Abstract
The substrate-specific deubiquitylating enzyme USP9X is a putative “stemness” gene expressed in many progenitor cell populations. To test its function in embryonic stem cell-derived neural progenitor/stem cells, we expressed USP9X from a Nestin promoter. Elevated USP9X levels resulted in two phenomena. First, it produced a dramatically altered cellular architecture wherein the majority (>80%) of neural progenitors was arranged into radial clusters. These progenitors expressed markers of radial glial cells and were highly polarized with adherens junction proteins (N-cadherin, β-catenin, and AF-6) and apical markers (Prominin1, atypical protein kinase C-ζ) as well as Notch, Numb, and USP9X itself, concentrated at the center. The cluster centers were also devoid of nuclei and so resembled the apical end-feet of radial progenitors in the neural tube. Second, USP9X overexpression caused a fivefold increase in the number of radial progenitors and neurons, in the absence of exogenous growth factors. 5-Bromo-2′-deoxyuridine labeling, as well as the examination of the brain lipid-binding protein:βIII-tubulin ratio, indicated that nestin-USP9X enhanced the self-renewal of radial progenitors but did not block their subsequent differentiation to neurons and astrocytes. nestin-USP9X radial progenitors reformed clusters after passage as single cells, whereas control cells did not, suggesting it aids the establishment of polarity. We propose that USP9X-induced polarization of these neural progenitors results in their radial arrangement, which provides an environment conducive for self-renewal.
INTRODUCTION
At early stages of vertebrate development, the future central nervous system (CNS) exists as a population of multipotent neural progenitor/neural stem cells (NP/NSC) arranged in a highly polarized epithelial sheet known as the neuroepithelium (NE). The progeny of these cells will ultimately be responsible for the production of all neurons, glia, and oligodendrocytes in the adult CNS (Gotz and Huttner, 2005). Initially, in most regions of the forming nervous system, these NE cells increase in number, resulting in the expansion and morphogenesis of the neural plate and neural tube. During mid-late embryogenesis, the NE cells switch from the expansive phase to a neurogenic phase, resulting in the production of neuronal cell types that migrate away from the proliferative zones of the neural tube (Huttner and Kosodo, 2005). At the onset of this process, the NE gives rise to a second class of NP/NSC, the radial glial cells (RGCs), which share the progenitor and stem cell characteristics of self-renewal and multipotency along with hallmarks of the astrocytic lineage (Gotz and Barde, 2005; Gotz and Huttner, 2005; Huttner and Kosodo, 2005). Lineage tracing experiments identified RGCs as the source of the majority of neurons at all axial levels studied (Malatesta et al., 2003; Anthony et al., 2004). The production of neurons is followed by a phase of gliogenesis, late in gestation, which continues through early postnatal development. The majority of multipotent stem cells in the murine CNS are depleted during these differentiation phases such that in the adult, NP/NSCs mainly exist in specialized niches, to supply new cells to the olfactory and hippocampal systems (Gregg and Weiss, 2003; Lledo et al., 2006). As a result, the regenerative capacity of the adult CNS is extremely limited. To understand how to identify and use cells with latent regenerative potential in the CNS, or realize the potentials of cell therapies, we must first identify the cellular and molecular pathways governing the processes of neurogenesis and gliogenesis. Central to such knowledge is a detailed understanding of the molecular mechanisms controlling the functions of NP/NSCs.
The inaccessibility of the embryonic CNS is a major impediment to unraveling how molecular and cellular pathways interconnect during neural development. In vitro systems have successfully complemented in vivo studies by providing a rapid, initial screening of candidate molecules. Several protocols exist for the differentiation of embryonic stem (ES) cells toward neural fates that replicate the in vivo chronological progression from neural progenitor to neuron and then later astrocytes and finally oligodendrocytes (reviewed in Cai and Grabel, 2007). With the sequential application of factors as they occur in vivo, mouse ES cells can be differentiated to subsets of neurons, indicating that in vivo molecular mechanisms can be activated in vitro (Wichterle et al., 2002; Salero and Hatten, 2007). It has been proposed that the reverse may also be true, that the in vitro generation of neural populations from ES cells may provide an avenue in which to test molecular aspects of neural differentiation (Bibel et al., 2004). The advantage of in vitro systems is the generation of relatively coordinated, homogeneous populations that are amenable to biochemical and molecular, and hence quantitative, analyses. The ease of genetic manipulation of mouse ES cells also makes them ideal for the molecular dissection of events underpinning neurogenesis (Bibel et al., 2004). We used these aspects to address putative functions of USP9X in the maintenance and expansion of ES-derived NPs.
Several studies have analyzed gene and protein expression in a range of progenitor/stem cells. The rationale being that, comparison of the transcriptome or proteome of stem cells such as embryonic, hemopoietic, neural, epidermal, and others might identify a common molecular pathway(s). For although stem cells differ widely in their proliferative rate and capacity, potency, location, and contribution to homeostasis and wound repair, they all share the defining characteristics of potency and self-renewal. The “stemness” hypothesis is controversial due to the apparent inconsistency in genes identified in these screens. However one gene/protein that has been identified in several independent studies is the substrate-specific deubiquitylating enzyme USP9X (also known as fat facets in mouse, FAM). USP9X was identified as a putative stemness gene in the initial microarray analyses of three mouse stem cell populations, including embryonic, hemopoietic, and neural (Ivanova et al., 2002; Ramalho-Santos et al., 2002). More recently, a comparison of gene expression in two populations of stem cells in the hair bulge with that published for embryonic, hemopoietic, and neural stem cells (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Blanpain et al., 2004) found USP9X to be one of 37 genes most highly expressed in all five populations. Proteomic (Van Hoof et al., 2006) and microarray (Sato et al., 2003) analyses have identified increased levels of USP9X in undifferentiated human and mouse embryonic stem cells and a screen for cancer stemness genes in hepatocellular carcinomas (Chiba et al., 2006).
The results from the stem cell analyses are consistent with what is known about USP9X in vivo. USP9X/FAM is highly expressed in the totipotent blastomeres of the preimplantation mouse embryo where it is required for cleavage and adhesion (Pantaleon et al., 2001). USP9X expression is also strong in the germ cells of males and females (Noma et al., 2002). During postimplantation embryogenesis, USP9X is most highly expressed in developing organs and expression is down-regulated as differentiation proceeds (Wood et al., 1997). Of particular note are the very high levels of USP9X/FAM mRNA in the neural progenitors of the developing CNS (Wood et al., 1997; Friocourt et al., 2005). The expression of USP9X in undifferentiated cell types was also observed in zebrafish embryos, where the protein is also very highly conserved (90% identity) (Khut et al., 2007). The function of USP9X as a deubiquitylating enzyme (Dub), which are major regulatory proteins, is also consistent with a potential role in progenitor/stem cell maintenance. Dubs by definition function downstream in the ubiquitin pathway, reversing ubiquitylation and potentially act as the final arbiter of substrate fate. Not surprisingly, Dubs are found to regulate all major facets of cell and developmental biology, by regulating protein stability, transport, localization, and activity (Nijman et al., 2005; Millard and Wood, 2006).
In this study, we used the directed differentiation of ES cells to neurons as a source of NP/NSCs and assayed the effect of transgenic USP9X expression in the NP/NSC population. We show that modest increases in USP9X had dramatic effects on the architecture of radial progenitor (RP) colonies resulting from polarization. The result of these changes was the enhanced self-renewing potential of in vitro-derived RPs.
MATERIALS AND METHODS
Immunofluorescence on Brain Sections
Embryos were collected at embryonic day (E) 14.5 from time mated Swiss mice. Embryos were fixed with 4% paraformaldehyde (PFA), embedded in paraffin, and 5-μm horizontal sections were cut as described previously (Junghans et al., 2005). Sections were subjected to antigen retrieval as described previously (Junghans et al., 2005) and blocked with phosphate-buffered saline (PBS) containing 0.2% Tween 20 (PBST) and 1% BSA before staining. Sections were incubated with primary antibodies O/N at 4°C diluted in PBST containing 0.01% bovine serum albumin (BSA); anti-USP9X-N1 (1:50, in-house), anti-Nestin (1:10; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), anti-Notch1 (1:500; Nyfeler et al., 2005), and anti-N-cadherin (1:1000; BD Biosciences Transduction Laboratories, Lexington, KY). Secondary antibodies were incubated for 2 h at room temperature (RT), diluted in PBST containing 0.01% BSA; goat anti-mouse immunoglobulin (Ig) G and donkey anti-rabbit antibodies conjugated with fluorescein isothiocyanate (FITC) or Cy3, respectively (1:600; The Jackson Laboratory, Bar Harbor, ME). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA), and coverslips were mounted with PBS-glycerol containing Anti-Fade reagent (Invitrogen). Effects of nonspecific staining were excluded by the use of secondary antibody-only controls (data not shown).
Transgene Construction
The vector pCAG-i-PURO (Miyahara et al., 2000) was linearized with Sca1 before electroporation into ES cells. Construction of the Nestin transgenes was performed as follows; the pNestin374-LacZ (a gift from U. Lendahl, Karolinska Institutet, Stockholm, Sweden) was digested with Not1 and Cla1 to excise the LacZ gene. The internal ribosome entry site (ires)-green fluorescent protein (GFP) cassette of pMSCV-ires-GFP (a gift from Arthur Nienhuis, St. Jude Children's Research Hospital, Memphis, TN) was removed by digestion with Not1 and EcoR1. Both the Nestin374 fragment and ires-GFP cassette were blunted before ligation to create pNestin374-ires-GFP. The vector pNestin374-GW-ires-GFP was generated by linearizing pNestin374-ires-GFP with Xho1 and generating blunt ends before ligating in the Gateway Cassette B (Invitrogen) to generate the Gateway Destination vector pNestin374-GW-ires-GFP. The vector pnestin-374-USP9X-ires-GFP was generated by conducting an LR reaction (Invitrogen) between the donor vector pDONR-USP9X (Murray et al., 2004) and the destination vector pNestin374-GW-ires-GFP. Both pNestin374-USP9X-ires-GFP and pNestin374-ires-GFP were linearized with AatII before electroporation into R1 ES cells.
Cell Culture
Feeder-independent R1 ES cells and 46EC Sox1-GFP knockin ES cells were routinely passaged every 3 d as described previously (Joyner, 2000). ES cell media consisted of DMEM (Invitrogen) supplemented with 15% fetal calf serum (JRH Biosciences, Lenexa, KS), nonessential amino acids (Sigma- Aldrich, St. Louis, MO), 2β-mercaptoethanol (Sigma-Aldrich), penicillin-streptomycin (JRH Biosciences), and 100 U/ml Leukemia Inhibitory Factor (LIF) (Adelaide University, Adelaide, SA, Australia). Cells were incubated in a humidified atmosphere containing 10% CO2. Electroporation of ES cells was as described previously (Joyner, 2000). Briefly, 6 × 106 R1 feeder-independent ES cells were mixed with 40 μg of linearized DNA containing a 10:1 M ratio of either pNestin374-USP9x-ires-GFP or pNestin374-ires-GFP:pCAGiPURO and electroporated using a Bio-Rad Gene-PulserII unit at 240 V and 500 μF in a 0.4-cm cuvette. Electroporated ES cells were plated onto 4 × 10-cm plates and incubated for 2 d in ES cell media before addition of 1 μg/ml puromyocin (Sigma-Aldrich).
For the directed differentiation of ES cells into neuronal lineages, a chemically defined medium was used in adherent monolayer conditions as described previously (Ying et al., 2003). Briefly, ES cells were passaged the day before differentiation and seeded at an equivalent density of 2 × 106 cells/10-cm plate. ES cells were then dissociated into single cells and washed twice to remove traces of LIF before plating into gelatinized dishes (Falcon; BD Biosciences Discovery Labware, Bedford, MA) at a density equivalent to 8.1 × 106 cells/10-cm dish in N2B27 media. N2B27 media consists of a 1:1 ratio of Neurobasal:DMEM/Hams F-12 (Invitrogen) supplemented with N2 (1:200; Invitrogen), B27 (1:100; Invitrogen), and 0.1% 2β-mercaptoethanol (Sigma-Aldrich). The day of plating in N2B27 media is referred to as day 0 of differentiation. N2B27 medium was replaced every second day during differentiation. For passaging of neural progenitors, day 5 cultures were dissociated with 0.05% trypsin (JRH Biosciences) and trituration to generate single cells that were replated onto dishes pretreated with 10 μg/ml fibronectin (Sigma-Aldrich), at a density equivalent to 2 × 106 cells/10-cm plate in N2B27 media.
Identification of Expressing Transgenic ES Cell Clones
Isolated puromyocin-resistant ES cell clones were genotyped by polymerase chain reaction (PCR) amplification of a 240-bp fragment of the IRES sequence with the AmpliTaq Gold PCR kit (Applied Biosciences, Foster City, CA; for primer sequences, see below). Transgenic ES cell lines were then screened for transgenic expression by northern analysis by using the 1.3-kb Not1–EcoR11 fragment of pMSCV-ires-GFP.
Immunoblot Analysis of ES Cell to Neural Differentiation Cultures
Total cell lysates were isolated and protein concentrations quantitated as described previously (Murray et al., 2004). Fifty micrograms of protein was loaded in each lane, and SDS-polyacrylamide gel electrophoresis was conducted using mini gel apparatus (Bio-Rad, Hercules, CA). Proteins were transferred to nitrocellulose membrane in Toubin's buffer containing 5–10% methanol for up to 2 h at 100 V using a mini-gel wet transfer apparatus (Bio-Rad). Ponceau staining was used to determine even loading before proceeding to immunoblot analysis. Western analysis was conducted as described previously (Murray et al., 2004). Antibody were used at the following dilutions: anti-USP9X-N1 (1:800), anti-β-catenin (1:4000; BD Biosciences Transduction Laboratories), anti-Nestin (1:400; Developmental Studies Hybridoma Bank), anti-N-cadherin (1:2000; BD Biosciences Transduction Laboratories), anti-βIII-tubulin (1:100; Sigma-Aldrich), anti-Neurofilament160 (NF-M subunit) (1:1000; Sigma-Aldrich), anti-glial fibrillary acidic protein (GFAP, 1:200; Sigma-Aldrich), anti-Oct3/4 (1:500; Santa Cruz Biotechnology, Santa Cruz, CA), anti-brain lipid-binding protein (BLBP) (1:2000; Millipore Bioscience Research Reagents, Temecula, CA), anti-β-tubulin (1:5000; Sigma-Aldrich), rabbit anti-mouse-horseradish peroxidase (HRP) (1:2000; Dako Denmark A/S, Glostrup, Denmark), goat anti-rabbit-HRP (1:2000; Dako Denmark A/S), and rabbit anti-goat-HRP (1:2000; Dako Denmark A/S). Enhanced chemiluminescence reagents were mixed 1:1 to detect HRP activity, and chemiluminescence was exposed to x-ray film (Fuji, Tokyo, Japan).
Real-Time Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was extracted from cell pellets using TRIzol (Invitrogen) and treated with rDNAse1 (Ambion, Austin, TX) to remove genomic contamination. cDNA was generated by reverse transcribing 1 μg of RNA with M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA) primed with random decamers (Geneworks, Adelaide, SA, Australia). The following primer sets were used in qPCR reactions: Nestin (327 bp; Chiba et al., 2006), Sox1 (151 bp; Jiang et al., 2005), βIII-tubulin (274 bp; Sonntag et al., 2005), USP9x (195 bp; F: AGATGACCAAGATGCCCCAGATGAG, R: TTCACCGCCTTCATAATTTTCTTGTGC), Transgenes: IRES (240 bp; F: GTCTTTTGGCAATGTGAGGG, R: GCAGGTGTATCTTATACACG), and β-actin (443 bp; F: ATGAAGATCCTGACCGAGCG, R: TACTTGCGCTCAGGAGGAGC). qPCR reactions used AmpliTaq-Gold kit (Applied Biosystems) with the addition of SYBR Green (Invitrogen), both as per the manufacturer's instructions. qPCR reactions were run on the Corbett Rotor-Gene 3000 Real-Time PCR machine, and data were collected using Rotor-Gene version 6 Software (Corbett Research, Sydney, Australia). All primer efficiencies were validated using serial template dilutions and applying the “delta-delta” methodology as described previously (Pfaffl, 2001), and they were further validated using Q-Gene software (http://www.biotechniques.com/softlib/qgene.html) as described previously (Muller et al., 2002). Normalized gene expression data were calculated with Q-Gene software using β-actin as a reference gene. All graphs were prepared using Excel (Micrsoft, Redmond, WA).
Immunofluorescence on Cultured Cells
Cells were grown in 35-mm dishes were fixed with 4% PFA for 20 min at RT, washed, and stored in PBS at 4°C until required. Cells were block/permeablized with PBST containing 10% normal horse serum (NHS; Sigma-Aldrich) or 3% BSA for at least 1 h at RT. Antibodies were diluted in PBST containing 3% NHS or 0.25% BSA. Primary antibodies were incubated overnight at 4°C at the following dilutions: anti-USP9X-N1 (1:200), anti-Nestin (1:10; Developmental Studies Hybridoma Bank), anti-βIII-tubulin (1:200; Sigma-Aldrich), anti-BLBP (1:3000; Millipore Bioscience Research Reagents), anti-GFAP (1:300; Sigma-Aldrich), RC2 (1:30; Developmental Studies Hybridoma Bank), anti-E-cadherin (1:1000; BD Biosciences Transduction Laboratories), anti-N-cadherin (1:1000; BD Biosciences Transduction Laboratories), anti-β-catenin (1:1000; BD Biosciences Transduction Laboratories), anti-AF6 (1:200), anti-Ki67-FITC (1:5; BD Biosciences, San Jose, CA) anti-protein kinase C (PKC) ζ (1:100; Santa Cruz Biotechnology), anti-GLAST (1:3000; Millipore Bioscience Research Reagents), anti-DCX (1:3000; Millipore Bioscience Research Reagents), anti-Prominin1 (1:100; Millipore Bioscience Research Reagents), anti-Notch1 (1:500; Nyfeler et al., 2005). Donkey anti-mouse IgG or goat anti-mouse IgM, donkey anti-rat IgG and goat anti-guinea pig antibodies conjugated with Alexa488 (Invitrogen) or sheep anti-rabbit and donkey anti-mouse antibodies conjugated with Cy3 (Sigma-Aldrich) were used as secondaries and incubated for 1 h at RT all at a 1:750 dilution. Cells were counterstained with DAPI (Invitrogen) for 5 min at RT. Coverslips were mounted with PBS-glycerol containing Anti-Fade reagent (Invitrogen). Effects of nonspecific staining were excluded by the use of secondary antibody-only controls (data not shown).
Microscopy
Fluorescence was observed with an Axioplan2 microscope (Carl Zeiss, Jena, Germany) fitted with an HBO 100 mercury lamp (Carl Zeiss). Images of brain sections were produced using this system fitted with the ApoTome slider module (Carl Zeiss). Images were captured on an Axiocam Mrm camera and Axio Vs40 version 4.5.0.0 software (AxioVision; Carl Zeiss). Phase contrast images were observed using an inverted Nikon Eclipse TE 2000-U microscope and captured with the F-View Soft Imaging System camera by using analySIS Software version 3.2 (Soft Imaging System, Muenster, Germany).
Cell Counts
For the quantitation of BLBP-positive and βIII-tubulin-positive percentages a number biological repeats were conducted which produced similar results. For each clone at each time point, five fields of view were chosen at random under UV light to visualize DAPI staining at a magnification of 200×. The number of βIII-tubulin-positive, BLBP-positive cells was counted and reported as a percentage of the total number of DAPI-stained nuclei. At each time point for each clone at least 1500 nuclei were counted. For the quantitation of the βIII-tubulin-positive:BLBP-positive cell ratio for each clone at each time point, 15 fields of view were selected at a magnification of 200× by seeking out BLBP-positive colonies. Total numbers of BLBP-positive and βIII-tubulin–positive cells were counted and ratios were calculated. At least 500 βIII-tubulin–positive cells were counted for each clone at each time point.
The percentage of polarized colonies was quantified by costaining seven nestin-USP9X and seven control cultures with a RP marker (RC2 or BLBP) and a cell adhesion or polarity marker (N-cadherin or β-catenin or AF6 or PKCζ or Prominin1). Colonies of RPs were scored as either polarized or nonpolarized based on the central or dispersed localization of the polarity markers, respectively. At least 700 colonies were counted for both nestin-USP9X and control lines. For the quantification of BrdU-positive RPs and neurons, 20 fields of view were chosen, at each time point, by screening plates for either BLBP or βIII-tubulin reactivity, and then capturing merged images of BrdU and DAPI staining at 200× magnification. At least 4000 BLBP-positive and 1000 βIII-tubulin–positive cells counted for each cell line, at each time point. For the quantitation of BLBP positive cells coexpressing Ki67 at day 6 of differentiation, BLBP-positive cells were located, and merged images of Ki67 and DAPI were captured at 200× magnification. At least 15 fields of view were captured for each clone, and at least 2000 BLBP-positive cells counted. For the quantification of apopotic cells, 15 fields of view were chosen, at each time point, by screening plates for either Oct4, BLBP, or βIII-tubulin expression and capturing merged images of DAPI and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining at 200× magnification. At least 3000 cells were counted at day 6 and 1000 cells at day 4 for each clone.
Statistics and Graphs
Statistics were conducted using single-tailed paired Students t tests, in which results from all controls and all transgenic samples were pooled to generate the samples to be compared. All error bars on graphs represent ±1 SD. All graphs were generated using Excel (Microsoft).
5-Bromo-2′-deoxyuridine (BrdU) and TUNEL Labeling and Staining
Cells were labeled by addition of BrdU to the culture media for 4 h. Cells were labeled, fixed, and stained for BrdU incorporation all as per manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Detection of apoptotic cells was conducted using the TUNEL assay as per the manufacturer's instructions (Roche Diagnostics).
Fluorescence-activated Cell Sorting (FACS)
When sorting for GFP-positive cells, single-cell suspensions at 1 × 106 were transferred to a flow cytometry tube (Falcon; BD Biosciences Discovery Labware) in 500 μl of ice-cold PBS and stained with 5 μl of a 5 μg/ml solution of propidium iodide (PI) for 5 min before being spun down and resuspended in PBS. Voltage was set using unstained R1 cells. All PI-positive cells were excluded from analysis, and GFP-positive cells were measured in comparison with equivalent nonfluorescent R1-derived populations. The sorting of CD133 high and low populations was performed on day 7 neural differentiation cultures as described previously (Kania et al., 2005). Cells were gated based on forward and side scatter to exclude cell debris. Low-voltage gates were set based on signal derived from unstained R1 cells. Positive staining was judged specific as secondary antibody-only (AlexaFluor488; Invitrogen) and isotype control (rat IgG2; BD Biosciences) staining were negative. High-voltage gates were arbitrarily set. Voltage gates were constant across all cell lines analyzed. Cell suspensions were analyzed using a FACScan (BD Biosciences), and data were collected using CellQuest Prosoftware (BD Biosciences) and manipulated using either CellQuest Proor FCS Express (Microsoft).
RESULTS
USP9X Is Enriched in Apical Domains of Forebrain Neural Progenitors
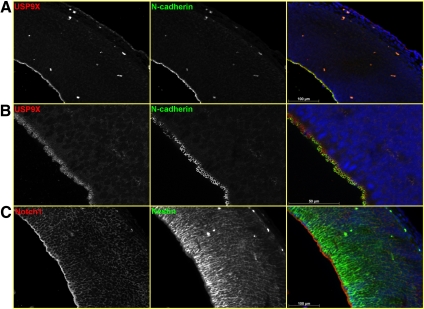
To establish whether USP9X might be a bona fide regulator of NP/NSC function, we first analyzed its expression in these cell types in vivo. We and others have reported previously high expression of USP9X mRNA in the proliferative zones of the developing CNS (Wood et al., 1997; Friocourt et al., 2005). Here, we analyzed the localization of the USP9X protein, by immunofluorescence using a polyclonal antibody (Kanai-Azuma et al., 2000), during the peak neurogenic period in the mouse forebrain at E14.5. USP9X was highly enriched in the apical region of the Nestin-positive ventricular-zone NPs (Figure 1A), a domain common to the fate determinant Notch1 (Figure 1C). Similar results were obtained using another polyclonal antibody raised against the putative Ubl domain of USP9X (amino acids 886-970) (data not shown). Within this region, USP9X partially colocalized with the adherens junction protein N-Cadherin (Figure 1B). Although concentrated toward the apical domain, USP9X protein was also detected throughout the length of the NP but at much lower levels. USP9X staining was punctuate, consistent with its association with vesicles and sites of protein trafficking in both polarized and nonpolarized cells (Murray et al., 2004). Additionally, within the adult brain, USP9X was enriched both in subgranular zone cells within the denate gyrus of the hippocampus and in subventricular zone cells lining the ventricle of the anterior lateral forebrain (Supplemental Figure 1). Because the fate of neuroepithelial cells is determined, at least in part, by apically localized proteins including those at the adherens junctions, we conclude that the temporal and spatial expression pattern of USP9X was consistent with it having a role in the regulation of NP/NSC function.
Figure 1.
USP9X localizes to the apical aspects of E14.5 forebrain neural progenitors. Immunofluorescence revealed USP9X to be highly enriched in apical regions of the neuroepithelia where it partially colocalized with N-cadherin (A and B). USP9X localized in puncta in the apical end-feet structures (B). This expression domain is shared by neural progenitor cell fate determinants such as Notch1 (C). Neural progenitors detected by expression of Nestin (C).
Transgenic Expression of nestin-USP9X in NP In Vitro Leads to Morphological Differences
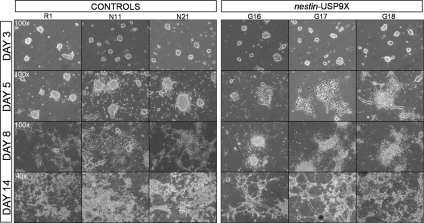
To test whether USP9X levels could alter the function of ES-derived NP, we introduced a vector containing the full-length USP9X cDNA under the transcriptional control of human nestin enhancer elements that are active in NPs (Lothian et al., 1999), into ES cells. Twelve nestin-USP9X transgenic and six empty-vector control ES cell clones were isolated and, along with the parental R1 ES cells, induced to differentiate toward neuronal lineages in an adherent monolayer culture (Ying et al., 2003). In this protocol, Nestin-positive NPs first arise ≈3 d after initiating differentiation of the ES cells (Ying et al., 2003). Consistent with this, the earliest detection of the USP9X transgene and endogenous Nestin mRNA expression was on day 3 of differentiation. At this time, no morphological differences were observed between empty vector (N11 and N21) and wild-type (R1) control cells, and nestin-USP9X clones (G16, G17, and G18) (Figure 2). At day 5, expression of the transgene was readily detected (Figure 3A) as were morphological differences between USP9X and control lines (Figure 2). The presence of bipolar cells, characteristic of in vitro NPs (Malatesta et al., 2003; Ying et al., 2003) arranged in rosettes, was readily detected in nestin-USP9X cultures but absent from controls at this time. The morphological differences between control and transgenic cultures became more dramatic with prolonged culture. At day 8, bipolar cells could be detected in cultures of control cells, often arranged in lattice-like arrays. At this time, the rosettes in nestin-USP9X cultures where replaced by radial clusters of cells consisting of a raised dome of central cells from which numerous long cellular extensions emanated radially, in many instances contacting neighboring colonies (Figure 2). After 14 d of culture, a striking architectural difference was apparent between nestin-USP9X and control cultures. Whereas control cell lines produced a lattice of neuronal-like and bipolar cells, in nestin-USP9X cultures pronounced, thick cellular extensions, interconnecting raised dome-shaped colonies dominated the cultures (Figure 2). In many instances, the extensions from these radial clusters had lost contact with the gelatin-coated culture dish surface but maintained a connection between clusters. The predominance of radial clusters was detected in the differentiated cultures of all clones expressing the nestin-USP9X transgene (n = 12).
Figure 2.
Transgenic USP9X expression induces dramatic morphological differences during the differentiation of ES cells to neurons. The morphology of the parental ES cell line (R1) and lines containing the empty transgene (N11 and N21) were compared with three lines containing the nestin-USP9X transgene (G16, G17, and G18) at days 3, 5, 8, and 14 by phase contrast. At day 3, no morphological differences were observed. At day 5, bipolar cells were detected only in G16, G17, and G18 cultures. At day 14, control cells differentiated to produce a carpet of neuronal-like cells, whereas nestin-USP9X lines generated large dome-shaped colonies with thick interconnecting cellular extensions.
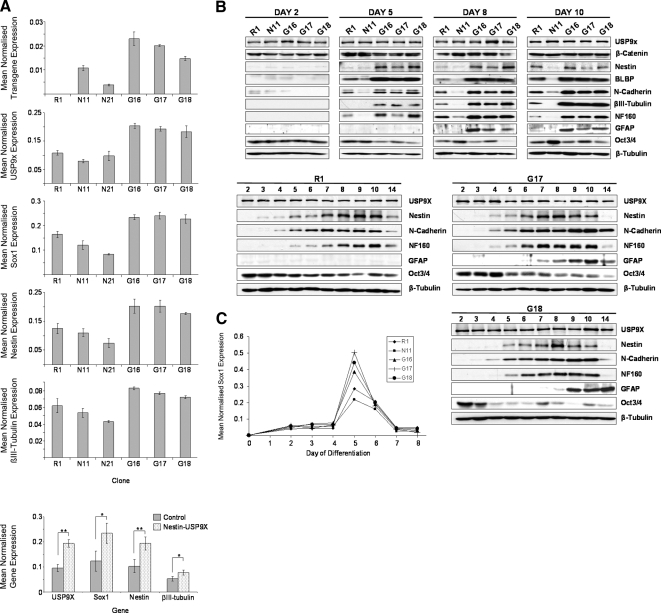
Figure 3.
nestin-USP9X expression elevated levels of progenitor, neuronal, and astrocytic marker genes, while decreasing that of the pluripotency marker Oct4. (A) qRT-PCR analyses of transgenic and total USP9X levels, as well as Sox1, nestin, and βIII-tubulin RNA levels from day 5 cultures. Transgene expression indicates the level of internal ribosome entry site sequences. *p < 0.02, **p < 0.005. (B) Immunoblot analyses of ES cell to neuronal differentiation. Marker proteins representative of pluripotent (Oct4), progenitor (Nestin, BLBP, and N-Cadherin), neuronal (βIII-tubulin and NF160), and astrocyte (GFAP) cells, as well as total levels of USP9X, β-catenin, and loading control β-tubulin were determined. (C) qRT-PCR levels of Sox1 expression during the first 8 d of differentiation. R1, parental ES cell line; N11 and N21, empty vector lines; G16, G17, and G18, nestin-USP9X lines.
Transgenic Expression of USP9X Increases Markers of NPs, Neurons, and Astrocytes
We next sought to determine the identity of the cells present in both the nestin-USP9X and control cultures. However, before doing so, we ascertained whether our culture conditions were generating neural cell types as reported previously (Ying et al., 2003). In our hands, the 46EC Sox1-GFP knockin ES cells, which express GFP in Sox1-positive NPs, generated GFP-positive cells to the same extent and duration (Supplemental Figure 2A) as Ying et al. (2003), confirming our culture conditions were appropriate. Additional preliminary studies detected the presence of neural progenitors and neurons marker genes and proteins, in a number of cell lines (Supplemental Figure 2, B and C).
To extend these analyses, we analyzed at least two, but more frequently three, independent nestin-USP9X clones (G16, G17, and G18) as well as the empty vector control lines (N11 and N21) and parental R1 cells. qRT-PCR at day 5 of differentiation, when the first morphological differences became evident, indicated that the Nestin promoter was active in USP9X transgenic and empty vector control cells (Figure 3A, top graph). Transgene expression resulted in an approximately twofold increase in the level of total USP9X transcripts (transgenic plus endogenous) in the G16, G17, and G18 clones (Figure 3A, second graph). At this time, both the early neural plate and neural tube progenitor markers Sox1 and Nestin were elevated (Figure 3A third and fourth graphs). The early pan-neuronal marker βIII-tubulin was also elevated in the nestin-USP9X clones (Figure 3A, second bottom graph). These analyses indicated that USP9X significantly increased the mRNA levels of NP and neural marker genes (Figure 3A, bottom graph). To more accurately map the induction of NPs, Sox1 mRNA was assessed by qRT-PCR over the first 8 d of differentiation (Figure 3C). In both nestin-USP9X and control cultures, Sox1 expression was markedly up-regulated at day 5, before dropping at day 6 to become almost absent by day 7. Although the kinetics of induction was identical, an average 1.8-fold increase in Sox1 levels was detected across the three nestin-USP9X clones. However, nestin-USP9X was unable to maintain Sox1-positive cells longer than controls.
To better define which cell types were present during the course of the differentiation, immunoblot analyses were performed on proteins collected each day from day 2 to day 10 and also at day 14 (Figure 3B and Supplemental Figure 3). We compared marker protein levels between the clones (R1, N11, G16, G17, G18) over time, and kinetics of marker protein induction and loss within single clones (R1, G17, G18). At day 2 of differentiation, the expression of marker proteins was similar comparing control and transgenic lines (Figure 3B). As early as day 3, however, higher levels of the early NP marker proteins Nestin and N-Cadherin, as well as the RGC and astrocyte marker BLBP were detected in the nestin-USP9X clones (G16, G17, G18) (Supplemental Figure 3). Higher levels of NP marker proteins in nestin-USP9X clones were maintained to day 10 (Figure 3B, top). Elevated levels of the immature and mature pan-neuronal markers βIII-tubulin and NF160, respectively, were also detected in nestin-USP9X clones (Figure 3B). In the ES cell-to-neuron differentiation system used, there is a concomitant loss of the pluripotency marker Oct 3/4 as ES and primitive ectoderm cells differentiate (Figure 3B), although ∼15% of the cells remain Oct3/4 positive (Ying et al., 2003). In the nestin-USP9X clones, there was a faster and more extensive loss of Oct3/4 during differentiation compared with R1 and N11 cells (Figure 3B). Although the differentiation protocol used specifies neuronal fates, nestin-USP9X clones expressed appreciable amounts of the astrocytic marker protein GFAP from day 8 onward. In the nestin-USP9X clones G17 and G18, Nestin expression peaked at day 8, 24–48 h before peak expression was detected in R1 control cultures. A similar 48-h precocious expression was observed for NF160, with peak expression occurring at day 8 in nestin-USP9X clones and at day 10 in R1 cells (Figure 3B, bottom). Overall, protein markers of neural progenitors and neurons occurred earlier and at increased levels in nestin-USP9X cells. Additionally, transgenic USP9X expression in NP resulted in expression of the astrocytic marker GFAP. Our control cells (R1 and N11) did not produce any GFAP reactivity, as detected by immunoblots, even at late stages of culture, suggesting a diminished capacity to differentiate into astrocytes under these conditions rather than simply a delay. However, a small number of GFAP-positive cells were detected by immunofluorescence in control cells (data not shown). This is consistent with the initial report (Ying et al., 2003) where replating and longer cultures were required to generate astrocytes and oligodendrocytes.
nestin-USP9X Expression Affects RP Colony Architecture and Subsequent Arrangement of Neurons
To confirm that the majority of cells were NPs as suggested by the immunoblot and qRT-PCR analyses, and to determine what type of NPs they most closely resembled, we performed immunofluorescent staining to determine whether individual cells expressed multiple markers of NPs. At early time points (days 6 and 8), cells displayed a bipolar morphology similar to RGCs. These bipolar cells, in both nestin-USP9X and control cultures, coexpressed the RGC markers BLBP, Nestin, RC2, and GLAST (Hockfield and McKay, 1985; Misson et al., 1988; Shibata et al., 1997; Malatesta et al., 2003; Anthony et al., 2004) (Figure 4A and Supplemental Figure 4). These cells were also negative for markers of differentiation such as the neuronal markers βIII-tubulin and NF160 (data not shown). The immunofluorescence confirmed that these radial glial-like cells, which we refer to as RPs, were the predominant cell type in the nestin-USP9X cultures.
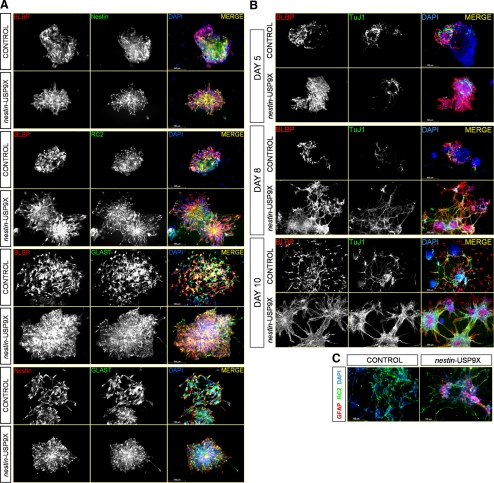
Figure 4.
Early morphological differences in nestin-USP9X transgenic cell lines correlate with differences in the number and arrangement of RPs. (A) Immunofluorescence of cultures at day 6 revealed BLBP-positive cells coexpressed GLAST and the progenitor markers Nestin and RC2, in both control and nestin-USP9X transgenic cell lines. (B) Immunofluorescence revealed a fundamentally different arrangement of RPs (BLBP positive) and immature neuronal cells (βIII-tubulin positive) at days 5, 8, and 10 of differentiation. (C) The astrocytic marker GFAP was readily detected in nestin-USP9X lines.
We next analyzed how RPs and neurons were arranged within the cultures. At day 5, a striking difference between nestin-USP9X and control cultures was observed. In control cultures, RPs were generally detected as individual cells, or small patches of BLBP-, RC2-, GLAST-positive cells dispersed throughout a predominantly BLBP-negative colony or in a lattice-like array adjacent to BLBP-negative cells (Figure 4B). However, in nestin-USP9X cultures, BLBP-positive cells were always arranged as contiguous blocks (Figure 4B). Even in colonies that were not composed entirely of RPs, in nestin-USP9X cultures, the BLBP-positive cells were adjacent to one another (Figure 4B). In addition, at day 5 the majority of nestin-USP9X RPs was arranged in rosettes. At day 8 of differentiation, radial clusters had developed in nestin-USP9X clones. These consisted of BLBP-positive cells at the center extending long cellular extensions in all directions, often forming connections with neighboring clusters (Figure 4B). βIII-Tubulin–positive neurons were also predominant in cultures of nestin-USP9X cells. The neurons were absent from the center of the clusters but rather encircled the RPs toward the periphery of the cluster. βIII-Tubulin–positive extensions also occurred between clusters but were generally found associated with BLBP-positive extensions (Figure 4B). In contrast, radial clusters or rosettes of RPs were infrequent in cultures of control cells (R1 and N11) at day 8 or later. Instead, both RPs and neurons were still predominantly present as dispersed cells contained within nonneural colonies, or arranged in lattice-like arrays (Figure 4B) and few BLBP- or βIII-tubulin–positive cellular extensions were detected in these cultures. By day 10, the architecture of the radial clusters in nestin-USP9X cultures was similar to that at day 8, except that both BLBP- and βIII-tubulin–positive extensions had consolidated. The βIII-tubulin–positive extension in particular had formed thick bundles interconnecting radial clusters giving rise to the striking architecture present at late stage nestin-USP9X cultures (Figures 2 and 4B). In cultures of control cells, far fewer BLBP- and βIII-tubulin–positive cells were observed, and they were predominantly arranged in lattice-like arrays (Figure 4B). These data suggested that the earliest morphological differences observed in nestin-USP9X cells were primarily due to differences in the number and organization of RPs and that this gave rise to the dramatic differences in the cellular architecture at later stages. We also detected GFAP-positive astrocytes (Figure 4C) and RIP-positive oligodendrocytes (data not shown) in nestin-USP9X cultures at later times (days 8 and 10) by immunofluorescence, confirming the immunoblot analyses.
nestin-USP9X Expression Increased Percentage of RPs and Neuronal Cells Early during Differentiation
The use of immunofluorescence also facilitated the direct counting of cells to determine whether the expression of nestin-USP9X increased the numbers of RPs, as was suggested, but not directly shown, by the qRT-PCR and Western analyses. Cultures were stained and five randomly selected fields were counted from several different plates. Because the whole plate was not counted, the results are presented as percentages and not absolute numbers. However there were generally the same, or an increased numbers of cells in the nestin-USP9X plates (Figure 6; data not shown), indicating that the percentages reflect absolute numbers of RPs.
Figure 6.
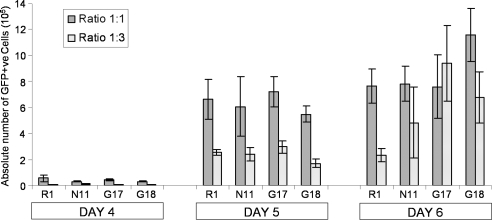
Cells expressing the nestin-USP9X transgene do not laterally induce Sox1-positive cells. Sox1-GFP knockin ES cells were mixed with either control (parental R1 or empty-vector N11) or nestin-USP9X (G17 or G18) cells at a ratios of either 1:1 or 1:3 (Sox1-GFP:Clone) and induced to differentiate into neurons. At days 4, 5, and 6, the absolute number of GFP-positive cells in each plate was calculated using FACS and total cell numbers.
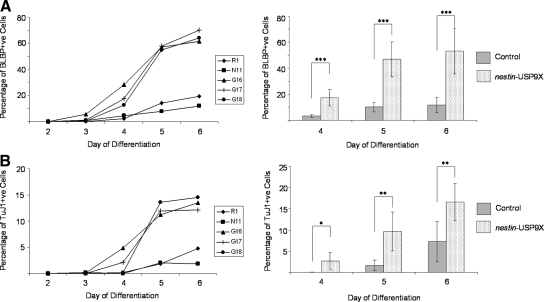
In agreement with the immunoblot analysis, nestin-USP9X clones (G16, G17, and G18) contained a significantly higher percentage of BLBP-positive cells compared with controls (R1 and N11) as early as day 4, increasing at day 5, and maintained to day 6 when the analysis was concluded (Figure 5A). The rate of increase was most prominent between days 4 and 5, during which the percentage of RPs cells in nestin-USP9X increased from an average of 2.4 to 56.5% (slope = 27.0), whereas in control cultures the percentage of RPs increased from an average of 0.5 to 10.9% (slope, 5.2). Thus, nestin-USP9X transgene expression resulted in an ∼fivefold increase in the percentage of RPs cells at day 5. A similar, significant increase in the percentage of βIII-tubulin–positive neurons was also observed in the nestin-USP9X clones (Figure 5B). The rate of increase was most prominent between day 4 and day 5, during which βIII-tubulin–positive neurons increased from 2.4 to 12.5% (slope, 9.8) in nestin-USP9X cells compared with controls (1.9 to 3.3%; slope, 1.8). From day 5 to day 6, however, the rate of increase between controls and nestin-USP9X was indistinguishable (slopes, 1.4). Absolute cell numbers, although incomplete as noted above, indicated that there was an approximate 10-fold increase in the total number of RPs and neurons in transgenic cultures (G16, G17, and G18). Together, the data show that nestin-USP9X expression in NPs can produce a measurable increase in RPs within 24 h of the first NP occurring. A similar, although slightly delayed increase in neurons suggests that USP9X neither hindered nor enhanced the subsequent differentiation of RPs.
Figure 5.
nestin-USP9X expression significantly increased the percentage of RPs and immature neural cells. The percentages of RPs and immature neuronal cells were counted during days 2–6 of neuronal differentiation of the parental ES cell line R1, the empty vector line N11 and lines housing the nestin-USP9X transgene G16, G17, and G18. Combined data from control and transgenic lines revealed that nestin-USP9X lines produced significantly higher percentages of RPs and neurons. *p < 0.05; **p < 0.01; ***p < 0.005.
We next determined which cells were expressing the USP9X transgene. The Nestin second intronic enhancer is well characterized and is most active in NPs (Lothian et al., 1999); however, many cell types were present in the cultures, raising the possibility of ectopic expression. Therefore, cultures were FAC sorted for the NP cell surface marker CD133 (Prominin1) (Kania et al., 2005) before being analyzed for marker gene expression by using qRT-PCR. Increased expression of Nestin and BLBP and diminished expression of Oct4 and βIII-tubulin in CD133hi cells versus CD133lo (Supplemental Figure 5A), confirmed that the FAC sort had enriched for NPs as reported previously (Kania et al., 2005). In the CD133hi cells, the transgene was highly expressed in both empty vector controls and Nestin-USP9X cells. This resulted in a threefold higher expression of USP9X in the transgenic lines (G16, G17, and G18) than empty vector control lines (N11 and N21) and wild-type R1 cells (Supplemental Figure 5B).
The Cellular Mechanism of Increased RP Generation by nestin-USP9X
We next sought to determine the cellular mechanism by which USP9X increased the number of RPs by determining whether USP9X acted in a cell autonomous and/or nonautonomous manner. The observation that BLBP-positive cells were present in contiguous blocks (G16, G17, and G18) and not individually dispersed (R1 and N11) suggested that a transgenic NP expressing higher levels of USP9X might recruit uncommitted neighboring cells to adopt a NP fate. Because some USP9X substrates such as epsin and Itch (Chen et al., 2002, 2003; Overstreet et al., 2004; Mouchantaf et al., 2006) regulate components of the Notch pathway, which influences cell fate by direct cell–cell contact, such a mechanism was plausible. Therefore, we cocultured nestin-USP9X cells (G16, G17, and G18) and control cells (R1 and N11) with 46EC Sox1-GFP knockin ES cells, which expresses GFP in Sox1-positive NPs in this differentiation system (Ying et al., 2003). Again ∼50–60% of 46EC cells became GFP-positive when differentiated (data not shown), which is comparable with that reported previously (Ying et al., 2003). To test the ability of nestin-USP9X to laterally induce an NP fate the 46EC Sox1-GFP, ES cells were cocultured with either control or nestin-USP9X cell lines at a ratios of either 1:1 or 1:3 (46EC:nestin-USP9X/control) and then induced to differentiate. FACS analyses coupled with cell counts were conducted at days 4, 5, and 6, and the absolute number of GFP-positive cells was quantified (Figure 6). There were no significant differences in the number of Sox1-positive cells detected in any of the cocultures, suggesting that lateral induction by nestin-USP9X cells could not explain the robust increase in Sox1 transcript levels at day 5 (Figure 3) nor the increased percentage of RPs (Figure 5A).
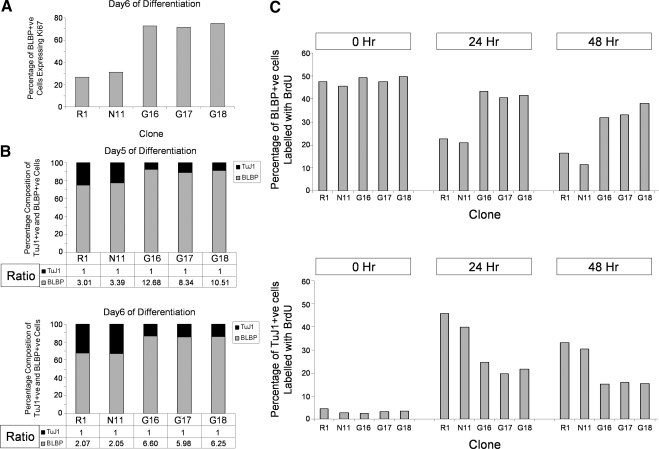
We next investigated whether transgenic USP9X expression affected the proliferation, apoptosis and/or self-renewal of RPs. We analyzed levels of apoptosis by staining cells for TUNEL (Supplemental Figure 6) and propidium iodide (data not shown). No differences in the level of apoptosis, either in BLBP- or Oct4-postive cells or in the total population in the Nestin-USP9X cells were detected (Supplemental Figure 6), indicating that the increase in RP number was not due to inhibition of their apoptosis by USP9X. To examine nestin-USP9X effects on early events we costained BLBP-positive cells with the proliferation marker Ki67 at day 6 (Figure 7A). In nestin-USP9X clones >60% of the BLBP-positive cells coexpressed high levels of Ki67, whereas only 20–30% of the BLBP cells were Ki67-positive in controls. Therefore, transgenic USP9X expression increased the proportion of in vitro-derived RPs in a proliferative state. However, this does not indicate the fate of these cells after mitosis. NPs, including RPs, are defined by their ability to self-renew as well give rise to differentiated progeny. Therefore, after mitosis there are three possible outcomes: an RP could give rise to 1) two RPs, 2) one RP plus one neuron, or 3) two neurons. If increased USP9X levels promoted self-renewal divisions, this should be reflected by an increased RP:neuron ratio irrespective of the initial number of RPs at any given time. Immunofluorescence analysis detected a 3:1 (BLBP:βIII-tubulin) ratio in control cells (R1 and N11) but a 10:1 ratio in the nestin-USP9X clones at day 5 (Figure 7B). At day 6, the average BLBP:βIII-tubulin ratio was 2:1 in control cultures, but 6:1 in nestin-USP9X cultures (Figure 7B). These data are consistent with transgenic USP9X expression promoting the self-renewal of RPs.
Figure 7.
Transgenic USP9X expression promotes self-renewal of RPs. (A) Immunofluorescence and cell counts quantified the percentage of BLBP-positive cells coexpressing the proliferation marker Ki67 at day 6 of differentiation. (B) At days 5 and 6, there was a higher BLBP:βIII-tubulin ratio in nestin-USP9X lines (G16, G17, and G18). (C) Percentage of BLBP (top) and βIII-tubulin (bottom) positive cells that costained for BrdU at 0, 24, and 48 h post-BrdU pulse.
To more directly test whether USP9X promoted RP self-renewal, we labeled the cultures at day 6 with a 4-h pulse of BrdU, to mark cells undergoing DNA synthesis, and we analyzed the percentage of BLBP-positive cells double positive for BrdU at 0, 24, and 48 h after pulse (Figure 7C). At the end of the initial labeling period, ∼50% of the BLBP-positive cells were labeled with BrdU in both control (45.6–47.6%) and nestin-USP9X (47.5–49.9%) cells. Twenty-four hours after the BrdU pulse, however, approximately twice as many nestin-USP9X cells (40.7–43.25%) were BLBP-positive compared with control cells (20.9–22.7%). By 48 h, this number approached a threefold increase (nestin-USP9X, 31.9–38% vs. 11.5–16.4% for controls). In contrast, there were ∼50% fewer BrdU-labeled, βIII-tubulin–positive neurons in nestin-USP9X cultures at both 24 and 48 h (Figure 7C). Together, these data strongly suggest that transgenic USP9X expression increased the self-renewal of RPs, either by symmetric proliferative and/or asymmetric divisions.
RPs in Radial Clusters Are Polarized
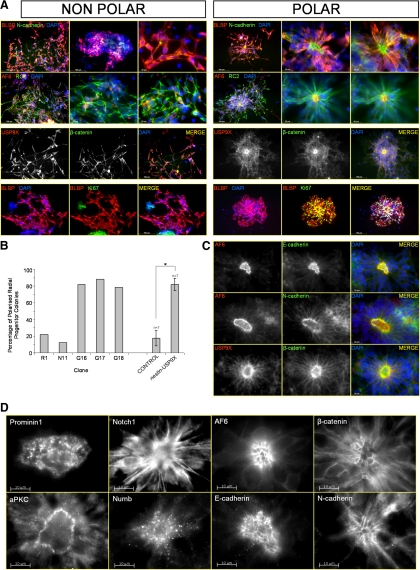
The most striking feature of the nestin-USP9X cultures was the arrangement of RPs into defined structures such as rosettes that, presumably, developed into radial clusters. Obvious corollaries from this arrangement were the potential for individual RPs to be polarized and indeed that transgenic USP9X expression promoted both polarity and radial cluster formation. Both in vivo (Gotz and Huttner, 2005) and in vitro (Liour and Yu, 2003; Liour et al., 2006; Elkabetz et al., 2008), the apical domain of RPs is delimited by the adherens junctions. Therefore, we investigated the localization of the adhesion junction proteins N-cadherin, β-catenin, and AF-6 (afadin) in RPs arranged in rosettes and radial clusters. RPs were identified by costaining with either BLBP or RC2. The adhesion-associated proteins were all concentrated at the center of the rosette, indicating that RPs in rosettes were highly polarized (Figure 8, A and C). The NP polarity proteins atypical PKCζ (Ghosh et al., 2008) and Prominin1 (Weigmann et al., 1997; Elkabetz et al., 2008) were also localized at the center of the clusters (Figure 8D and Supplemental Figures 7 and 8). The center of the cluster was composed of long, thin cellular extensions devoid of nuclei, suggesting that these regions were akin to apical end-feet structures observed in RGCs in vivo (Figure 8D). As occurs in vivo, these end-feet structures were enriched in Numb and Notch1 (Figure 8D). USP9X was also enriched in a punctate pattern, in these apical-like regions of RPs cells, and partially colocalized with β-catenin as it does in vivo (Figure 8, A and C). In contrast, RPs arranged in lattice structures or randomly within colonies did not display polarized location of these adhesion proteins nor USP9X. In control cultures, components of the cadherin complex displayed a generalized cytoplasmic distribution, with perinuclear enrichment likely to be Golgi associated, or at occasional sites of cell-cell contact, thus revealing a lack of cellular polarity. Using markers of RGCs together with either cell adhesion components or apical markers, we scored the number of polarized RP colonies at day 8 of differentiation (Figure 8B). In nestin-USP9X cultures, 82% of RP-containing colonies were polarized, whereas only 17% of RP-containing colonies in control cultures were polarized (p < 0.001). However, polarized RP clusters in control cultures closely resembled those in nestin-USP9X cultures.
Figure 8.
nestin-USP9X results in polarization and proliferation of RP colonies. (A) Adherens junction associated proteins (N-cadherin, β-catenin, and AF-6) were concentrated at the center of rosettes indicating polarization of RPs (BLBP-positive and RC2-positive). In nonpolarized cells, these proteins were evenly distributed throughout the RPs. USP9X was also concentrated at the rosette center. The majority of RPs arranged in rosettes were positive for the proliferation marker Ki67. (B) A significantly higher percentage of rosettes and radial clusters were present in nestin-USP9X transgenic cultures. Results are from combined control and transgenic lines from two experiments. *p < 0.001. (C and D) The center of clusters resembled in vivo apical end-feet. Adherens junction associated proteins (N-cadherin, E-cadherin, β-catenin, and AF-6) were detected in honeycomb-like pattern at the apical end of the RPs, which were devoid of nuclei. USP9X was highly concentrated at the apical domain as it is in vivo.
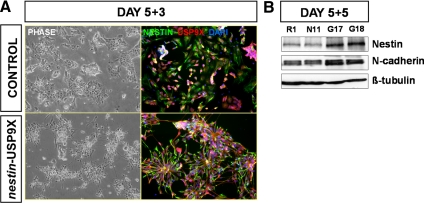
To investigate whether transgenic USP9X expression could promote the establishment of RP polarity, we dissociated the colonies at day 5 into single cells to disrupt existing sites of cell adhesion. The majority of cells which survive the passage were Nestin-positive as reported previously (Ying et al., 2003). After reattachment to a fibronectin substrate at low density and a further 3 d of culture, we assessed the ability of cells to form polarized colonies (Figure 9A). R1 control cells were arranged randomly and did not display bipolar morphology, whereas in nestin-USP9X clones the Nestin-positive cells were both bipolar and arranged in rosettes. These data suggest that transgenic USP9X expression helps establish RP polarity in vitro. Five days after replating nestin-USP9X lines G17, G18 expressed higher levels of NP marker Nestin (Figure 9B).
Figure 9.
nestin-USP9X promoted neural progenitor polarity in dissociated cultures. Control (R1 and N11) or nestin-USP9X (G17 and G18) ES cell lines were differentiated for 5 d before dissociation to single cells and reseeded onto fibronectin. (A) After a further 3-d culture, phase contrast and immunofluorescence was used to identify progenitor cells (Nestin positive), colony architecture, and USP9X expression. (B) After 5-d after seeding, immunoblot analysis was used to compare levels of Nestin and N-cadherin.
RPs within Rosettes and Radial Clusters Are Proliferating
Finally, we analyzed whether the USP9X-induced increase in RP self-renewal and polarity were linked. Immunofluorescence against Ki67 was used to determine the proliferative state of both polarized and nonpolarized colonies of RPs (Figure 8A). Although only a small proportion of cells expressing high levels of Ki67 were detected in nonpolarized RG colonies, the majority of RPs arranged in polarized rosettes and radial clusters, whether transgenic or controls, expressed high levels of Ki67 (Figure 8A). Thus, a higher proportion of RPs in clusters were proliferating, and because most RPs in G16, G17, and G18 cultures were arranged in rosettes/radial clusters, this may have been sufficient to account for the increased numbers of RPs induced by the nestin-USP9X transgene.
DISCUSSION
The principal findings of these studies are that elevated expression of USP9X in ES cell-derived NP resulted in two phenomena, namely, 1) a dramatically altered cellular architecture, wherein the majority of RPs are polarized and arranged into radial clusters; and 2) generation of a fivefold increase in the number of RPs and neurons. We also show that USP9X promotes the self-renewal of RPs and propose that polarization of the RPs precedes and engenders the radial arrangement of RPs, which was conducive to their self-renewal and hence expansion. Although we cannot rule out that it was rather an increase in RP numbers that resulted in the differences in architecture, four observations suggest this was not the case. First, seeding ES cells across a range of cell densities did not influence the frequency of rosette and/or radial cluster formation (data not shown). Second, BrdU labeled a similar percentage of nestin-USP9X and control RPs undergoing S phase at day 6 (Figure 7C). Third, polarized RPs in clusters in control cultures were equally labeled for the proliferation marker Ki67 as those in transgenic cultures. Finally, after passage of RPs as single cells, only nestin-USP9X colonies regenerated rosettes with higher levels of NP markers Nestin and N-cadherin (Figure 9). These observations strongly suggest that the polarized arrangement of RPs precedes and promotes an increase in their number. Irrespective, increased USP9X expanded the population of RPs in an in vitro differentiation system in the absence of exogenously added growth factors.
USP9X Increases Rosette Formation by Radial Progenitors
The arrangement of NP/RPs into rosettes and radial clusters occurs in a number of ES-derived neurogenic systems containing exogenous growth factors (Liour and Yu, 2003; Ying et al., 2003; Liour et al., 2006; Elkabetz et al., 2008). Our control lines generated NPs at similar percentages to those reported previously (Ying et al., 2003), some of which (10–20%) were arranged in clusters (Figure 8C) in this chemically defined differentiation system. However, the expression of nestin-USP9X resulted in the majority (∼80%) of RPs growing in clusters (Figure 8C), which had a subsequent dramatic effect on the overall architecture of the cultures, including the arrangement of neurons (Figures 2 and 4). The RPs in radial clusters, whether in control or nestin-USP9X cultures, displayed several features of their in vivo counterparts. They were bipolar cells with characteristics of apical end-feet at the center of clusters, including concentration of adherens junction and polarity marker proteins and both Notch and Numb (Figure 8). These end-feet were also devoid of nuclei. The arrangement of immature and migrating neurons, and the detection of BrdU stained nuclei, immediately after label, at the periphery of the cluster (Figure 4B and Supplemental Figure 9) indicated that the basal compartment of RPs were located at the outer edge as in other systems (Liour et al., 2006; Elkabetz et al., 2008). The detection of S phase nuclei at the periphery and G2/M phase nuclei, marked by strong Ki67 staining toward the center (Supplemental Figure 9) suggested interkinetic nuclear migration was occurring. It has been proposed that radial arrangement of NPs/RPs creates a community effect (Ying et al., 2003), which is critical for the maintenance and expansion NPs (Elkabetz et al., 2008). Arrangement of human and mouse ES cell-derived NPs into rosettes (so called neural rosette cells (R-NSC) is associated an increased differentiation potential and is promoted by activation of the Notch and SHH pathways (Elkabetz et al., 2008). USP9X may also control aspects of the Notch pathway in our cells. USP9X's Drosophila homologue, fat facets, activates the Notch ligand Delta via its regulation of the endocytic accessory protein liquid facets (epsin homologue) (Overstreet et al., 2004; Wang and Struhl, 2004). Mammalian USP9X also activates epsin by deubiquitylation (Chen et al., 2003). Because the differentiation system used here included no exogenous growth factors, increased expression of USP9X in NP/RPs may function by enhancing endogenously produced extrinsic factors such as Notch. That nestin-USP9X enhanced albeit significantly, what is a normal behavior of RPs, also suggests that it acted upon endogenous pathways, perhaps already regulated by endogenous USP9X.
USP9X Promoted Polarization and Self-Renewal of Radial Progenitors
In vivo, the polarity, cell adhesion, self-renewal, and proliferation of RGCs are all interdependent (Kosodo et al., 2004; Gotz and Huttner, 2005; Huttner and Kosodo, 2005). Determinants of symmetric or asymmetric cell division, and hence cell fate, reside apically, and it has been proposed that retention of the apical plasma membrane or regions adjacent to it, after mitosis results in daughter cells retaining a neural stem cell fate (Gotz and Huttner, 2005; Huttner and Kosodo, 2005). Polarization of RGCs also affects their proliferation (Klezovitch et al., 2004; Huttner and Kosodo, 2005). Our experiments have shown that the fivefold increase in RP numbers in nestin-USP9X lines is best explained by USP9X-induced self-renewal divisions of RP, which in turn were a consequence of increased RP polarization. Three arguments support self-renewal rather than increased cell cycle rate as the cause of increased RP numbers. First, the three- to fourfold increased BLBP:βIII-tubulin ratio in nestin-USP9X cells at both day 5 and day 6 indicates transgenic USP9X favored self-renewal over differentiation. A similar rationale has been used during the examination of the roles of the polarity genes lethal giant larvae and pins in Drosophila neuroblast self-renewal (Lee et al., 2006). Second, BLBP and βIII-tubulin staining after BrdU pulse labeling also revealed a more rapid loss of BLBP-positive cells and faster acquisition of a neural fate (βIII-tubulin positive) in control cells (Figure 7C). Although, based on this data alone, it might be suggested that USP9X slowed RP cell cycle times, hence the increased retention of BrdU in BLBP-positive RPs and slower rate of appearance in neuroblasts, several other data indicate that this is not the case. The immediate post-BrdU data showed no difference in the percentage of nestin-USP9X and control RPs in S phase (Figure 7C). Although this does not necessarily correlate with cell cycle lengths, it is consistent with this interpretation. Also, nestin-USP9X cultures also generate more neurons and at earlier time points then control cells, therefore a slowing of RP proliferation and/or differentiation due to increased USP9X expression is counterintuitive. Finally, our data suggest that the increased self-renewal might be a secondary effect due to USP9X-induced polarity. The 10–20% of RPs in clusters in control cultures stained just as strongly with Ki67 as the transgenic lines (Figure 8A); therefore, expression of the proliferation marker correlated with cell architecture rather than the presence of the transgene per se.
As well as enhancing RP self-renewal, USP9X also maintained a broader differentiation capacity. nestin-USP9X RPs differentiated into neurons as efficiently as control cells; indeed, the increase in neurons can be accounted for by the increase in RP number. However, only nestin-USP9X cultures generated glial cells efficiently. GFAP-positive astrocytes were readily detected by immunoblot by day 7 in G16, G17, and G18 cells (Figure 3) but not from control cultures through all stages. However, a few GFAP-positive cells were detected by immunofluorescence in control cultures (data not shown). This suggested not simply a delay but rather a possible inability of control cells to generate significant numbers of astrocytes. RIP-positive oligodendrocyte were also detected in cultures expressing transgenic USP9X, albeit at very low frequencies (<0.1%), but not in controls. Although the majority of cells differentiate to neurons in this differentiation system both astrocytes and oligodendrocytes have also been detected, although their frequency was not reported and this requires additional culturing steps (Ying et al., 2003). It is unclear at present whether this ability of USP9X to promote the formation of astrocytes also stems from its ability to polarize the RGP.
Possible Molecular Mechanisms Regulated by USP9X in Radial Progenitors
In addition to activation of Notch ligands, as noted above, USP9X may also regulate other components of the Notch pathway. Recently, it has been reported that USP9X/FAM interacts with the ubiquitin ligase Mind Bomb-1, which is a positive regulator of the Notch pathway (Choe et al., 2007). Increasingly, examples of interactions between Dubs and ubiquitin ligases are coming to light (reviewed in Nijman et al., 2005; Millard and Wood, 2006). In general, the Dubs stabilize the ligase thereby increasing its activity. USP9X also stabilizes and activates the Itch (Mouchantaf et al., 2006) and MARCH7 (Nathan et al., 2008) ubiquitin ligases, raising the possibility that it might act similarly in its interaction with Mind Bomb-1.
Molecular pathways important in the trafficking and localization of cadherin–catenin complexes to adherens junctions are also necessary for the maintenance of RGC polarity in vivo. Changing levels of the endocytic adaptor protein Numb have direct effects on RGC polarity and proliferation and do so in a cadherin-dependent manner (Rasin et al., 2007). In RGCs, Numb is required for the correct localization of cadherins rather than increasing overall levels (Rasin et al., 2007). USP9X has also been proposed to facilitate the exocytic trafficking of the E-cadherin/β-catenin dimer to the plasma membrane at the time at which adherens junctions are being established in epithelial cells (Murray et al., 2004). The trafficking of another adherens associated protein AF-6, which also localized to the end-feet–like structures (Figure 8) is also thought to be regulated by USP9X. In polarized Madin-Darby canine kidney epithelia, AF-6 and USP9X/FAM are recruited to nascent adherens junctions with similar kinetics after repolarization (Taya et al., 1998). Depletion of USP9X/FAM levels in preimplantation mouse embryos resulted in the mislocalization of AF-6 to the apical domain of blastomeres (Pantaleon et al., 2001). It may be that increased USP9X in these in vitro-derived RPs facilitates and maintains intact adherens junctions which in turn give rise to polarization. AF-6 is critical for the establishment of polarization in embryonic NPs in vivo (Ikeda et al., 1999; Zhadanov et al., 1999). Other lines of evidence are also consistent with a role for USP9X in the establishment and/or maintenance of polarity. USP9X-mediated deubiquitylation activates the MARK4 and NUAK1 polarity-associated proteins that are central to polarization of many mammalian cells, including NPs (Brajenovic et al., 2004; Al-Hakim et al., 2005; Al-Hakim et al., 2008). These observations raise the possibility that USP9X may regulate polarity in NPs by several mechanisms, as part of a polarity complex and also by regulating the trafficking of cadherin complexes.
As well as regulation of the Notch pathway and trafficking of cadherin–catenin complex, several other molecular mechanisms may play a role in mediating USP9X's effect. USP9X in its role as Dub has the potential to interconnect and coordinate several pathways. To date, we and others have identified 15 proteins that bind USP9X and of these at least nine are substrates (Taya et al., 1998, 1999; Bouwmeester et al., 2004; Brajenovic et al., 2004; Al-Hakim et al., 2005; Friocourt et al., 2005; Vong et al., 2005; Mouchantaf et al., 2006; Kaltenbach et al., 2007; Nathan et al., 2008; Wood, unpublished data). Cytoplasmic β-catenin can also be stabilized by USP9X (Taya et al., 1999; Pantaleon et al., 2001). Therefore, as well as regulating β-catenin at the adherens junction, USP9X may also facilitate its role as a signaling molecule in the Wnt pathway. Stabilized β-catenin has been shown to increase the numbers of embryonic NPs in vivo (Chenn and Walsh, 2002). In the embryonic NPs, β-catenin did not prevent their differentiation into neurons but simply delayed their exit from the cell cycle (Chenn and Walsh, 2002). Our data are consistent with USP9X exerting a similar effect on RPs in vitro. Still other substrates of USP9X such as Survivin (Vong et al., 2005), which is a member of the inhibitor of apoptosis family, are required to prevent apoptosis in NPs in vivo (Jiang et al., 2005).
USP9X Acts as a Neural Stemness Gene
Stemness genes are a proposed shared regulatory network, necessary for the self-renewal and/or potency of most, if not all, stem cells. The number of genes/proteins in these networks has generally been estimated to be between 100 and 200 (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Van Hoof et al., 2006), although one analysis of gene expression across five mouse stem cell populations narrowed the number of common genes to 37, including USP9X (Blanpain et al., 2004). One conclusion from these analyses is that stemness does not rely on a single master regulator but rather that networks afford stem cells flexibility to respond to extrinsic influences or intrinsic perturbations. Here, we show that modest elevation of a single stemness gene, the substrate-specific Dub USP9X, significantly increased the number of NP/NSC generated during the differentiation of ES cells to neurons. It will be of interest to identify which USP9X substrate is critical for the self-renewal of RPs and this in vitro system will facilitate these endeavors. However, as USP9X has the capacity to integrate and regulate a number of pathways, via its range of substrates, it may be that the positive effect of USP9X on NP function cannot be replaced by a single substrate, but rather a coordinated subset.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to U. Lendahl and A. Nienhuis for plasmids. This work was supported by funding from the National Health and Medical Research Council (to S.A.W.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-06-0596) on January 28, 2009.
REFERENCES
- Al-Hakim A. K., Goransson O., Deak M., Toth R., Campbell D. G., Morrice N. A., Prescott A. R., Alessi D. R. 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J. Cell Sci. 2005;118:5661–5673. doi: 10.1242/jcs.02670. [DOI] [PubMed] [Google Scholar]
- Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem. J. 2008;411:249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- Anthony T. E., Klein C., Fishell G., Heintz N. Radial Glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Bibel M., Richter J., Schrenk K., Tucker K. L., Staiger V., Korte M., Goetz M., Barde Y. A. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Lowry W. E., Geoghegan A., Polak L., Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T., et al. A physical and functional map of the human TNF-a/NF-kB signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Brajenovic M., Joberty G., Kuster B., Bouwmeester T., Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 2004;279:12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- Cai C., Grabel L. Directing the differentiation of embryonic stem cells to neural stem cells. Dev. Dyn. 2007;236:3255–3266. doi: 10.1002/dvdy.21306. [DOI] [PubMed] [Google Scholar]
- Chen H., Polo S., Di Fiore P. P., De Camilli P. V. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc. Natl. Acad. Sci. USA. 2003;100:14908–14913. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhang B., Fischer J. A. A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of fat facets. Genes Dev. 2002;16:289–294. doi: 10.1101/gad.961502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A., Walsh C. A. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chiba T., Kita K., Zheng Y., Yokosuka O., Saisho H., Iwama A., Nakauchi H., Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- Choe E. A., Liao L., Cheng D., Duong D. M., Jin P., Tsai L. H., Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and mind bomb 1. J. Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N. D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G., et al. Doublecortin interacts with the ubiquitin protease DFFRX, which associates with microtubules in neuronal processes. Mol. Cell. Neurosci. 2005;28:153–164. doi: 10.1016/j.mcn.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Marquardt T., Thaler J. P., Carter N., Andrews S. E., Pfaff S. L., Hunter T. Instructive role of aPKCzeta subcellular localization in the assembly of adherens junctions in neural progenitors. Proc. Natl. Acad. Sci. USA. 2008;105:335–340. doi: 10.1073/pnas.0705713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Barde Y. A. Radial glial cells defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Gotz M., Huttner W. B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gregg C., Weiss S. Generation of functional radial glial cells by embryonic and adult forebrain neural stem cells. J. Neurosci. 2003;23:11587–11601. doi: 10.1523/JNEUROSCI.23-37-11587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockfield S., McKay R. D. Identification of major cell classes in the developing mammalian nervous system, J. Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner W. B., Kosodo Y. Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 2005;17:648–657. doi: 10.1016/j.ceb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Ikeda W., et al. Afadin: a key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 1999;146:1117–1131. doi: 10.1083/jcb.146.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N. B., Dimos J. T., Schaniel C., Hackney J. A., Moore K. A., Lemischka I. R. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jiang Y., de Bruin A., Caldas H., Fangusaro J., Hayes J., Conway E. M., Robinson M. L., Altura R. A. Essential role for Survivin in early brain development. J. Neurosci. 2005;25:6962–6970. doi: 10.1523/JNEUROSCI.1446-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner A. L. Gene Targeting: A Practical Approach. Oxford, NY: Oxford University Press; 2000. [Google Scholar]
- Junghans D., Hack I., Frotscher M., Taylor V., Kemler R. Beta-catenin-mediated cell-adhesion is vital for embryonic forebrain development. Dev. Dyn. 2005;233:528–539. doi: 10.1002/dvdy.20365. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L. S., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:689–708. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M., Mattick J. S., Kaibuchi K., Wood S. A. Co-localization of FAM and AF-6, the mammalian homologues of Drosophila faf and canoe, in mouse eye development. Mech. Dev. 2000;91:383–386. doi: 10.1016/s0925-4773(99)00294-4. [DOI] [PubMed] [Google Scholar]
- Kania G., Corbeil D., Fuchs J., Tarasov K. V., Blyszczuk P., Huttner W. B., Boheler K. R., Wobus A. M. Somatic stem cell marker Prominin-1/CD133 is expressed in embryonic stem cell-derived progenitors. Stem Cells. 2005;23:791–804. doi: 10.1634/stemcells.2004-0232. [DOI] [PubMed] [Google Scholar]
- Khut P.-Y., Tucker B., Lardelli M., Wood S. A. Evolutionary and expression analysis of the zebrafish deubiquitylating enzyme, Usp9. Zebrafish. 2007;4:95–101. doi: 10.1089/zeb.2006.0502. [DOI] [PubMed] [Google Scholar]
- Klezovitch O., Fernandez T. E., Tapscott S. J., Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y., Roper K., Haubensak W., Marzesco A.-M., Corbeil D., Huttner W. B. Asymmetric distribution of the apical membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Robinson K. J., Doe C. Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Liour S. S., Kraemer S. A., Dinkins M. B., Su C.-Y., Yanagisawa M., Yu R. K. Further characterisation of embryonic stem cell-derived radial glial cells. Glia. 2006;53:43–56. doi: 10.1002/glia.20257. [DOI] [PubMed] [Google Scholar]
- Liour S. S., Yu R. K. Differentiation of radial glia-like cells from embryonic stem cells. Glia. 2003;42:109–117. doi: 10.1002/glia.10202. [DOI] [PubMed] [Google Scholar]
- Lledo P.-M., Alonso M., Grubb M. S. Adult neurogenesis and functional plasticity in neuronal circuits. Nat. Rev. Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- Lothian C., Prakash N., Lendahl U., Wahlstrom G. M. Identification of both general and region-specific CNS enhancer elements in the nestin promoter. Exp. Cell Res. 1999;248:509–519. doi: 10.1006/excr.1999.4417. [DOI] [PubMed] [Google Scholar]
- Malatesta P., Hack M. A., Hartfuss E., Kettenmann H., Klinkert W., Kirchhoff F., Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Millard S. M., Wood S. A. Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J. Cell Biol. 2006;173:463–468. doi: 10.1083/jcb.200602082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J. P., Edwards M. A., Yamamoto M., Caviness V.S.J. Identification of radial glial cells within the developing murine central nervous system: studies based upon a new immunohistochemical marker. Brain Res. Dev. Brain Res. 1988;44:95–108. doi: 10.1016/0165-3806(88)90121-6. [DOI] [PubMed] [Google Scholar]
- Miyahara M., Nakanishi H., Takahashi K., Satoh-Horikawa K., Tachibana K., Takai Y. Interaction of Nectin with Afadin is necessary for its clustering at cell-cell contact sites but not for its cis dimerization or trans interaction. J. Biol. Chem. 2000;275:613–618. doi: 10.1074/jbc.275.1.613. [DOI] [PubMed] [Google Scholar]
- Mouchantaf R., Azakir B. A., McPherson P. S., Millard S. M., Wood S. A., Angers A. The ubiquitin ligase Itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–1374. [PubMed] [Google Scholar]
- Murray R. Z., Jolly L. A., Wood S. A. The FAM deubiquitylating enzyme locates to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and b-catenin. Mol. Biol. Cell. 2004;15:1591–1599. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., Lehner P. J. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:249–260. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M.B., Luna-Vargas M.P.A., Velds A., Brummelkamp T. R., Dirac A.M.G., Sixma T. K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Noma T., Kanai Y., Kanai-Azuma M., Ishii M., Fujisawa M., Kurohmaru M., Kawakami H., Wood S. A., Hayashi Y. Stage- and sex-dependent expressions of Usp9x, an X-linked mouse ortholog of Drosophila Fat facets, during gonadal development and oogenesis in mice. Mech. Dev. 2002;119:1–5. doi: 10.1016/s0925-4773(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Nyfeler Y., Kirch R. D., Mantei N., Leone D. P., Radtke F., Suter U., Taylor V. Jagged1 signals in the postnatal subventricular zone are required for neural stem cell self-renewal. EMBO J. 2005;24:3504–3515. doi: 10.1038/sj.emboj.7600816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet E., Fitch E., Fischer J. A. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 2004;131:5355–5366. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- Pantaleon M., Kanai-Azuma M., Mattick J. S., Kaibuchi K., Kaye P. L., Wood S. A. FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech. Dev. 2001;109:151–160. doi: 10.1016/s0925-4773(01)00551-2. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. A new mathematical model for relative quantification in real-time PCR. Nucleic Acids Res. 2001;29:345. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rasin M.-R., et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nature Neuroscience. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Salero E., Hatten M. E. Differentiation of ES cells into cerebellar neurons. Proc. Natl. Acad. Sci. USA. 2007;104:2997–3002. doi: 10.1073/pnas.0610879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Sanjuan I. M., Heke M., Uchida M., Naef F., Brivanlou A. H. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Shibata T., Yamada K., Watanabe M., Ikenaka K., Wada K., Tanaka K., Inoue Y. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J. Neurosci. 1997;17:9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag K. C., et al. Context-dependent neuronal differentiation and germ layer induction of Smad4−/− and Cripto−/− embryonic stem cells. Mol. Cell. Neurosci. 2005;28:417–429. doi: 10.1016/j.mcn.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Taya S., Yamamoto T., Kanai-Azuma M., Wood S. A., Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes β-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- Taya S., et al. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J. Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D., Pasier R., Ward-van Osstwaard D., Pinkse M.W.H., Heck A.J.R., Mummery C., Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- Vong Q. P., Cao K., Li H. Y., Iglesias P. A., Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- Wang W., Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Weigmann A., Corbeil D., Hellwig A., Huttner W. B. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc. Natl. Acad. Sci. USA. 1997;94:12425–12430. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J. A., Jessell T. M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wood S. A., Pascoe W. S., Ru K., Yamada T., Hirchenhain J., Kemler R., Mattick J. S. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech. Dev. 1997;63:29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Zhadanov A. B., Provance D.W.J., Speer C. A., Coffin J. D., Blixt J. A., Reichert C. M., Mercer J. A. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr. Biol. 1999;9:880–888. doi: 10.1016/s0960-9822(99)80392-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.