Abstract
Membrane type-1 matrix metalloproteinase (MT1-MMP) supports tumor cell invasion through extracellular matrix barriers containing fibrin, collagen, fibronectin, and other proteins. Here, we show that simultaneous knockdown of two or three members of the tetraspanin family (CD9, CD81, and TSPAN12) markedly decreases MT1-MMP proteolytic functions in cancer cells. Affected functions include fibronectin proteolysis, invasion and growth in three-dimensional fibrin and collagen gels, and MMP-2 activation. Tetraspanin proteins (CD9, CD81, and TSPAN2) selectively coimmunoprecipitate and colocalize with MT1-MMP. Although tetraspanins do not affect the initial biosynthesis of MT1-MMP, they do protect the newly synthesized protein from lysosomal degradation and support its delivery to the cell surface. Interfering with MT1-MMP-tetraspanin collaboration may be a useful therapeutic approach to limit cancer cell invasion and metastasis.
INTRODUCTION
Matrix metalloproteinases (MMPs) are a family of soluble and membrane-anchored proteolytic enzymes that can remodel the extracellular matrix (ECM) and cleave several other substrates, including cell–cell and cell–matrix adhesion molecules, chemokines, cytokines, latent growth factors, and cell surface receptors (Egeblad and Werb, 2002). Membrane type-1 MMP (MT1-MMP; MMP-14) is of particular interest because MT1-MMP knockout mice show severe defects, including skeletal abnormalities and fibrosis (Holmbeck et al., 1999). MT1-MMP also plays critical roles during tumor malignancy and is one of the best-validated proteolytic enzyme targets on cancer cells. First, MT1-MMP is up-regulated in several tumor types, including breast, cervical, and ovarian cancer (Drew et al., 2004; Zhai et al., 2005; Jiang et al., 2006) and is significantly associated with adverse outcome (Jiang et al., 2006). Second, overexpression of MT1-MMP in the mammary gland of mice results in hyperplasia and spontaneous adenocarcinomas (Ha et al., 2001). Third, ectopic expression of MT1-MMP and MT2-MMP, but not soluble MMPs, enables tumor cell penetration of three-dimensional (3D) collagen gels (Hotary et al., 2000). Similarly, introduction of MT1-MMP into poorly invasive MCF-7 breast cancer cells renders those cells more invasive in vitro and more tumorigenic in vivo (Borrirukwanit et al., 2007). Finally, silencing of endogenous MT1-MMP markedly decreases invasive and migratory properties of breast cancer cells (Jiang et al., 2006).
MT1-MMP is regulated at the level of gene transcription, proenzyme activation, subcellular localization, internalization/recycling, dimerization, shedding, inhibition by natural inhibitors (tissue inhibitor of metalloproteinases, RECK), and posttranslational modifications (Itoh and Seiki, 2006). In addition MT1-MMP associates with cell surface molecules such as β1 integrins (Galvez et al., 2002), CD44 (Mori et al., 2002), and tetraspanin proteins such as CD63 (Takino et al., 2003), CD151 (Yanez-Mo et al., 2008), CD81, and others (Kolesnikova et al., 2009). Tetraspanins are a family of cell surface proteins (33 human members), each with four conserved transmembrane domains, characteristic extracellular loops, and short cytoplasmic domains (Hemler, 2003). These relatively small proteins (typically 22–30 kDa) generally do not function as classical cell surface receptors. Rather, they serve as molecular organizers of multiprotein membrane complexes, thereby influencing cell proliferation, fusion, signaling, and migration (Stipp et al., 2003b). Tetraspanins can associate with each other and with other proteins such as integrins, immunoglobulin (Ig) superfamily members, proteoglycans, ligands, and growth factor receptors to form specialized membrane structures called tetraspanin-enriched microdomains (TEMs) (Hemler, 2005). The tendency of different tetraspanins to associate closely within TEMs probably underlies the ability of distinct tetraspanins to provide functional compensation for each other (Fradkin et al., 2002; Kaji et al., 2002). On cancer cells, some tetraspanins (CD151 and CO-029) promote invasion (Claas et al., 1998; Yang et al., 2008), whereas others (NET-6, CD82, and CD9) behave more as tumor suppressors (Liu and Zhang, 2006; Huang et al., 2007; Takeda et al., 2007).
Given their tendency to localize into cellular lamellipodia and filopodia (Penas et al., 2000), tetraspanins are well positioned on the cell surface to orchestrate events such as pericellular proteolysis by membrane-anchored proteases. In this regard, tetraspanin CD151 association with endothelial cell MT1-MMP may inhibit MT1-MMP–dependent MMP-2 activation, while paradoxically supporting MT1-MMP–dependent collagen degradation (Yanez-Mo et al., 2008). In another study, association with tetraspanin CD63 was suggested to accelerate lysosomal degradation of MT1-MMP in HeLa cells (Takino et al., 2003). Given the importance of MT1-MMP as a proinvasive protease in cancer cells, we set out to identify the functional importance of tetraspanin-MT1-MMP associations in cancer cells. Among seven different tetraspanins analyzed, CD9, CD81, and TSPAN12 showed the most readily detectable associations with MT1-MMP. Hence, functional studies focused mostly on those tetraspanins. To ensure sufficient disruption of TEMs and to minimize functional compensation, we knocked down these tetraspanins two to three at a time. Furthermore, because tetraspanins had been shown to have conflicting effects on MT1-MMP functions, we undertook a comprehensive functional analysis, by using four different assays for MT1-MMP function. In addition, we provide mechanistic insight into how tetraspanins regulate MT1-MMP function.
MATERIALS AND METHODS
Cell Culture
Cell lines were from American Type Culture Collection (Manassas, VA). MCF-7 cells were stably transfected with MT1-MMP-FLAG (MCF-7-MT1), MT1-MMP-green fluorescent protein (GFP) (MCF-7-MT1-GFP, GFP at C terminus), or vector only using FuGENE6 (Roche Diagnostics, Indianapolis IN). FLAG-MT1-MMP from M. Seiki (University of Tokyo, Tokyo, Japan), was cloned into pcDNA3.1 (Invitrogen, Carlsbad CA). MT1-MMP-GFP is from wild type (wt) MT1-MMP cloned into phosphorylated enhanced green fluorescent protein (BD BioSciences, Palo Alto CA) and subcloned into pcDNA3.1. Stable cell lines were sorted as a pool population by flow cytometry. HT1080-FLAG–tagged MT1-MMP cells were generated as described above. TSPAN12-FLAG was cloned into pLXIZ and transfected into the pT67 packaging cell line. HT1080 cells were infected with viral supernatants to generate stable cell lines. All cells were maintained in DMEM (Invitrogen) with 10% fetal bovine serum (FBS).
Reagents and Antibodies
Bafilomycin A1 (vacuolar type H+ ATPase inhibitor), cytochalasin D, thrombin, pepstatin A (aspartyl peptidase inhibitor), E64, 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), anti-FLAG (M2) antibody, anti-β actin antibody, and secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody were from Sigma-Aldrich (St. Louis, MO). ALLM and MG132 were obtained from BIOMOL Research Laboratories (Plymouth Meeting, PA). Plasminogen-depleted fibrinogen was from Calbiochem (San Diego, CA). Protein G-agarose beads, aprotinin, and leupeptin (serine and thiol protease inhibitor) were from Roche Diagnostics. MT1-MMP antibodies (AB815 and LEM2/15.8) and GM6001 were from Millipore Bioscience Research Reagents (Billerica, MA). MT1-MMP antibody (28209) was from Abcam (Cambridge, MA). Anti-GFP antibody was from Clontech (Mountain View, CA). Anti-CD71 and anti-E-cadherin (G-10) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Fibronectin and anti-fibronectin antibody were from BD Biosciences. Secondary HPR-conjugated goat anti-rabbit light chain was from Zymed Laboratories (South San Francisco, CA). Alexa 546-fibrinogen, Alexa 546-phalloidin, ProLong Gold antifade mounting media with 4,6-diamidino-2-phenylindole (DAPI), Alexa 488-donkey anti-rabbit, Alexa 488-donkey anti-mouse, and Alexa 546 goat anti-mouse were from Invitrogen. Type I collagen was from Inamed (Fremont, CA). Antibodies to CD9 included MM2/57 (BioSource International, Camarillo, CA), MEM-61 (GeneTex, San Antonio, TX) and ALB6 (Immunotech, Marseille, France). Other antibodies are TSPAN4 (Tachibana et al., 1997), MHC I (W6/32) (Barnstable et al., 1978), CD98 (4F2) (Hemler and Strominger, 1982), CD147 (8G6), α3 integrin (A3X8), CD63 (6H1), CD81 (M38), CD82 (M104), and CD151 (5C11) (Yang et al., 2002).
Immunoprecipitation, Western Blotting, and Zymography
Cells were lysed in 25 mM HEPES, 150 mM NaCl, 5 mM MgCl2, and 5 mM CaCl2 with protease inhibitor cocktail (Roche Diagnostics) and 1% detergent (Triton X-100, Brij 96, Brij 99, or 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate [CHAPS]) at 4°C. Radioimmunoprecipitation assay contained the above-mentioned components with 1% deoxycholate, 1% Triton X-100, and 0.1% SDS. In some cases, lysates were ultracentrifuged (Beckman L8-80M) at 100,000 × g. Equal lysate volumes were precleared with protein G-agarose beads and then immunoprecipitated with indicated antibodies and protein G overnight at 4°C. Beads were washed and eluted with SDS-sample buffer (50 mM Tris-HCl, pH 6.8, 1% SDS, 0.025% bromophenol blue, and 10% glycerol). Western blot analysis for MT1-MMP and antibody validation was performed as described previously (Lafleur et al., 2006). Western blot analysis for CD9, CD81, and actin and antibody validation was performed as described previously (Stipp et al., 2003a; Kovalenko et al., 2007). For zymography, samples were separated on a 10% polyacrylamide gel copolymerized with 1 mg/ml gelatin (Sigma-Aldrich) and developed as described previously (Lafleur et al., 2006).
Small Interfering RNA (siRNA) Transfection
Cells were transfected with 10 nM (MCF-7 cells) or 25 nM (HT1080 cells) of indicated siRNAs (Dharmacon RNA Technologies, Lafayette, CO) with Lipofectamine RNAiMAX (Invitrogen). The sense sequences for the siRNAs are as follows: CD9#1, CCAAGAAGGACGUACUCGAUU; CD9#2, UUAAGGAAGUCCAGGAGUU; CD81#1, CCACCAACCUCCUGUAUCUUU; CD81#2, CCAACAACGCCAAGGCUGU; CD151#1, CCUCAAGAGUGACUACAUCUU; TSPAN12#1, GCAAACAGCUUUAAUACACUU; TSPAN12#2, GUACAAUGGUCAGAUAUGGUU; and control#1, UAGCGACUAAACACAUCAA.
3D Collagen and Fibrin Gels
Two days after siRNA transfection, cells were embedded into fibrin or collagen gels. For collagen gels, 5 × 104 cells were resuspended in 400 μl of neutralized and buffered collagen gels (2 mg/ml) and polymerized for 1 h at 37°C. Finally, cells were covered in growth media. For fibrin gels, 5 × 104 cells were resuspended in 400 μl of 2.5 mg/ml fibrinogen in serum-free media and polymerized with thrombin (1 U/ml) for 30 min at 37°C. Then, they were covered with growth media containing 50 μg/ml aprotinin.
Fibronectin Immunofluorescence
siRNA-treated cells were cultured on fibronectin coated (10 μg/ml) Lab-Tek chamber slides (Nalge Nunc International, Rochester, NY) for 24 h in serum-free media with protease inhibitors: 100 μg/ml aprotinin, 5 μM leupeptin, 20 μM E64, 20 μM pepstatin A, and 100 μM AEBSF. Cells were fixed with 4% paraformaldehyde, blocked/permeabilized with 5% bovine serum albumin (BSA)/phosphate-buffered saline (PBS)/0.25% Triton X-100, and immunostained with an anti-fibronectin antibody for 1 h at 20°C. Cells were then incubated with an Alexa 488 secondary antibody and Alexa 546-phalloidin for 1 h at 20°C. Cells were mounted with ProLong Gold antifade mounting media containing DAPI. Specificity of the anti-fibronectin antibody was obtained by negative staining of collagen coated slides (data not shown).
MTT Proliferation and Flow Cytometry
MCF-7-MT1 cells transfected with indicated siRNAs were plated in 96-well plates (2000 cells/well) in triplicate for 1–7 d. An MTT proliferation assay (Roche Diagnostics) was performed each day using an Opsys MR Thermo Lab Systems plate reader. For flow cytometry, cells were blocked with 5% goat serum/1% BSA/0.02% NaN3 in PBS at 4°C and then stained with primary antibodies for 1 h at 4°C. Cells were washed and incubated with Alexa 488-conjugated donkey anti-rabbit or anti-mouse secondary antibodies for 1 h at 4°C and washed before analysis.
Internalization
MCF-7-MT1-GFP cells were incubated with 0.5 mg/ml EZ-link Sulfo-NHS-SS-biotin (Pierce Chemical, Rockford, IL) at 4°C for 1 h. Cells were washed in PBS and incubated in serum-free media for 0, 10, or 30 min at 37°C. Cells were washed and incubated with reducing solution (42 mM glutathione, 75 mM NaCl, 1 mM EDTA, 1% BSA, and 75 mM NaOH) for 40 min at 4°C. Cells were washed with PBS, lysed as described above for immunoprecipitation with 1% Triton X-100 lysis buffer, and MT1-MMP was immunoprecipitated with an anti-GFP antibody. Eluates were analyzed by Western blotting using ExtrAvidin-HRP.
Pulse-Chase
MCF-7-MT1-FLAG cells were incubated with DMEM lacking l-cysteine and l-methionine (Invitrogen), 10% dialyzed FBS, 25 mM HEPES, l-glutamine, and penicillin/streptomycin for 30 min at 37°C. Cells were pulsed (30 min) with the above-mentioned media with 0.2 mCi/ml EasyTag Expression Protein Labeling Mix [35S]l-methionine and [35S]l-cysteine (PerkinElmer Life and Analytical Sciences, Waltham, MA). Cells were chased with DMEM lacking l-cysteine and l-methionine, 10% dialyzed FBS, 25 mM HEPES, l-glutamine, penicillin/streptomycin, 0.2 mM l-cysteine, and 0.2 mM l-methionine. Cells were lysed as described above for immunoprecipitation with 1% Triton X-100 lysis buffer with 10 mg/ml BSA. Lysates were precleared and immunoprecipitated using 28209 anti-MT1-MMP antibody. Eluates were separated on a 4–20% gradient gel (Invitrogen), transferred to polyvinylidene difluoride membrane, and exposed to a PhosphorImager screen (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) overnight. Cell surface pulse-chase experiments were as described above, except that before immunoprecipitation, intact cells were cooled to 4°C and then incubated with the M2 anti-FLAG antibody for 2 h at 4°C, and then lysed.
RNA Isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RNA was harvested using RNeasy mini columns (QIAGEN, Valencia CA). Reverse transcription was performed with the SuperScript first-strand synthesis system (Invitrogen). PCR was performed with PCR buffer (1.5 mM MgCl2), 200 μM dNTPs (Roche Diagnostics), 200 nM forward and reverse primers, 5 U of Taq DNA polymerase (Roche Diagnostics), and template. PCR conditions were as follows: 94°C for 2 min and then 35 cycles (TSPAN12) or 25 cycles (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) of 94°C 15 s, 60°C 30 s, and 72°C 1 min by using a DNA Engine Peltier thermal cycler (Bio-Rad, Hercules, CA).
Primers used were as follows: TSPAN12 5′, CTCTTGATAAAGGTCACTGAGATC; TSPAN12 3′, TGTTGCTTCTTGCATGGTACTTTG; GAPDH 5′, CGGAGTCAACGGATTTGGTCGTAT; and GAPDH 3′, AGCCTTCTCCATGGTGGTGAAGAC.
Statistical Analysis
Where indicated, a t test (2-tailed, unequal variance) was performed when comparing two groups; control versus tetraspanin knockdown. The p values were obtained for each tetraspanin knockdown condition (single, double, or triple) compared with the control.
Image and Data Acquisition, Including Microscopy
For zymograms and Western blots, original gels or films, respectively, were scanned using an Epson perfection 1650 scanner. Images were cropped using Adobe Photoshop 6.0 (Adobe Systems, Mountain View, CA) and imported into Canvas 9.0 for labeling. For 35S pulse-chase experiments, images were obtained by PhosphorImager (Scanner Control, version 4.1; GE Healthcare). For all RT-PCR, DNA images were acquired with a Gene Genius Bio Imaging System and GeneSnap software version 4.0. Where indicated, densitometry was performed with ImageQuant version 5.2 (GE Healthcare). Phase-contrast images were acquired using an Axiovert 135 microscope (Carl Zeiss, Jena, Germany) and RT monochrome Spot camera and software, version 3.3 (Diagnostic Instruments, Sterling Heights, MI) with a 40× objective (Figure 2D) or a 10× objective (Figure 3A). Growth area in Figure 3 was quantitated using Scion Image, version 1.62, while focused on the same plane for each sample. Immunofluorescent images were acquired with a Nikon Eclipse TE300 fluorescent microscope and RT SE. Spot camera and software, version 4.6 (Diagnostic Instruments) with a 40× objective. Fibronectin degradation was quantitated by measuring pixel density (using ImageQuant 5.2) within comparably sized areas of fibronectin proteolysis. Confocal images were acquired with a Zeiss LSM 510 META confocal microscope (Harvard NeuroDiscovery Center, Boston, MA). Z-stack reconstructions (143.4 × 143.4 × 22.1 μm) were performed from images taken every 1.1 μm with a 63× objective, and images were viewed with a Zeiss LSM Image Browser, version 3.5.0.223. Flow cytometry data acquisition was performed using a FACSCalibur flow cytometer with CellQuest software, version 3.3, with data analyzed using FlowJo software, version 6.4.4 (BD Biosciences).
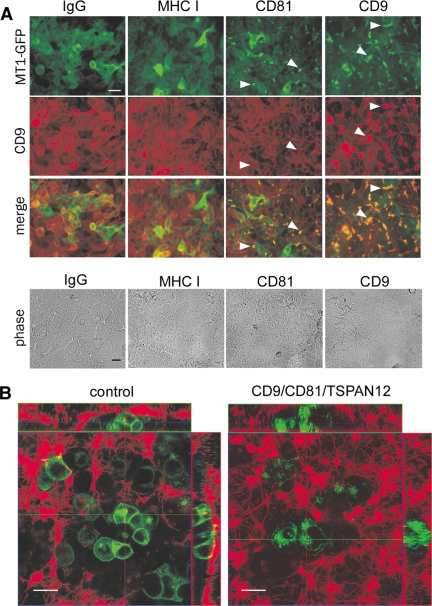
Figure 2.
Tetraspanin effects on MT1-MMP subcellular localization. (A) MCF-7-MT1-GFP cells were treated with the indicated control (IgG and MHC I) and tetraspanin (CD9 and CD81) antibodies (50 μg/ml for 2 d), fixed (in 4% paraformaldehyde), blocked in 5% BSA/PBS, incubated at 20°C with Alexa Fluor 546-conjugated anti-CD9 antibody (ALB6), and analyzed by fluorescence or phase-contrast microscopy. Bar, 20 μm. White arrowheads indicate coclustering of MT1-MMP-GFP with CD9 or CD81. (B) MCF-7-MT1-GFP cells were transfected with control or CD9/CD81/TSPAN12 siRNAs and plated within fibrin gels 2 d after siRNA transfection. Fibrin gels were spiked with 1% Alexa 546-labeled fibrinogen before addition of thrombin to form the gel. After 2 d, cells were visualized by confocal microscopy. Green, MT1-MMP-GFP; red, fibrin matrix. Laser power was adjusted to show similar green intensities between control and tetraspanin knockdown cells. Bar, 20 μm. Horizontal (green) and vertical (red) lines within the central XY image correspond to the planes chosen for XZ (up) and YZ (right) images, respectively.
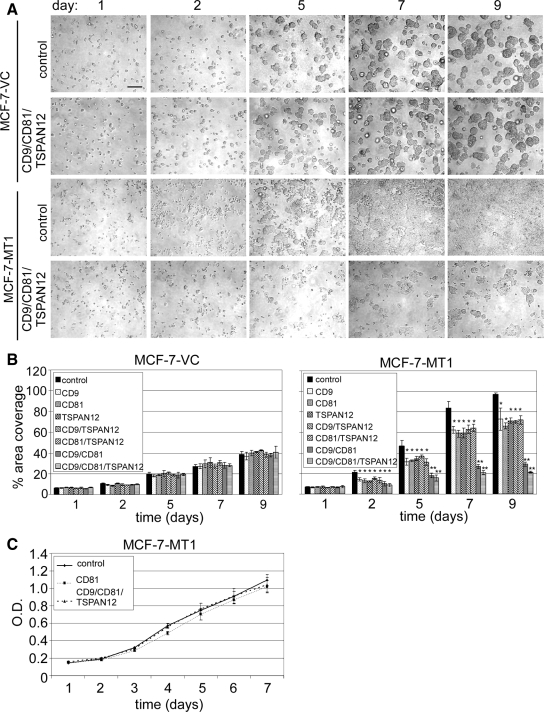
Figure 3.
Quantitation of tetraspanin effects on MT1-MMP-dependent cell growth/invasion within 3D fibrin gels. (A) MCF-7-MT1 and MCF-7-VC cells were treated for 2 d with siRNAs, and then cells were embedded within fibrin gels for the indicated times. Representative images are shown. Bar, 200 μm. (B) The percentage of area of cell coverage was determined to yield mean ± SD (n = 3; *p < 0.05, **p < 0.005 compared with control at each time point). (C) MCF-7-MT1 cells were treated with siRNAs as indicated, and then 2D growth was assessed using an MTT proliferation assay. Results are mean ± SD (n = 3).
RESULTS
MT1-MMP Association with Tetraspanin Proteins
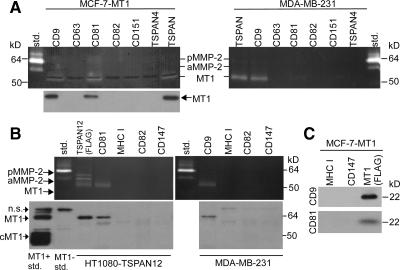
MT1-MMP may associate with at least four different tetraspanin proteins (Takino et al., 2003; Yanez-Mo et al., 2008; Kolesnikova et al., 2009). To clarify which might associate best with MT1-MMP on tumor cell lines, we immunoprecipitated CD9, CD63, CD81, CD82, CD151, and TSPAN4 from MCF-7-MT1 and MDA-MB-231 cells (Figure 1A). CD9 and CD81 yielded the most MT1-MMP, as seen by gelatin zymography and Western blot. TSPAN12 also associated with MT1-MMP, as seen upon immunoprecipitating TSPAN12-FLAG from HT1080 cells (Figure 1B). Antibodies to other tetraspanins (CD63, CD82, and CD151) or other abundant cell surface proteins (major histocompatibility complex I [MHC I], CD147) yielded little or no MT1-MMP (Figures 1, A and B). All tetraspanins and negative control proteins analyzed were well expressed by MCF-7-MT1, HT1080, and MDA-MB-231 cells with the exception of CD9 in HT1080 cells (Supplemental Table 1). In a reciprocal experiment, immunoprecipitation of MT1-MMP-FLAG but not control proteins (MHC I and CD147) yielded CD9 and CD81 (Figure 1C).
Figure 1.
Specific tetraspanins associating with MT1-MMP. (A) MCF-7-MT1 and MDA-MB-231 cells were lysed in 1% Brij 99 lysis buffer, and the indicated tetraspanins were immunoprecipitated (TSPAN, cocktail of tetraspanin antibodies against CD9, CD63, CD81, CD82, CD151, and TSPAN4). Eluates were analyzed by gelatin zymography and by Western blotting for MT1-MMP, using the anti-catalytic domain LEM2/15.8 antibody. Std, MMP-2 standard; pMMP-2, pro-MMP-2; aMMP-2, active MMP-2; n.s., nonspecific. All antibodies used in these and other immunoprecipitation experiments have been previously shown to immunoprecipitate their target antigens (Barnstable et al., 1978; Tachibana et al., 1997; Yang et al., 2002). (B) HT1080 cells stably expressing TSPAN12 and MDA-MB-231 cells were lysed in 1% Brij 99, cell surface proteins were immunoprecipitated, and eluates were analyzed by gelatin zymography and by MT1-MMP Western blotting using the anti-hinge AB815 antibody. cMT1, cleaved MT1. MT1 Standards are from MCF-7 cells (negative for MT1) or MCF-7 cells stably expressing MT1-MMP. (C) FLAG-MT1-MMP, MHC1, and CD147 were immunoprecipitated from MCF-7-MT1 cells (lysed in 1% Brij 99) and eluates were Western blotted for CD9 and CD81.
CD81 immunoprecipitation from HT1080 cells yielded MT1-MMP regardless of whether lysates were centrifuged at 100,000 × g (Supplemental Figure 1A). Hence, MT1-MMP-tetraspanin associations are not a protein insolubility artifact. MT1-MMP-tetraspanin association was also observed in MDA-MB-231, BT549, 293, U87, and NT2 cancer cells. Cells with minimal MT1-MMP expression (HeLa, LN827, MCF-7, A549, and K562) did not yield MT1-MMP zymogram activity (Supplemental Figure 1B).
Choice of lysis buffer is critical in these experiments. Immunoprecipitation of TSPAN12 yielded MT1-MMP in relatively stringent conditions (1% Brij 96 or TX-100), whereas CD9 and CD81 associations were seen in less stringent conditions (1% Brij 99 and CHAPS) (Supplemental Figure 2A and Supplemental Table 2). To confirm that tetraspanin-MT1-MMP associations are not postlysis artifacts, HT1080 1% Brij 99 lysates, containing TSPAN12-FLAG or MT1-MMP-GFP, were either mixed 1:1 or kept separate. Immunoprecipitation of FLAG yielded TSPAN12-FLAG and immunoprecipitation of GFP yielded MT1-MMP-GFP. There was no evidence of postlysis association between FLAG- and GFP-tagged proteins (Supplemental Figure 2B). As a positive control, immunoprecipitation of TSPAN12-FLAG yielded endogenous MT1-MMP (Supplemental Figure 2B, bottom left). Immunoprecipitation of CD81 yielded comparable amounts of both endogenous MT1-MMP and MT1-MMP-GFP (Supplemental Figure 2B, right). Hence, the GFP moiety does not interfere with MT1-MMP-GFP-tetraspanin association.
We also treated MCF-7-MT1-GFP cells with anti-CD9, anti-CD81, or control antibodies. Cells were then fixed and assessed for MT1-MMP-GFP and CD9 expression. Antibodies to CD9 or CD81, but not control antibodies (IgG or anti-MHC I), triggered coclustering of MT1-MMP-GFP (green) and CD9 (red) into overlapping punctate complexes (Figure 2A, white arrowheads). Cell morphology was not noticeably altered by antibody treatment (Figure 2A, bottom). These results support the conclusion that MT1-MMP associates with tetraspanins CD9 and CD81.
Tetraspanins Promote MT1-MMP–dependent Functions
In previous studies, tetraspanins CD63 and CD151 were suggested to inhibit MT1-MMP–dependent MMP-2 activation (Takino et al., 2003; Yanez-Mo et al., 2008), while paradoxically promoting collagen degradation (Yanez-Mo et al., 2008). Here, we undertook a comprehensive assessment of the effects of tetraspanins (CD9, CD81, and TSPAN12) on tumor cell MT1-MMP, by using four different functional assays. To focus attention on MT1-MMP, we mostly compared the functions of MCF-7-MT1 and MCF-7-VC cells. The former cells are stably transfected to express moderate levels of MT1-MMP, whereas the latter are vector control cells, with minimal background MMP activity. To efficiently disrupt the tetraspanin network in MCF-7-VC and MCF-7-MT1 cells, we silenced tetraspanins (CD9, CD81, and TSPAN12) in combinations of two or three at a time. Evidence for efficient knockdown of CD9, CD81, and TSPAN12 is shown in Supplemental Figure 3, A–C.
Tetraspanin effects on MT1-MMP function were first analyzed using Alexa 546-labeled 3D fibrin gels. During a 2-d growth interval, control siRNA-treated MCF7-MT1-GFP cells created large fibrin-depleted boroughs, as seen in both XY planes and in Z stacks (Figure 2B, left). By contrast, CD9/CD81/TSPAN12 siRNA-treated cells showed smaller regions of fibrin proteolysis (Figure 2A, right). Also evident were striking differences in MT1-MMP-GFP localization. In control siRNA-treated cells, MT1-MMP-GFP was localized mainly at the periphery, with some intracellular signal (Figure 2B, left). On tetraspanin knockdown, MT1-MMP-GFP became predominantly intracellular (Figure 2B, right).
In another 3D fibrin gel experiment, embedded MCF-7-VC and MCF-7-MT1 cells were monitored for changes in invasion and growth, due to tetraspanin knockdown (Figure 3). Using MT1-MMP to escape from fibrin growth constraints (Hotary et al., 2003), control siRNA-treated MCF-7-MT1 cells efficiently spread, invaded, and proliferated within fibrin gels (Figure 3A, row 3). By contrast, CD9/CD81/TSPAN12 knockdown cells showed minimal invasion and growth (Figure 3A, row 4). Double knockdown (CD9/CD81) MCF-7-MT1 cells also were markedly impaired, whereas single knockdown cells were affected to a lesser extent (Figure 3B). Because MCF-7-VC cells are minimally fibrinolytic, they were unable to escape matrix growth constraints. Consequently, they grew as immobile cysts, regardless of tetraspanin knockdown (Figure 3A, rows 1 and 2; and B). Proliferation of MCF-7-MT1 cells was not significantly affected by tetraspanin knockdown in a 2D proliferation/viability assay (Figure 3C).
Similar experiments were then carried out using 3D collagen gels. Once again, control siRNA-treated MCF7-MT1 cells, but not tetraspanin knockdown cells, escaped from a 3D growth restraint (Supplemental Figure 4B). Again, triple knockdown and CD9/CD81 double knockdown were most effective, whereas other double and single knockdowns were less effective (Supplemental Figure 4B). By contrast, tetraspanin knockdown had little effect on control MCF-7 cells (Supplemental Figure 4A).
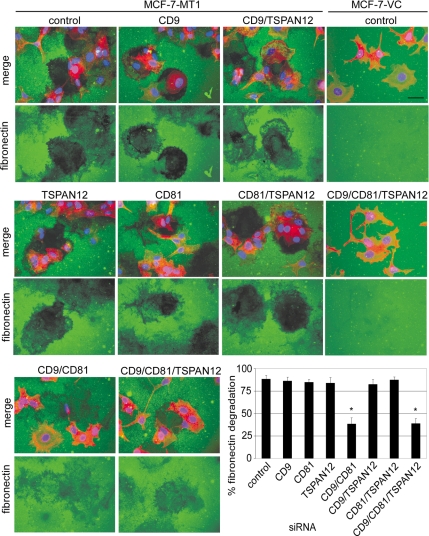
A third type of ECM degradation assay was then carried out, involving MT1-MMP-dependent pericellular fibronectin degradation. On plating on fibronectin overnight, MCF-7-MT1 cells efficiently cleared fibronectin, leaving dark patches in the fluorescent green lawn (Figure 4, top left). However, fibronectin degradation was significantly reduced after knockdown of CD9/CD81 or CD9/CD81/TSPAN12 (Figure 4, bottom). Other single or double knockdowns had minimal effect on MCF-7-MT1 cells (Figure 4). As expected, control MCF-7-VC cells did not degrade fibronectin, with or without tetraspanin knockdown (Figure 4, far right).
Figure 4.
Tetraspanin effects on MT1-MMP-dependent fibronectin degradation. MCF-7-VC and MCF-7-MT1 cells were treated with siRNAs, plated 4 d later on fibronectin (FN)-coated slides, and then incubated overnight in serum-free media with protease inhibitors (50 μg/ml aprotinin, 2 μM leupeptin, 20 μM E64, and 20 μM pepstatin A) to prevent non-MMP proteolysis of the FN. The following day, cells were washed, fixed, and FN was detected using an anti-FN antibody followed by Alexa 488-conjugated anti-mouse secondary antibody (green). The position of the cells was determined by staining the cytoskeleton with Alexa Fluor 546-phalloidin (red) and the nuclei with DAPI (blue). Representative fluorescent images (merge green/red/blue or green only) are shown. Bar, 20 μm. For the bar graph, 100% fibronectin degradation is defined as 0 pixel density within defined proteolyzed areas, whereas 0% degradation is pixel density in the unperturbed fibronectin layer. Results are mean of percentage of fibronectin degradation ± SD (n = 3; *p < 0.01 compared with control). Efficient tetraspanin knockdown was obtained similarly to that shown in Supplemental Figure 3.
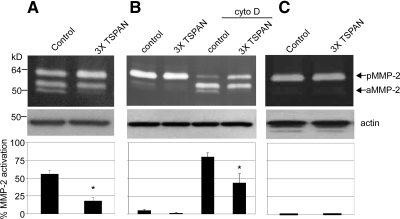
We next analyzed pro-MMP-2 activation, which is principally mediated by MT1-MMP (Sato et al., 1994). Compared with control knockdown, CD9/CD81/TSPAN12 knockdown in MCF-7-MT1 cells caused activation of pro-MMP-2 to be ∼78% inhibited (Figure 5A). We also analyzed tetraspanin knockdown effects on endogenous MT1-MMP in HT1080 cells. Because HT1080 cells do not express CD9, we knocked down CD81, TSPAN12, and CD151 (which weakly associates with MT1-MMP). Again, tetraspanin knockdown decreased pro-MMP-2 activation, this time by ∼40% (Figure 5B). The effect was particularly obvious when cytochalasin D (cyto D) was added to increase MT1-MMP protein levels in HT1080 cells. Cyto D has been shown previously to increase the transcription of MT1-MMP (Yan et al., 2000). Little MMP-2 activation was seen in MCF-7-VC cells (Figure 5C), thus confirming that MMP-2 activation is almost entirely MT1-MMP dependent in our MCF-7 cell system.
Figure 5.
Tetraspanin effects on MT1-MMP–dependent pro-MMP-2 activation. (A) MCF-7-MT1 cells were treated with control or CD9/CD81/TSPAN12 (3xTSPAN) siRNAs and after 4 d, cells were plated and incubated overnight in serum-free media supplemented with conditioned media from MCF-7 cells stably expressing MMP-2. Eighteen hours later, conditioned media and lysates were collected and analyzed for pro-MMP-2 activation by gelatin zymography or actin expression by Western blotting (as loading control), respectively. Densitometry was performed on zymograms and results displayed as percentage of active MMP-2 ± SD (n = 3; *p < 0.01 compared with control). (B) HT1080 cells were transfected with control or CD81/CD151/TSPAN12 (3xTSPAN) siRNAs. After 4 d, cells were plated and then incubated overnight in serum-free media with or without 4 μM cyto D. Conditioned media were then collected and analyzed by gelatin zymography for pro-MMP-2 activation. Zymogram densitometry was performed (n = 4; *p < 0.05 compared with control). (C) MCF-7-VC cells were treated and analyzed as described in A (n = 2).
Mechanistic Insights: How Do Tetraspanins Affect MT1-MMP Functions?
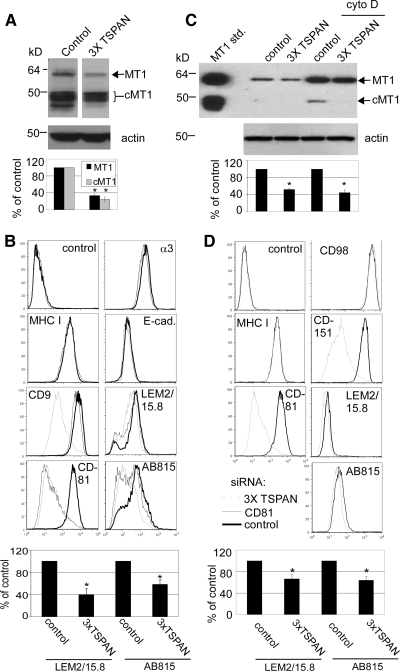
Tetraspanin knockdown (CD9/CD81/TSPAN12) in MCF-7-MT1 cells decreased levels of both full-length and cleaved MT1-MMP (cMT1) by 75 and 80%, respectively, as seen by densitometric quantitation (Figure 6A). Cell surface expression of MT1-MMP was also diminished, by 40–60%, as seen by flow cytometry using two different anti-MT1-MMP antibodies (Figure 6B). Surface expressions of other abundant cell surface proteins (MHC I, E-cadherin, and α3 integrin) were mostly unaffected, whereas CD9 and CD81 were appropriately diminished (Figure 6B). To confirm CD9/CD81/TSPAN12 knockdown results above, a second set of CD9, CD81, and TSPAN12 siRNAs were tested. Again, substantial knockdown of CD9, CD81, and TSPAN12 was accompanied by ∼50% reduction in expression of MT1-MMP (Supplemental Table 3).
Figure 6.
Tetraspanin effects on MT1-MMP expression. (A) TX114 cell lysates from MCF-7-MT1 samples in Figure 5A were blotted for MT1-MMP (AB815 antibody) and actin. Densitometry was performed on Western blots to yield indicated percent values relative to control lanes ± SD (n = 3; *p < 0.05 compared with control); cMT1 is cleaved MT1 lacking the catalytic domain (generated via an autocatalytic cleavage event). (B) MCF-7-MT1 cells were treated with control, CD81, or CD9/CD81/TSPAN12 (3xTSPAN) siRNAs. After 4 d, cell surface expression of MHC I, E-cadherin, α3 integrin, CD9, CD81, and MT1-MMP was determined by flow cytometry. For MT1-MMP, AB815 is anti-hinge antibody (detects both full-length and cleaved MT1-MMP) and LEM2/15.8 is anti-catalytic domain antibody (detects full-length MT1-MMP only). Quantitation of flow cytometry mean fluorescent intensity (MFI) values of MT1-MMP by using either AB815 or LEM2/15.8 antibodies is shown and expressed as percentage of control ± SD (n = 4; *p < 0.05 compared with control for each antibody). (C) Cell lysates from HT1080 samples in Figure 5B were blotted for MT1-MMP (AB815 antibody) or actin. Densitometry of total MT1-MMP was expressed as percentage relative to controls ± SD (n = 3; *p < 0.01 compared with control). cMT1 is cleaved MT1-MMP. MT1-MMP standard was prepared as described in Figure 1B. (D) HT1080 cells were transfected with control or CD81/CD151/TSPAN12 (3xTSPAN) siRNAs, and flow cytometry was performed 4 d after transfection for MHC I, CD98, CD81, CD151, and MT1-MMP (AB815 and LEM2/15.8) with cyto D stimulation 1 d before flow cytometry. Quantitation of flow cytometry MFI values of MT1-MMP is shown and expressed as percentage of control ± SD (n = 5; *p < 0.05 compared with control for each antibody).
Tetraspanin knockdown (CD81/TSPAN12/CD151) also decreased expression of total endogenous MT1-MMP (full length and cMT1) in HT1080 cells. Expression decreased by 45–55%, depending on whether cytochalasin D was absent or present (Figure 6C). Cell surface expression of endogenous HT1080 MT1-MMP also showed a significant decrease (∼30%), as detected by flow cytometry using two different anti-MT1-MMP antibodies. Control experiments confirmed knockdown of CD81, CD151, and TSPAN12 (Figure 6D and Supplemental Figure 4D), whereas other abundant cell surface proteins (MHC I and CD98) were unaffected (Figure 6D).
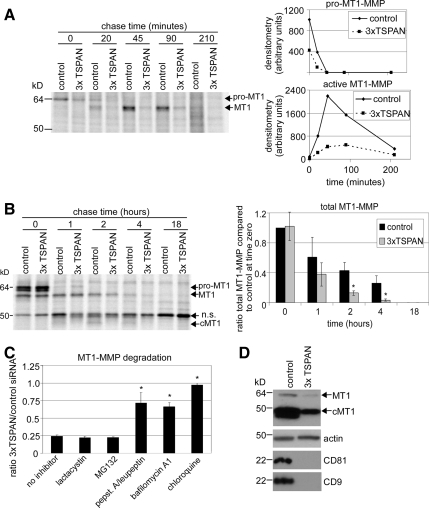
To address the mechanism whereby tetraspanins affect MT1-MMP protein levels, we first analyzed cell surface internalization. Knockdown of CD9/CD81/TSPAN12 did not affect MT1-MMP internalization in MCF-7-MT1-GFP cells, as determined using a reducible biotin accessibility assay (Supplemental Figure 5). Next, we used pulse-chase [35S]Met/Cys labeling to assess MT1-MMP delivery to the cell surface. After siRNA treatment, MCF-7-MT1 cells were pulsed (30 min) and then chased for 0, 20, 45, 90, or 210 min. Before lysis, cells were incubated with anti-FLAG antibody, which bound to extracellular pro- and active MT1-MMP-FLAG. After lysis, cell surface MT1-MMP-FLAG proteins were immunoprecipitated and analyzed (Figure 7A). Already at 30-min pulse, 0-min chase, there was less newly synthesized MT1-MMP delivered to the surface of CD9/CD81/TSPAN12 knockdown cells (Figure 7A), despite there being similar amounts of total MT1-MMP synthesized in control and tetraspanin knockdown cells (Figure 7B, time zero). Later time points (20-, 45-, and 90-min chase) confirmed the deficit in cell surface delivery caused by tetraspanin knockdown (Figure 7A). Figure 7D confirmed CD9 and CD81 knockdown and decreased levels of total MT1-MMP. Pulse-chase analysis of total MT1-MMP (Figure 7B) showed that tetraspanin knockdown did not diminish total MT1-MMP synthesis rates nor affect much the rapid conversion of pro- to active form. However, total MT1-MMP was degraded at a faster rate, and very little of the autocatalytically generated cMT1 was produced upon CD9/CD81/TSPAN12 knockdown. These results indicate that silencing of CD9/CD81/TSPAN12 causes less MT1-MMP to reach the cell surface. Instead, it seems to be rerouted to an intracellular compartment and degraded at a faster rate.
Figure 7.
Tetraspanins affect MT1-MMP trafficking and stability. (A) MCF-7-MT1 cells were transfected with control or CD9/CD81/TSPAN12 (3xTSPAN) siRNAs. After 4 d, the cell surface subset of MT1-MMP was analyzed by [35S]Met/Cys pulse-chase (see Materials and Methods). Appearance and processing of cell surface MT1-MMP was quantitated by densitometry and plotted as raw densitometric values for both pro- and active MT1-MMP as line graphs. (B) Four days after siRNA treatment, total MT1-MMP was analyzed by [35S]Met/Cys pulse-chase, and densitometry was performed on total labeled MT1-MMP protein and expressed as ratio compared with control at time zero ± SD (n = 3; *p < 0.05 compared with control at each time point). cMT1 is cleaved MT1. (C) MCF-7-MT1-GFP cells were transfected with control or CD9/CD81/TSPAN12 (3xTSPAN) siRNAs. Two days after siRNA treatment, cells were treated with either lactacystin (10 μM), MG132 (2.5 μM), pepstatin A (50 μM)/leupeptin (10 μM), bafilomycin A1 (25 nM), or chloroquine (50 μM) for 2 d. Cell lysates were then collected, and MT1-MMP was analyzed by Western blotting. Densitometry was performed on total MT1-MMP and represented as ratio of total MT1-MMP for 3xTSPAN/control for each inhibitor ± SD (n = 3; *p < 0.05 compared with no inhibitor). Note that all the lysosomal inhibitors stimulated MT1-MMP expression in control siRNA-treated cells compared with no inhibitor, consistent with normal degradation of MT1-MMP in lysosomes (data not shown). (D) Tetraspanin knockdown was confirmed by Western blotting of CD9 and CD81 from cell lysates. Decreased MT1-MMP expression with tetraspanin knockdown is shown by Western blotting. TSPAN12 mRNA was also appropriately decreased (data not shown).
To gain further insight into MT1-MMP degradation, we treated control or CD9/CD81/TSPAN12-silenced MCF-7-MT1-GFP cells with lysosome and proteasome inhibitors. Lysosome inhibitors (chloroquine, bafilomycin A1, and pepstatin A/leupeptin combination) largely protected total MT1-MMP from CD9/CD81/TSPAN12 knockdown-induced degradation (Figure 7C). By contrast, proteasome inhibitors (MG132 and lactacystin) did not protect MT1-MMP from degradation.
DISCUSSION
The role of MT1-MMP as a proinvasive protease during tumor cell invasion has been well established (Osenkowski et al., 2004; Itoh and Seiki, 2006). MT1-MMP associates with various tetraspanin proteins (Takino et al., 2003; Yanez-Mo et al., 2008; Kolesnikova et al., 2009), resulting in possible negative regulation of expression (Takino et al., 2003), and both negative and positive functional regulation (Yanez-Mo et al., 2008). Here, we describe how multiple tetraspanins associate with newly synthesized MT1-MMP in cancer cells, thereby preventing its lysosomal degradation, supporting cell surface expression and uniformly enhancing MT1-MMP functions in multiple proteolysis assays.
Functional Consequences of Tetraspanin Association
Among seven tetraspanins tested, CD9, CD81, and TSPAN12 showed the most robust MT1-MMP association, as seen by reciprocal coimmunoprecipitation and antibody-induced colocalization. Removal of tetraspanins CD9, CD81, and/or TSPAN12 markedly diminished all MT1-MMP–dependent functions tested. MT1-MMP promotes fibronectin proteolysis, leading to enhanced cell migration and invasion (Takino et al., 2007). Tetraspanin knockdown impaired MT1-MMP–dependent proteolysis of two-dimensional fibronectin, which should lead to impaired cell migration and invasion. MT1-MMP also supports tumor proliferation by relieving growth constraints imposed by 3D ECM (Hotary et al., 2003). Tetraspanin knockdown markedly diminished cancer cell invasion and proliferation within 3D fibrin and collagen gels, consistent with relief of growth constraints. These results may help to explain why loss of tetraspanin-MT1-MMP association correlated with diminished glioblastoma growth within a 3D in vivo setting (Kolesnikova et al., 2009). Another important function of MT1-MMP is to activate MMP-2 (Sato et al., 1994). Tetraspanin knockdown markedly diminished activation of MMP-2 by MT1-MMP (either endogenous or overexpressed) in two different tumor cell lines. Given that MMP-2 also can be a key player during tumor cell invasion/tumor progression (Itoh et al., 1998), loss of MMP-2 activation likely contributes to impaired tumor cell functions caused by tetraspanin knockdown.
Deletion of individual tetraspanins in mice and flies has yielded relatively minor developmental effects (Hemler, 2005), likely due to functional compensation among different tetraspanins (Fradkin et al., 2002; Kaji et al., 2002). Here, compared with single tetraspanin knockdowns, depletion of two or three tetraspanins more notably diminished MT1-MMP expression and functions. Among combinations tested, CD9/CD81 knockdown was most effective, consistent with these molecules providing overlapping (i.e., partly compensating) contributions toward MT1-MMP expression and function. In this regard, the structurally similar CD9 and CD81 molecules partially compensate for each other during oocyte fertilization (Rubinstein et al., 2006). CD9 and CD81 are not known to support tumor invasion and growth, perhaps because they are usually studied one at a time. Notably, CD9/CD81 null and MT1-MMP null mice show striking similarities. Both strains show diminished size, abnormal bone-associated phenotypes (e.g., osteopenia), and alveolar airspace enlargement (Holmbeck et al., 1999; Hemler, 2003; Takeda et al., 2003; Atkinson et al., 2005). Hence, the CD9/CD81 null mouse phenotype could arise, at least partly, from altered MT1-MMP regulation.
Compared with CD9/CD81 double knockdown results, TSPAN12/CD9/CD81 triple knockdown effects were only slightly more obvious, possibly because endogenous TSPAN12 protein is less abundant than CD9 and CD81. Nonetheless, knockdown of endogenous TSPAN12 individually did have a small but significant effect on MT1-MMP functions within 3D fibrin and collagen matrices, suggesting that TSPAN12 can be a contributor. Its wide expression (Serru et al., 2000) should enable collaboration with MT1-MMP on many cancer cell types. Further study of endogenous TSPAN12 protein awaits the development of appropriate antibody reagents.
Our CD81/CD151/TSPAN12 knockdown diminished MT1-MMP-dependent MMP-2 activation in HT1080 cells. Elsewhere, CD151 knockdown stimulated MT1-MMP–dependent MMP-2 activation on endothelial cells (Yanez-Mo et al., 2008). These results are not necessarily contradictory because 1) knockdown of CD151 alone may be insufficient to disrupt MT1-MMP function within TEMs, and 2) regulation of MT1-MMP may differ in primary endothelial cells compared with tumor cell lines. In addition, in tumor cell lines MT1-MMP associations are much more robust with CD9 and CD81 (compared with CD151).
Tetraspanin Effects on MT1-MMP: Mechanistic Insights
Tetraspanin knockdown decreased MT1-MMP expression (both cell surface and total) in both overexpressed and endogenous systems. Hence, combinations of tetraspanins (especially CD9, CD81, TSPAN12, and CD151) support MT1-MMP expression. Elsewhere, knockdown of CD151 alone did not affect MT1-MMP expression (Yanez-Mo et al., 2008), and CD63 knockdown increased rather than decreased MT1-MMP expression (Takino et al., 2003). Because we did not observe CD63 association with MT1-MMP, we did not pursue CD63 further. MT1-MMP association with other cell surface proteins, including CD44, syndecan, and αV integrins (Barbolina and Stack, 2008), leads to shedding and/or proteolytic processing. However, MT1-MMP does not seem to cause tetraspanin proteolysis.
For total MT1-MMP, initial synthesis and stability were unaltered upon tetraspanin removal—decreased expression was not seen until later time points (e.g., 2- to 4-h chase). However, cell surface MT1-MMP expression was greatly diminished even at early time points. Rather than going to the cell surface, newly synthesized MT1-MMP is apparently diverted to an intracellular compartment and subsequently degraded at an accelerated rate. Consistent with this, confocal microscopy showed that MT1-MMP was not only diminished in tetraspanin knockdown cells but also shifted away from the surface and into intracellular compartments. We considered that tetraspanin knockdown might cause MT1-MMP to be rapidly removed soon after it arrives at the cell surface. However, we did not observe increases in MT1-MMP internalization or shedding into exosome/microvesicle particles (unpublished data), again consistent with MT1-MMP not reaching the cell surface. After tetraspanin silencing, MT1-MMP disappearance was inhibited by lysosomal protease inhibitors chloroquine, bafilomycin A1, and pepstatin A/leupeptin combination. These results suggest that MT1-MMP is diverted into lysosomal compartments and degraded.
There is precedent for tetraspanins supporting partner protein surface expression. CD81 facilitates EWI-2 (Stipp et al., 2003a) and CD19 (Shoham et al., 2006) surface expression, and tetraspanins UP1a and UP1b enable UPII and UPIII surface expression (Hu et al., 2005). However, in these cases, tetraspanin absence impairs intracellular maturation. By contrast, diminished cell surface MT1-MMP is not accompanied by delayed maturation (Figure 7B). Hence, tetraspanins may affect MT1-MMP at a post-endoplasmic reticulum/Golgi stage, by a novel mechanism.
Because tetraspanins such as CD9, CD81, CD151 (Berditchevski, 2001), and TSPAN12 (our unpublished data) associate with integrins, MT1-MMP should be brought into proximity with integrins, as demonstrated in endothelial cells for CD151 and MT1-MMP (Yanez-Mo et al., 2008). Hence, tetraspanins may coordinate processes of cell adhesion and ECM proteolysis during cell invasion and migration. Within invading cells, MT1-MMP in lamellipodia and invadopodia is well positioned for localized ECM degradation (Itoh, 2006). Indeed, silencing of proteins such as TI-VAMP/VAMP-7, Sec3/Sec8/IQGAP1, or RAb8 leads to disruption of invadopodia formation and/or loss of MT1-MMP-dependent tumor cell invasion (Bravo-Cordero et al., 2007; Sakurai-Yageta et al., 2008; Steffen et al., 2008). Tetraspanin protein complexes contribute to the organization of lamellipodia, filopodia, and uropodia (Shigeta et al., 2003; Stipp et al., 2003a) and therefore should also contribute to invadopodia. However, it remains to be seen whether ablation of tetraspanins, and loss of cell surface MT1-MMP expression, is accompanied by disruption of invadopodia.
Implications
Broad-spectrum synthetic inhibitors, targeting MMP active sites, have undergone extensive clinical testing. Unfortunately, these agents failed to provide useful cancer therapy in humans (Overall and Kleifeld, 2006). Hence, the challenge remains to target MMPs successfully with minimal harmful side effects. For a membrane-bound protease, interfering with subcellular localization may be a beneficial alternative to typical active site inhibition approaches. Anti-tetraspanin antibody effects on MT1-MMP localization may represent a promising step in this direction. Furthermore, because adult mice survive well without CD9 and CD81 expression (Takeda et al., 2003), therapies targeting these molecules should not be excessively toxic. This approach could potentially provide alternative treatment options for tumor invasion and metastasis and other pathological conditions dependent on pericellular proteolysis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant GM-38903 and a Canadian Institutes of Health Research (CIHR) fellowship (to M.A.L.).
Abbreviations used:
- GFP
green fluorescent protein
- MMP
matrix metalloproteinase
- MT1-MMP
membrane type-1 matrix metalloproteinase
- TEM
tetraspanin-enriched microdomain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-11-1149) on February 11, 2009.
REFERENCES
- Atkinson J. J., Holmbeck K., Yamada S., Birkedal-Hansen H., Parks W. C., Senior R. M. Membrane-type 1 matrix metalloproteinase is required for normal alveolar development. Dev. Dyn. 2005;232:1079–1090. doi: 10.1002/dvdy.20267. [DOI] [PubMed] [Google Scholar]
- Barbolina M. V., Stack M. S. Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin. Cell Dev. Biol. 2008;19:24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- Borrirukwanit K., Lafleur M. A., Mercuri F. A., Blick T., Price J. T., Fridman R., Pereira J. J., Leardkamonkarn V., Thompson E. W. The type I collagen induction of MT1-MMP-mediated MMP-2 activation is repressed by alphaVbeta3 integrin in human breast cancer cells. Matrix Biol. 2007;26:291–305. doi: 10.1016/j.matbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Marrero-Diaz R., Megias D., Genis L., Garcia-Grande A., Garcia M. A., Arroyo A. G., Montoya M. C. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas C., Seiter S., Claas A., Savelyeva L., Schwab M., Zoller M. Association between the rat homologue of CO-029, a metastasis-associated tetraspanin molecule and consumption coagulopathy. J. Cell Biol. 1998;141:267–280. doi: 10.1083/jcb.141.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew A. F., Blick T. J., Lafleur M. A., Tim E. L., Robbie M. J., Rice G. E., Quinn M. A., Thompson E. W. Correlation of tumor- and stromal-derived MT1-MMP expression with progression of human ovarian tumors in SCID mice. Gynecol. Oncol. 2004;95:437–448. doi: 10.1016/j.ygyno.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Fradkin L. G., Kamphorst J. T., DiAntonio A., Goodman C. S., Noordermeer J. N. Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc. Natl. Acad. Sci. USA. 2002;99:13663–13668. doi: 10.1073/pnas.212511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez B. G., Matias-Roman S., Yanez-Mo M., Sanchez-Madrid F., Arroyo A. G. ECM regulates MT1-MMP localization with beta1 or alphavbeta3 integrins at distinct cell compartments modulating its internalization and activity on human endothelial cells. J. Cell Biol. 2002;159:509–521. doi: 10.1083/jcb.200205026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H. Y., et al. Overexpression of membrane-type matrix metalloproteinase-1 gene induces mammary gland abnormalities and adenocarcinoma in transgenic mice. Cancer Res. 2001;61:984–990. [PubMed] [Google Scholar]
- Hemler M. E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- Hemler M. E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell. Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Hemler M. E., Strominger J. L. Characterization of antigen recognized by the monoclonal antibody (4F2): different molecular forms on human T and B lymphoblastoid cell lines. J. Immunol. 1982;129:623–628. [PubMed] [Google Scholar]
- Holmbeck K., et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000;149:1309–1323. doi: 10.1083/jcb.149.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- Hu C. C., Liang F. X., Zhou G., Tu L., Tang C. H., Zhou J., Kreibich G., Sun T. T. Assembly of urothelial plaques: tetraspanin function in membrane protein trafficking. Mol. Biol. Cell. 2005;16:3937–3950. doi: 10.1091/mbc.E05-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Sossey-Alaoui K., Beachy S. H., Geradts J. The tetraspanin superfamily member NET-6 is a new tumor suppressor gene. J. Cancer Res. Clin. Oncol. 2007;133:761–769. doi: 10.1007/s00432-007-0221-1. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tanioka M., Yoshida H., Yoshioka T., Nishimoto H., Itohara S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998;58:1048–1051. [PubMed] [Google Scholar]
- Itoh Y. MT1-MMP: a key regulator of cell migration in tissue. IUBMB Life. 2006;58:589–596. doi: 10.1080/15216540600962818. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J. Cell. Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- Jiang W. G., Davies G., Martin T. A., Parr C., Watkins G., Mason M. D., Mansel R. E. Expression of membrane type-1 matrix metalloproteinase, MT1-MMP in human breast cancer and its impact on invasiveness of breast cancer cells. Int. J. Mol. Med. 2006;17:583–590. [PubMed] [Google Scholar]
- Kaji K., Oda S., Miyazaki S., Kudo A. Infertility of CD9-deficient mouse eggs is reversed by mouse CD9, human CD9, or mouse CD81; polyadenylated mRNA injection developed for molecular analysis of sperm-egg fusion. Dev. Biol. 2002;247:327–334. doi: 10.1006/dbio.2002.0694. [DOI] [PubMed] [Google Scholar]
- Kolesnikova T., Kazarov A., Lemieux M. E., Lafleur M. A., Kesari S., Kung A. L., Hemler M. E. Glioblastoma Inhibition by Cell Surface Immunoglobulin Protein EWI-2, In Vitro and In Vivo. Neoplasia. 2009;11:77–86. doi: 10.1593/neo.81180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko O. V., Yang X. H., Hemler M. E. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol. Cell Proteomics. 2007;6:1855–1867. doi: 10.1074/mcp.M700183-MCP200. [DOI] [PubMed] [Google Scholar]
- Lafleur M. A., Mercuri F. A., Ruangpanit N., Seiki M., Sato H., Thompson E. W. Type I collagen abrogates the clathrin-mediated internalization of membrane type 1 matrix metalloproteinase (MT1-MMP) via the MT1-MMP hemopexin domain. J. Biol. Chem. 2006;281:6826–6840. doi: 10.1074/jbc.M513084200. [DOI] [PubMed] [Google Scholar]
- Liu W. M., Zhang X. A. KAI1/CD82, a tumor metastasis suppressor. Cancer Lett. 2006;240:183–194. doi: 10.1016/j.canlet.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Mori H., Tomari T., Koshikawa N., Kajita M., Itoh Y., Sato H., Tojo H., Yana I., Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenkowski P., Toth M., Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J. Cell. Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- Overall C. M., Kleifeld O. Tumour microenvironment–opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- Penas P. F., Garcia-Diez A., Sanchez-Madrid F., Yanez-Mo M. Tetraspanins are localized at motility-related structures and involved in normal human keratinocyte wound healing migration. J. Invest. Dermatol. 2000;114:1126–1135. doi: 10.1046/j.1523-1747.2000.00998.x. [DOI] [PubMed] [Google Scholar]
- Rubinstein E., Ziyyat A., Prenant M., Wrobel E., Wolf J. P., Levy S., Le Naour F., Boucheix C. Reduced fertility of female mice lacking CD81. Dev. Biol. 2006;290:351–358. doi: 10.1016/j.ydbio.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Sakurai-Yageta M., Recchi C., Le Dez G., Sibarita J. B., Daviet L., Camonis J., D'Souza-Schorey C., Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- Serru V., Dessen P., Boucheix C., Rubinstein E. Sequence and expression of seven new tetraspans. Biochim. Biophys. Acta. 2000;1478:159–163. doi: 10.1016/s0167-4838(00)00022-4. [DOI] [PubMed] [Google Scholar]
- Shigeta M., Sanzen N., Ozawa M., Gu J., Hasegawa H., Sekiguchi K. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J. Cell Biol. 2003;163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoham T., Rajapaksa R., Kuo C. C., Haimovich J., Levy S. Building of the tetraspanin web: distinct structural domains of CD81 function in different cellular compartments. Mol. Cell. Biol. 2006;26:1373–1385. doi: 10.1128/MCB.26.4.1373-1385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., Chavrier P. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 2008;18:926–931. doi: 10.1016/j.cub.2008.05.044. [DOI] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 2003a;163:1167–1177. doi: 10.1083/jcb.200309113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp C. S., Kolesnikova T. V., Hemler M. E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003b;28:106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- Tachibana I., Bodorova J., Berditchevski F., Zutter M. M., Hemler M. E. NAG-2, a novel transmembrane-4 superfamily (TM4SF) protein that complexes with integrins and other TM4SF proteins. J. Biol. Chem. 1997;272:29181–29189. doi: 10.1074/jbc.272.46.29181. [DOI] [PubMed] [Google Scholar]
- Takeda T., Hattori N., Tokuhara T., Nishimura Y., Yokoyama M., Miyake M. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res. 2007;67:1744–1749. doi: 10.1158/0008-5472.CAN-06-3090. [DOI] [PubMed] [Google Scholar]
- Takeda Y., et al. Tetraspanins CD9 and CD81 function to prevent the fusion of mononuclear phagocytes. J. Cell Biol. 2003;161:945–956. doi: 10.1083/jcb.200212031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takino T., Miyamori H., Kawaguchi N., Uekita T., Seiki M., Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem. Biophys. Res. Commun. 2003;304:160–166. doi: 10.1016/s0006-291x(03)00544-8. [DOI] [PubMed] [Google Scholar]
- Takino T., Saeki H., Miyamori H., Kudo T., Sato H. Inhibition of membrane-type 1 matrix metalloproteinase at cell-matrix adhesions. Cancer Res. 2007;67:11621–11629. doi: 10.1158/0008-5472.CAN-07-5251. [DOI] [PubMed] [Google Scholar]
- Yan L., Moses M. A., Huang S., Ingber D. E. Adhesion-dependent control of matrix metalloproteinase-2 activation in human capillary endothelial cells. J. Cell Sci. 2000;113:3979–3987. doi: 10.1242/jcs.113.22.3979. [DOI] [PubMed] [Google Scholar]
- Yanez-Mo M., et al. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112:3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 2002;13:767–781. doi: 10.1091/mbc.01-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. H., Richardson A. L., Torres-Arzayus M. I., Zhou P., Sharma C., Kazarov A. R., Andzelm M. M., Strominger J. L., Brown M., Hemler M. E. CD151 accelerates breast cancer by regulating alpha 6 integrin function, signaling, and molecular organization. Cancer Res. 2008;68:3204–3213. doi: 10.1158/0008-5472.CAN-07-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Hotary K. B., Nan B., Bosch F. X., Munoz N., Weiss S. J., Cho K. R. Expression of membrane type 1 matrix metalloproteinase is associated with cervical carcinoma progression and invasion. Cancer Res. 2005;65:6543–6550. doi: 10.1158/0008-5472.CAN-05-0231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.