regulated autophagy processes by enhancing the catalytic activity of ULK and allowing ULK to phosphorylate FIP200, the protein crucial for autophagosome formation (Hara et al., 2008).
DISCUSSION
Our study has revealed that the mammalian homologue of Atg13 interacts with ULK1 and ULK2 and participates in the processes leading to the autophagosome formation. While preparing this article, another group reported that Atg13 interacts with ULK1 and ULK2 and regulates autophagy (Chan et al., 2009). However, we have extended these observations to report that Atg13 is also a direct substrate of mTOR. Furthermore, our studies provide several important findings regarding the function of Atg13. According to our results, Atg13 mediates the interaction between ULK and FIP200, thereby forming two protein complexes: ULK1-Atg13-FIP200 and ULK2-Atg13-FIP200. We observed that ULK1 and ULK2 have different binding affinities toward Atg13 and FIP200 (Figure 1, a and b). In contrast to the direct interaction between ULK1 and Atg13, the interaction between ULK2 and Atg13 was only detected in the presence of FIP200 (Figure 2, e and f). This may indicate that the affinity of Atg13 toward binding ULK1 and ULK2 is different and that the ULK2-Atg13 interaction depends highly on FIP200. Possibly, the two complexes might have distinct functions in autophagy regulation. Given that only one Atg1 exists in yeast and Drosophila (Kamada et al., 2000; Scott et al., 2004), our results suggest that a higher level of complexity has evolved with regard to the Atg1-Atg13-Atg17 complex machinery in mammals. Furthermore, our blast searches showed that there are more than three splicing variants of Atg13 in humans. We confirmed that at least two of the variants interact with both ULK1 and ULK2, implying that more than two ULK-Atg13 complexes may exist in humans.
Through in vitro kinase assay and analysis of mTOR-dependent phosphorylations of ULK and Atg13, we have determined that mTOR is the kinase phosphorylating Atg13, ULK1, and ULK2. We have provided several lines of evidence to demonstrate that mTOR-mediated phosphorylations of Atg13 and ULKs have negative effects on the ULK activity. First, rapamycin or leucine deprivation increased the kinase activity of ULK1 and ULK2 in vitro (Figure 6, a–d, g, and h); second, Rheb overexpression reduced the kinase activity of ULK1 in vitro (Figure 6e and f); third, ULK1 autophosphorylation was increased under mTOR inhibitory conditions (Figure 6i and Supplemental Figure S4); and fourth, ULK-dependent phosphorylation of FIP200 was induced by mTOR inhibition (Figure 6j). Recent studies have shown that ULK or Atg1 kinase activity is important for the regulation of autophagy (Tekinay et al., 2006; Scott et al., 2007; Hara et al., 2008), which agrees with our model that mTOR regulates autophagy through phosphorylation of the ULK complexes and inhibition of the kinase activity of ULK (Figure 7f). Given that starvation does not induce any significant alteration in the binding affinity between ULK and Atg13 (Figure 6i), we assume that the phosphorylation of Atg13 and/or ULK by mTOR may not induce a drastic conformational change in Atg13 and ULK. Rather, the phosphorylation may induce a local perturbation of the protein–protein interaction that interferes with ULK recognition of phosphorylation sites of FIP200 or other unidentified substrates.
Our study provides solid evidence that mTOR inhibition activates ULK and triggers phosphorylations of Atg13, ULK, and FIP200 under starvation conditions. Given that ULK1 interacts with multiple proteins (Okazaki et al., 2000; Tomoda et al., 2004), the starvation-induced phosphorylations might alter a protein–protein interaction mediated by ULK. Alternatively, the phosphorylations might have effects on the catalytic activity or localization to autophagosomes of ULK. Consistent with the latter possibility, a recent study showed that ULK1 and ULK2 localize to autophagosomal structures in a starvation-dependent manner (Hara et al., 2008). We also observed that rapamycin induces accumulation of ULK1 in membrane fractions of HeLa cells (Supplemental Figure S3). The regulation of ULK localization by starvation-induced phosphorylations is further supported by the demonstrated roles of FIP200, the substrate of ULKs, in ULK localization to autophagosomal structures (Hara et al., 2008).
Our study has indicated that Atg13 is an important component of the autophagy machinery regulated by mTOR. Atg13 plays a positive role in the regulation of ULK1 activity. Atg13 enhances the kinase activity of ULK1 in vitro (Figure 7, a–d). Atg13 is important for the stability of ULK1 (Figures 3, a and c, and 7e, and Supplemental Figure S2). Atg13 mediates the interaction between ULKs and FIP200 and is required for the rapamycin-induced phosphorylation of FIP200 by ULKs (Figures 2, f and g, and 7e). Our result also supports that Atg13, like ULK1 and ULK2, is important for autophagosome formation but may not be essential for the lipidation of LC3 (Figure 3). This is consistent with the roles of FIP200 that were demonstrated to be crucial for autophagosome formation but not for the LC3 lipidation (Hara et al., 2008).
Chang and Neufeld (2009) and Hosokawa et al. (2009) in this issue of the journal have reported findings, which are similar to those that we made, on the Atg1-Atg13 complex in Drosophila and the ULK1-Atg13-FIP200 complex in mammalian systems. Consistent with our findings, both studies showed that high nutrition increases the phosphorylation state of Atg1/ULK1 and rapamycin or that nutrient starvation suppresses the phosphorylation. However, the results of the three groups show that the regulation of Atg13 phosphorylation seems to be different between Drosophila and mammalian systems. Atg13 phosphorylation in Drosophila cells was much highly induced by starvation compared with mammalian Atg13 phosphorylation (Figures 5–7; Chang and Neufeld, 2009). This implies that the phosphorylation state of Atg13 may be more dependent on Atg1 than TOR in Drosophila. Hosokawa et al. (2009) showed that mTOR interacts with the ULK1-Atg13-FIP200 complex in a nutrient-dependent manner and phosphorylates ULK1 and Atg13. All of these reports together with ours provide comprehensive characterization of Atg1/ULK-Atg13 complexes defining the link between TOR and the autophagy machinery.
In addition to the autophagy regulation, the ULK complexes are involved in the regulation of S6K1 phosphorylation (Figure 3c and Supplemental Figure S2). This additional function is supported by the previous findings that ULK1 and ULK2 and their Drosophila Atg1 homolog negatively regulate S6K1 and cell growth (Lee et al., 2007; Scott et al., 2007). The up-regulation of S6K1 in ULK-silenced cells is mimicked by Atg13 silencing, implying that Atg13 functions together with ULKs for the S6K1 regulation. These results suggest a conserved mechanism between Drosophila and mammals with regard to the cross-talk between Atg1-Atg13 and S6K1. Although it is unclear how the ULK complexes negatively regulate S6K1 phosphorylation, the ULK complexes may contribute favorably to autophagy induction by inhibiting the kinase activity of mTORC1 or S6K1. Alternatively, the regulation of S6K1 by the ULK complexes may occur independently of autophagy regulation.
It is intriguing that the C-terminus of ULKs binds both Atg13 and FIP200. The ULK1 C-terminus is also involved in binding SynGAP and syntenin the proteins regulating the neuronal axon outgrowth, and is important for the autophagosomal localization of ULK1 (Tomoda et al., 2004; Chan et al., 2007). ULK1 also interacts with GATE-16 and GABARAP, the mammalian homologues of Atg8 that localize to autophagosome (Okazaki et al., 2000; Kabeya et al., 2004). Given that ULK1 binds multiple proteins that localize to autophagosomes, it may be possible that ULK1 regulates the autophagosomal localization of the binding proteins. Alternatively, the ULK1 binders may recruit ULK1 to autophagosomal membranes where ULK1 can phosphorylate proteins involved in the downstream events of autophagy. Future studies on mTOR-regulated phosphorylations of the ULK complexes may increase further our knowledge on the roles of the mTOR-ULK pathway in autophagy induction and autophagy-related human diseases such as cancer and neurodegeneration.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Neufeld, Y.-Y. Chang, and N. Mizushima for discussion and sharing unpublished data; T. Shimizu for the mouse ULK1 clone; Kim lab members, S. Woo, B. Vang, and Y.S. Yun, for helpful comments; BIPL for the confocal microscope facility; and L. Potter for the in vivo labeling facility. This study was supported by the National Institutes of Health Grant DK072004, the Tuberous Sclerosis Alliance, the American Diabetes Association Grant 7-07-CD-08, the American Heart Association Grant 0655706Z, and Minnesota Obesity Center Grant P30DK50456.
Abbreviations used:
- Atg
Autophagy-related gene
- FIP200
focal adhesion kinase (FAK) family interacting protein of 200 kDa
- GST
glutathione S-transferase
- LC3
microtubule-associated protein 1 light chain 3
- MBP
myelin basic protein
- MEF
mouse embryonic fibroblast
- mTOR
mammalian target of rapamycin
- Rheb
Ras homolog enriched in brain
- shRNA
short hairpin RNA
- ULK1
Unc-51-like kinase 1
- ULK2
Unc-51-like kinase 2
- S6K1
S6 kinase 1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1249) on February 18, 2009.
REFERENCES
- Chan E. Y., Kir S., Tooze S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. Kinase-inactivated ULK1 proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-Y., Neufeld T. P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S.-I., Natsume T., Guan J.-L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., et al. Nutrient-dependent mTORC1 association with ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. mTOR interacts with raptor to form a nutrient-regulated complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Kundu M., Lindsten T., Yang C.-Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A., Thompson C. B. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Kim S., Lee J., Park J., Lee G., Kim Y., Kim J. M., Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J. A. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Meléndez A., Tallóczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Neufeld T. P., Baehrecke E. H. Eating on the fly: function and regulation of autophagy during cell growth, survival, and death in Drosophila. Autophagy. 2008;4:557–562. doi: 10.4161/auto.5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Okazaki N., Yan J., Yuasa S., Ueno T., Kominami E., Masuho Y., Koga H., Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res. Mol. Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- Otto G. P., Wu M. Y., Kazgan N., Anderson O. R., Kessin R. H. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- Qu X., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Saucedo L. J., Gao X., Chiarelli D. A., Li L., Pan D., Edgar B. A. Rheb promotes cell growth as a component of the insulin/TOR signaling network. Nat. Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Hall M. N. TOR, a central controller for cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Scott R. C., Juhász G., Neufeld T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P., Daram P., Breuer S., Thomas G., Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Sugimoto S. A novel vacuolar myopathy with dilated cardiomyopathy. Autophagy. 2007;3:638–639. doi: 10.4161/auto.4931. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tekinay T., Wu M. Y., Otto G. P., Anderson O. R., Kessin R. H. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot. Cell. 2006;5:1797–1806. doi: 10.1128/EC.00342-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda T., Kim J. H., Zhan C., Hatten M. E. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.-i., Bandhakavi S., Griffin T. J., Kim D.-H. Insulin signaling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Vellai T., Bicsak B., Tóth M. L., Takács-Vellai K., Kovács A. L. Regulation of cell growth by autophagy. Autophagy. 2008;4:507–509. doi: 10.4161/auto.5670. [DOI] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A. J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.