Abstract
Cell shape can influence cell behavior. In Saccharomyces cerevisiae, bud emergence can influence cell cycle progression via the morphogenesis checkpoint. This surveillance pathway ensures that mitosis always follows bud formation by linking degradation of the mitosis-inhibitory kinase Swe1p (Wee1) to successful bud emergence. A crucial component of this pathway is the checkpoint kinase Hsl1p, which is activated upon bud emergence and promotes Swe1p degradation. We have dissected the large nonkinase domain of Hsl1p by using evolutionary conservation as a guide, identifying regions important for Hsl1p localization, function, and regulation. An autoinhibitory motif restrains Hsl1p activity when it is not properly localized to the mother-bud neck. Hsl1p lacking this motif is active as a kinase regardless of the assembly state of cytoskeletal septin filaments. However, the active but delocalized Hsl1p cannot promote Swe1p down-regulation, indicating that localization is required for Hsl1p function as well as Hsl1p activation. We also show that the septin-mediated Hsl1p regulation via the novel motif operates in parallel to a previously identified Hsl1p activation pathway involving phosphorylation of the Hsl1p kinase domain. We suggest that Hsl1p responds to alterations in septin organization, which themselves occur in response to the local geometry of the cell cortex.
INTRODUCTION
Cells can detect and respond to physical signals such as tension, shear stress, surface topography, gravity, or even their own cell shape in a variety of circumstances (Vogel and Sheetz, 2006). With the notable exception of mechanosensitive ion channels, the molecular mechanisms by which physical cues are detected remain largely unknown. In the budding yeast Saccharomyces cerevisiae, the cell's shape changes in every cell cycle as buds emerge, grow, and then separate from the mother cell. However, a variety of everyday environmental stresses, including fluctuations in temperature and osmolarity, can interrupt this process and delay bud emergence (Chowdhury et al., 1992; Delley and Hall, 1999). Remarkably, yeast cells in such conditions compensate by delaying mitosis, so that bud formation and the nuclear cell cycle always remain coordinated (Lew, 2003). The pathway that responds to delayed bud emergence by blocking nuclear division is called the morphogenesis checkpoint, and it provides an experimentally tractable system to investigate how a change in cell geometry is detected and used to regulate the cell cycle engine.
The morphogenesis checkpoint delays mitosis by promoting inhibitory tyrosine phosphorylation of the cyclin-dependent kinase (CDK) Cdc28p in complex with the mitosis-inducing cyclin Clb2p, catalyzed by the Wee1-family kinase Swe1p (Lew and Reed, 1995; Sia et al., 1996; Keaton et al., 2007). In unperturbed cells, Swe1p is degraded after bud emergence (Sia et al., 1998), through a pathway that involves Swe1p phosphorylation by both Clb2p/Cdc28p and the Polo-family kinase Cdc5p (Sakchaisri et al., 2004; Asano et al., 2005). However, if bud emergence is delayed by environmental stresses (or experimentally by mutations or drugs), then Swe1p degradation is halted, contributing to a delay in mitosis (Sia et al., 1998; Lew, 2003).
Swe1p, Cdc5p, and Clb2p are all concentrated at the mother-bud neck in unperturbed cells, in a manner dependent on a family of cytoskeletal proteins called septins (Longtine et al., 2000; Bailly et al., 2003; Sakchaisri et al., 2004). Septins are evolutionarily conserved guanine nucleotide-binding proteins that assemble into oligomeric complexes which polymerize to form filaments (Weirich et al., 2008). In S. cerevisiae, septins assemble in a cortical ring in late G1, at the site of future bud emergence (Ford and Pringle, 1991; Gladfelter et al., 2001). As the bud emerges, the ring expands to form an hourglass-shaped collar, which remains at the mother-bud neck during S, G2, and M phases. The collar splits to form two rings during cytokinesis, and the split rings disassemble in the next G1. The transitions between ring and collar structures are accompanied by changes in septin filament organization that remain poorly understood.
Targeting of Swe1p to the septin collar occurs via two linking proteins: Hsl1p and Hsl7p (Longtine et al., 2000). Hsl1p is a Nim1-family protein kinase that binds to both septins and Hsl7p, and Hsl7p binds to both Hsl1p and Swe1p (Barral et al., 1999; McMillan et al., 1999; Shulewitz et al., 1999). Mutations that abrogate Hsl1p-Hsl7p or Hsl7p-Swe1p interactions prevent both Swe1p neck localization and Swe1p degradation (Cid et al., 2001; McMillan et al., 2002), suggesting that targeting of Swe1p to the septin collar triggers Swe1p degradation. When bud emergence is blocked, Hsl1p accumulates at the septin ring but does not recruit Hsl7p or Swe1p to that site (Theesfeld et al., 2003). Furthermore, whereas Hsl1p at the septin collar promotes extensive autophosphorylation and Hsl7p phosphorylation, neither protein becomes phosphorylated when bud emergence is blocked (Theesfeld et al., 2003). These observations suggest that Hsl1p at the septin ring of unbudded cells is inactive, whereas Hsl1p at the septin collar of budded cells is active, both in terms of its kinase catalytic activity and its ability to recruit Hsl7p and Swe1p.
The studies introduced above indicate that Hsl1p is responsive to the presence or absence of a bud and that Hsl1p is ideally situated within the cell to detect the presence or absence of a bud. Here, we present a structure–function analysis of Hsl1p, guided by identification of evolutionarily conserved regions, which reveals both functional and regulatory regions of the protein. Of particular interest, we found a short conserved regulatory motif which is important for Hsl1p regulation by the septins.
MATERIALS AND METHODS
Yeast Strains and Plasmids
The S. cerevisiae strains that were used in this study are listed in Table 1. All yeast strains are in the BF264-15DU background (ade1, his2, leu2-3112, trp1-1a, and ura3Δns) (Richardson et al., 1989). The creation of the following alleles has been described previously: hsl1::URA3 (Ma et al., 1996); cdc12-6:LEU2 and HSL7-HA:kanR (Longtine et al., 2000); mih1::LEU2 (Russell et al., 1989); CDC3-GFP:URA3 (Caviston et al., 2003); HSL1-myc:TRP1, HSL1T273E-myc:TRP1, hsl1T273A-myc:TRP1, elm1::LEU2, GAL1-GST, and GAL1-GST-HSL1987-1518 (Szkotnicki et al., 2008).
Table 1.
Strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| DLY1 | abar1 | Richardson et al. (1989) |
| DLY5844 | ahsl1::URA3 HSL7-HA:kanR | Szkotnicki et al. (2008) |
| DLY6900 | aGAL1-GST-HSL1951-1100:TRP1 | This study |
| DLY6902 | aGAL1-GST-HSL11138-1307:TRP1 | This study |
| DLY6903 | aGAL1-GST-HSL11379-1518:TRP1 | This study |
| DLY7166 | aGAL1-GST-HSL11138-1307:TRP1 HSL7-HA:kanR HSL1-13myc:URA3 | This study |
| DLY7382 | ahsl1::URA3 hsl1Δ5b-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY7383 | ahsl1::URA3 hsl1Δ5b-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY7402 | ahsl1::URA3 HSL1Δ1-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY7403 | ahsl1::URA3 HSL1Δ1-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY7405 | ahsl1::URA3 HSL1Δ2-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY7462 | ahsl1::URA3 HSL1Δ2-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8113 | ahsl1::URA3 HSL1-13myc:TRP1 HSL7-HA:kanR | Szkotnicki et al. (2008) |
| DLY8117 | ahsl1::URA3 hsl1K110R-13myc:TRP1 HSL7-HA:kanR | Szkotnicki et al. (2008) |
| DLY8140 | ahsl1::URA3 HSL1Δ4-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8141 | ahsl1::URA3 HSL1Δ8c-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8142 | ahsl1::URA3 hsl1Δ9a-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8143 | ahsl1::URA3 hsl1Δ9b-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8146 | ahsl1::URA3 hsl1Δ5-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8147 | ahsl1::URA3 hsl1Δ9c-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8148 | ahsl1::URA3 hsl1Δ8a-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8182 | ahsl1::URA3 hsl1Δ6-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8223 | ahsl1::URA3 HSL1Δ8b-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8356 | ahsl1::URA3 hsl1Δ6-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8389 | ahsl1::URA3 hsl1Δ5-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8391 | ahsl1::URA3 HSL1Δ4-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8603 | ahsl1::URA3 HSL1Δ3-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8674 | ahsl1::URA3 HSL1Δ3-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8682 | aGAL1-GST:TRP1 HSL7-HA:kanR HSL1-13myc:URA3 | This study |
| DLY8683 | ahsl1::URA3 hsl1Δ9-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8684 | ahsl1::URA3 hsl1Δ7a-13myc:TRP1 HSL7-HA:kanR | Szkotnicki et al. (2008) |
| DLY8685 | ahsl1::URA3 hsl1Δ8-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8686 | aHSL1::URA3 hsl1Δ7-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8695 | ahsl1::TRP1:GAL1-GST-HSL1987-1518-13myc:URA3 HSL7-HA:kanR | This study |
| DLY8707 | ahsl1::URA3 hsl1Δ9-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8711 | ahsl1::URA3 hsl1Δ8-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8713 | aHSL1::URA3 hsl1Δ7-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY8745 | ahsl1::URA3 HSL1-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | Szkotnicki et al. (2008) |
| DLY8775 | ahsl1::URA3 HSL1Δ5a-13myc:TRP1 HSL7-HA:kanR | This study |
| DLY8777 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ9c-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY8778 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ9-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY8779 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ9b-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY8786 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ8a-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY8787 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ9a-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY8850 | ahsl1::URA3 HSL1Δ5a-13myc:TRP1 HSL7-HA:kanR cdc12-6:LEU2 | This study |
| DLY9273 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ8c-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY9335 | ahsl1::LEU2:GAL1-GST-HSL1987-1518 Δ8b-13myc:TRP1 HSL7-HA:kanR hsl1::URA3 | This study |
| DLY9406 | aelm1::LEU2 mih1::LEU2 hsl1T273A-13myc:TRP1 pELM1-URA3 | Szkotnicki et al. (2008) |
| DLY9407 | aelm1::LEU2 mih1::LEU2 HSL1T273E-13myc:TRP1 pELM1-URA3 | Szkotnicki et al. (2008) |
| DLY9409 | aelm1::LEU2 mih1::LEU2 HSL1-13myc:TRP1 pELM1-URA3 | Szkotnicki et al. (2008) |
| DLY9411 | aelm1::LEU2 mih1::LEU2 HSL1Δ5a-13myc:TRP1 pELM1-URA3 | This study |
| DLY9527 | aGAL1-GST-HSL1663-914:TRP1 | This study |
| DLY9537 | ahsl1::LEU2:GAL1-GST-HSL1987-1518-13myc:TRP1 | This study |
| DLY9587 | aGAL1-GST:TRP1 | This study |
| DLY9814 | aelm1::LEU2 mih1::LEU2 HSL1T273E Δ5a-13myc:TRP1 pELM1-URA3 | This study |
| DLY9817 | aelm1::LEU2 mih1::LEU2 HSL1T273A Δ5a-13myc:TRP1 pELM1-URA3 | This study |
| DLY9851 | ahsl1::LEU2:GAL1-GST-HSL1470-1518-13myc:TRP1 | This study |
| DLY10097 | aGAL1-GST:TRP1 CDC3-GFP:URA3 | Szkotnicki et al. (2008) |
| DLY10099 | ahsl1::LEU2:GAL1-GST-HSL1470-1518-13myc:TRP1 CDC3-GFP:URA3 | This study |
| DLY11000 | aGAL1-GST-HSL1663-914:TRP1 CDC3-GFP:URA3 | This study |
| DLY11049 | a/α HSL1Δ2-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11050 | a/α HSL1Δ8c-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11051 | a/α hsl1Δ9a-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11052 | a/α hsl1Δ9b-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11053 | a/α hsl1Δ9c-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11054 | a/α hsl1Δ8a-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11055 | a/α HSL1Δ8b-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11056 | a/α HSL1Δ5a-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11057 | a/α HSL1-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11058 | a/α hsl1Δ6-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11059 | a/α hsl1Δ4-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11060 | a/α hsl1Δ5-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11061 | a/α HSL1Δ1-13myc:TRP1/trp1 mih1::LEU2/MIH1 HSL7-HA:kanR/HSL7 hsl1::URA3/HSL1 | This study |
| DLY11244 | α hsl1::URA3 hsl1Δ5Δ9-13myc:TRP1 | This study |
HSL1 regional deletion mutants were created by overlap polymerase chain reaction (PCR) by using wild-type HSL1 as a template. The PCR product was cloned into pDLB2548 (integrating TRP1-marked plasmid expressing HSL1-myc from the HSL1 promoter) (Szkotnicki et al., 2008). In some cases, short intervening sequences introducing diagnostic restriction sites occur in place of the deleted region, as listed in Table 2. The double-deletion mutant HSL1Δ5Δ9 was made by cloning a 0.6-kb StuI/ApaI fragment from HSL1Δ5 into the corresponding sites in the plasmid harboring HSL1Δ9. All mutants were sequenced to confirm the change and that there were no other mutations. The linker between the HSL1 and the 13myc tag sequences encodes RIPGLIN. The plasmids containing the mutants were targeted for integration at TRP1 by digestion at the unique BstXI site.
Table 2.
HSL1 regional deletion mutants
| Mutant | Deleted residues | Inserted residues |
|---|---|---|
| Δ1 | 15-76 | GS |
| Δ2 | 116-145 | GS |
| Δ3 | 389-465 | V |
| Δ4 | 537-610 | None |
| Δ5 | 663-743 | None |
| Δ5a | 663-684 | GS |
| Δ5b | 721-735 | None |
| Δ6 | 879-914 | None |
| Δ7 | 951-1100 | RIW |
| Δ8 | 1138-1307 | RIW |
| Δ8a | 1144-1200 | L |
| Δ8b | 1206-1234 | None |
| Δ8c | 1253-1302 | I |
| Δ9 | 1379-1518 | None |
| Δ9a | 1419-1451 | None |
| Δ9b | 1457-1474 | S |
| Δ9c | 1481-1518 | None |
To express glutathione transferase (GST)-Hsl1p fragments, HSL1 sequences were amplified by PCR with primers containing flanking BamHI and SacI sites, as follows: GAL1-GST-HSL1663-914, CGGGATCCAGAAGAACACTACAAAACTCGGCT and GCGAGCTCTTATTTCAGTCCCTGCAATAGTGATTC; GAL1-GST-HSL1951-1100, CGGGATCCAGTTCATATGTACCTTTAACAACAG and GCGAGCTCTTATTTGGGCGAAGTATTGATTTTATT; GAL1-GST-HSL11138-1307, CGGGATCCACATATGATGAAAAAGCCATTAAT and GCGAGCTCTTATTTTTTAGATTCTTCCTCCTCCTC; GAL1-GST-HSL11379-1518, CGGGATCCAAAAGTGTCCAAAATGTTGAAGTTG and GCGAGCTCTTATGAACGTCCGGCATTTCGAATTA; and GAL1-GST-HSL1470-530, CGGGATCCTCAAGCGAAAACAAGAAATCAGCAAC and GCGAGCTCTTACCCTGTAATTGCGTTGTTCTGATTC. The fragments were cloned into BamHI and SacI sites of pDLB2492 or pDLB2493 (integrating TRP1-marked and LEU2-marked plasmids expressing GST from the GAL1 promoter) (Szkotnicki et al., 2008). Plasmids were targeted to integrate at TRP1 by digesting with either EcoRV (HSL1951-1100, HSL11138-1307, HSL11379-1518) or BstXI (HSL1663-914). To express GAL1-GST-HSL1470-1518, a LEU2-marked GAL1-GST-HSL1470-530 was targeted to integrate at HSL1-myc:TRP1 by digesting with StuI. In addition to expressing GST-Hsl1p470-1518-myc from the GAL1 promoter, this strain expresses a truncated Hsl1p1-530 from the HSL1 promoter. To express GST-Hsl1p987-1518 with internal deletions (Figure 3A), we targeted integration of pDLB2506 (GAL1-GST-HSL1987-1100; Szkotnicki et al., 2008) to the desired HSL1ΔN-myc:TRP1 mutant by digestion with BsaBI. These strains also express a truncated Hsl1p1-1100 from the HSL1 promoter.
Figure 3.
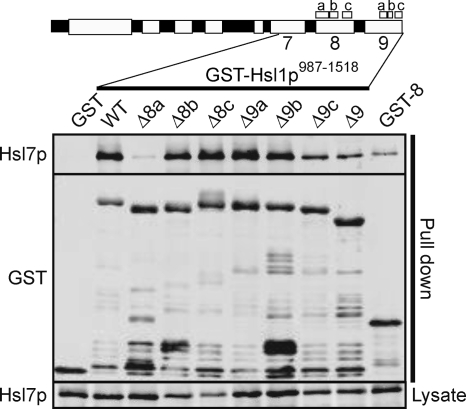
Regional requirements for Hsl7p binding. GST-Hsl1p987-1518 fragments containing the indicated regional deletions were overexpressed from the GAL1 promoter in strains expressing Hsl7p-HA. The left-most lane expressed a GST-only control, and the right-most lane expressed a GST-Hsl1p1138-1307 fragment containing only region 8. After precipitation with glutathione beads and washing, bead-bound proteins were detected by Western blot. Top, bound Hsl7p-HA; middle, GST fusions; and bottom, Hsl7p-HA in the starting lysate.
Media and Growth Conditions
Yeast strains were grown in 1% yeast extract 2% Bacto Peptone, 2% dextrose, 0.012% adenine, and 0.01% uracil (YEPD) or in synthetic dextrose complete media lacking relevant amino acids at 30°C unless otherwise noted. Bacterial strains were grown in Difco LB broth (BD Biosciences, San Jose, CA) supplemented with 50 μg/ml ampicillin at 37°C with the exception of those bacteria used to produce recombinant proteins, which were grown at 18°C.
For expression of GST-Hsl1p fragments under GAL1 promoter control, yeast were grown at 24°C in YEPS (as YEPD but with 2% sucrose and 0.05% dextrose) for 12 h, galactose was added to a final concentration of 2%, and cells were collected 5 h (Figure 3A) or 20 h (all other experiments) later.
GST Precipitation
Cells were lysed by bead beating in cold lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.1% Triton X-100, 5 mM EDTA, 1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), plus inhibitors (1 μg/ml pepstatin A, 1 μg/ml leupeptin, 2.7 μg/ml aprotinin, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 20 mM NaF). The lysates were then cleared by centrifugation. To remove endogenous glutathione which might interfere with the precipitation, the supernatant was passed over two BioSpin 6 columns (Bio-Rad, Hercules, CA). Glutathione beads (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom) were washed four times with cold lysis buffer and then resuspended in an equal volume of lysis buffer to make a 50% slurry. Five milligrams of lysate was diluted to a final concentration of 10 mg/ml with cold lysis buffer, and 60 μl of bead slurry was added to the mixture and allowed to incubate at 4°C for 2 h while rotating. Beads were washed four times with cold lysis buffer and resuspended in and equal volume of hot (95°C) 2× SDS loading buffer (62.5 mM Tris-HCl, pH 6.8, 1% SDS, 25% glycerol, 355 mM β-mercaptoethanol, and 0.01% bromophenol blue) and boiled for 10 min to release the bound proteins.
Western Blotting
Lysis was performed by either the trichloroacetic acid (TCA) method (Keaton et al., 2008) for most Western blots or the NP-40 method (Bose et al., 2001) for immunoprecipitation and for Western blots in Figure 1 and Supplemental Figure S2. Samples were then subjected to polyacrylamide gel electrophoresis (PAGE). To resolve Hsl1p-myc phosphoforms, lysate was run on a 6% low-bis polyacrylamide gel (Barral et al., 1999). To resolve Hsl7p-hamagglutinin (HA), Cdc11p, and the GST-Hsl1p fragments, lysates were run on 8% polyacrylamide gels, and for Cdc28p detection a 10% polyacrylamide gel was used. Proteins were transferred to a nitrocellulose membrane (Pall, East Hills, NY) and blocked for 1 h with either phosphate-buffered saline (PBS) containing 3% nonfat dry milk (for staining with anti-myc, anti-HA, or anti-Cdc11), a 1:1 solution containing Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE) and Tris-buffered saline (TBS) (for staining with anti-PSTAIRE and anti-phospho-Cdc2), or a solution of TBS containing 2% fish gelatin (Sigma-Aldrich) and 0.5% egg albumin (Sigma-Aldrich) (for staining with anti-GST). Antibodies were diluted in blocking buffer supplemented with 0.1% Tween 20 (Bio-Rad), and membranes were probed overnight with 1:250 mouse anti-myc 9E10, 1:10,000 mouse anti-PSTAIRE (for Cdc28p), 1:2000 rabbit anti-phospho-tyr15-Cdc2 (for phospho-Cdc28p; Cell Signaling Technology, Danvers, MA), 1:50,000 rabbit anti-Cdc11p (Santa Cruz Biotechnology, Santa Cruz, CA), or for 1 h with 1:1000 mouse anti-HA 12CA5 (Roche Diagnostics, Indianapolis, IN), 1:3000 rabbit anti-GST, or 1:2000 mouse anti-GST (Santa Cruz Biotechnology). Membranes were then washed three times with PBS/0.1% Tween 20 (myc, HA, Cdc11p, GST) or TBS/0.1% Tween 20 (PSTAIRE and phospho-Cdc2) and then probed with 1:7500 goat anti-mouse IRdye800 (Rockland Immunochemicals, Gilbertsville, PA) or 1:7500 goat anti-rabbit AlexaFluor680 (Invitrogen, Carlsbad, CA) for 1 h in blocking buffer containing 0.1% Tween 20 and 0.01% SDS. Blots were then washed as above and visualized using an Odyssey scanner (Li-Cor Biosciences). Fluorescence was quantified using Odyssey software (Li-Cor Biosciences).
Figure 1.
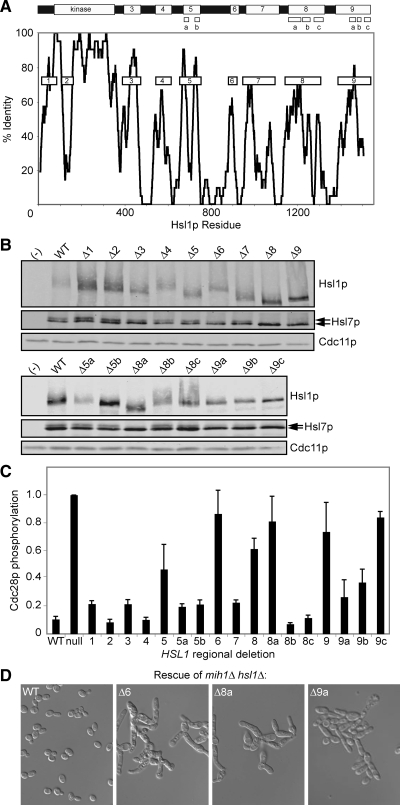
Identification of Hsl1p regions by phylogenetic comparison. (A) Top, schematic of Hsl1p. Bottom, homology scan across the protein. To identify potential functional motifs, Hsl1p sequences from S. cerevisiae, S. bayanus, and S. castelli were aligned using ClustalW, and the alignment was scanned using a sliding 21-residue window, scoring only those residues that were identical in all three species. Conserved regions (boxes) were numbered 1 and 3–9 and targeted for deletion analysis. Region 2 is a region of rapid sequence divergence within the kinase domain, which was targeted based on the possibility that regulatory interactions with the kinase domain may use regions that are free to evolve more rapidly than the structurally constrained kinase domain itself. For regions 5, 8, and 9, we also generated smaller deletions as indicated in the schematic. (B) Western blots of myc-tagged Hsl1p, HA-tagged Hsl7p, and Cdc11p (a septin, used as loading control), in strains expressing regional deletions of HSL1 at single copy instead of endogenous HSL1 (see Table 2 for sequence). Lanes labeled (−) contain lysates from a strain lacking tagged Hsl1p/7p. Arrows indicate Hsl7p phosphoforms. (C) Cdc28p tyrosine phosphorylation in strains expressing Hsl1p regional deletions was determined by measuring the ratio of anti-phospho-Cdk1 to anti-total-Cdk1 signal in two-color Western blots (Supplemental Figure S2). Mean ratios (± SEM) from at least three independent blots were normalized to the phosphorylation in the hsl1Δ null mutant. (D) Double-mutant strains containing Hsl1p regional deletions and mih1Δ illustrate the elongated bud G2 delay phenotype that accompanies impaired Hsl1p function. See Supplemental Figure S2 and Table 3 for other mutants.
A quantitative measure (Figure 1C) of Cdc28p tyr19 phosphorylation from gels including those in Supplemental Figure S2A was derived from the ratio of anti-phospho-Cdc2 staining to anti-PSTAIRE staining in two-color blots, as described previously (Keaton et al., 2007).
Microscopy and Immunofluorescence
To analyze cell morphology, cells were fixed in 70% ethanol for 1 h at room temperature, washed with PBS, sonicated, resuspended in mounting medium (90% glycerol, 9.2 mM p-phenylenediamine in PBS [Sigma-Aldrich], and 0.06 μg/ml 4,6-diamidino-2-phenylindole [Sigma-Aldrich]), and spotted onto a 1% agarose pad.
For imaging Cdc3p-green fluorescent protein (GFP), cells were fixed in 2% paraformaldehyde (Sigma-Aldrich) for 10 min at room temperature, washed once with solution B (0.1 M KPO4 pH 7.5, and 1.2 M sorbitol) and once with PBS, resuspended in mounting medium, and spotted onto a 1% agarose pad.
For immunofluorescence, cells were fixed in 3.7% formaldehyde at 30°C for 1 h. Cells were then resuspended in 500 μl of solution B. To digest the cell walls, 5 μl of lyticase (Sigma-Aldrich) and 5 μl of 14.3 M β-mercaptoethanol (Sigma-Aldrich) were added to the cells, and the mixture was incubated at room temperature for 10–15 min. After washing twice with solution B and once with TBS, the spheroplasts were resuspended in 5% goat serum (Invitrogen) in TBS and applied to precleaned 10-well microscope slides (Polysciences, Warrington, PA) coated with 2% polyethylenimine (Sigma-Aldrich), and incubated with either mouse anti-myc 9E10 (1:200) (Santa Cruz Biotechnology), mouse anti-HA 12CA5 (1:100) (Roche Diagnostics), or rabbit anti-Cdc11 (1:200) (Santa Cruz Biotechnology) for 1 h. The wells were then washed 10 times with 5% goat serum in TBS and incubated with goat anti-mouse AlexaFluor 488 (1:150) (Invitrogen) and/or goat anti-rabbit AlexaFluor 568 (1:150) (Invitrogen), followed by 10 more washes. Mounting medium was applied to the wells and the slide was topped with a coverslip and sealed with clear nail polish.
Cells were examined on an Axioimager (Carl Zeiss, Jena, Germany) and photographed with an Orca ER monochrome cooled-charge-coupled device camera (Hamamatsu, Bridgewater, NJ). Images were captured using MetaMorph software (Millipore, Billerica, MA).
Kinase Assays
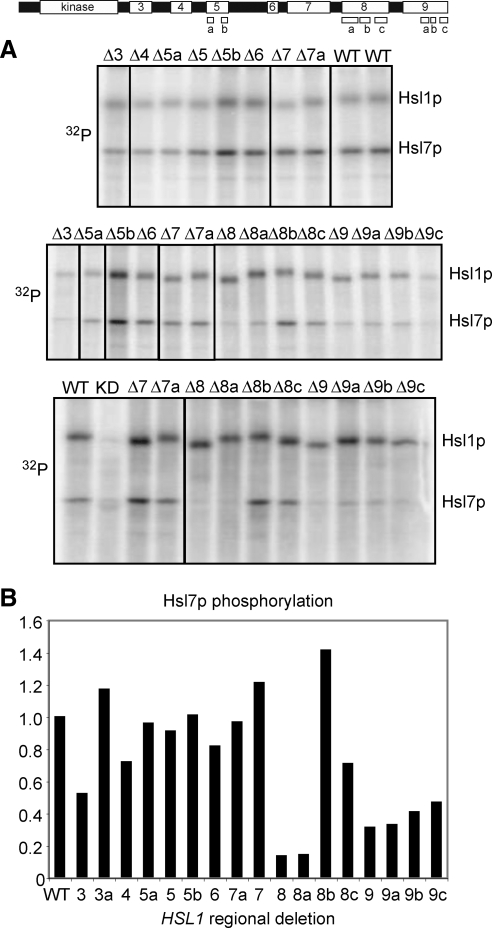
Hsl1p-myc immunoprecipitation, production of recombinant GST-Hsl7p, and in vitro kinase assays were performed as described previously (Szkotnicki et al., 2008). To derive a quantitative measure of Hsl7p phosphorylation by the Hsl1p deletion mutants by autoradiography (including the gels in Figure 4), we first calculated the ratio of 32P incorporation into Hsl7p to the 32P incorporation into Hsl1p, and normalized that ratio to the value for wild-type Hsl1p from the same gel. In gels lacking a wild-type Hsl1p lane, the normalization was carried out with Hsl1pΔ987-1100, which was identical to wild-type in a large number of trials. Because GST-Hsl7p preps varied, all normalizations were performed within the same experiment. Normalized values were averaged from two to four experiments to yield the data in Figure 3B.
Figure 4.
Hsl1p regions 8a and 9 are important for Hsl7p phosphorylation. Top, schematic of Hsl1p with regions indicated. (A) Myc-tagged Hsl1p proteins expressed at endogenous levels were immunoprecipitated and incubated with [γ-32P]ATP and recombinant GST-Hsl7p, separated by SDS-PAGE, and phosphorylation was detected by autoradiography. WT, wild type; K110R, kinase-dead control; and Δ7a, Δ987-1100 mutant (Hanrahan and Snyder, 2003). Additional lanes from the gels were removed using Photoshop (Adobe Systems, Mountain View, CA), as indicated by black lines. (B) The ratio of 32P incorporation into Hsl7p versus Hsl1p was quantitated and normalized to wild type. Averages from at least two kinase assays (including but not limited to the gels in A) are shown.
Homology Scanner Program
This program calculates the percentage of amino acids that are identical among all sequences of an alignment within a window of defined width. Starting from a given alignment, the program first removes any gaps in the “base” sequence (in our case S. cerevisiae Hsl1p) and then iteratively calculates the percentage of identity by using a sliding window. In our case, the alignments were first generated using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and then scanned with a 21-residue window (10 amino acids on either side of a central amino acid). The program can be found at http://oreo.biology.duke.edu/alignProtienSequence/slidingWindowAlignment2.pl.
RESULTS
Regional Requirements for Hsl1p Function
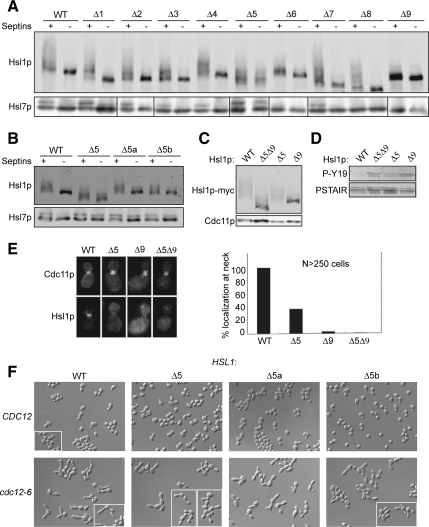
Hsl1p is a 1518-residue protein with a Nim1-family kinase domain near the N terminus (Figure 1A). Outside of the kinase domain, Hsl1p orthologues and paralogues have diverged rapidly (Supplemental Figure S1). Alignment of Hsl1p from closely related Saccharomyces species, however, identified several conserved motifs, which we targeted for deletion analysis (Figure 1A). Each of the mutants was expressed at levels roughly comparable with wild type (Figure 1B). Hsl1p-dependent Hsl7p phosphorylation, detected as a slower mobility species in blots (McMillan et al., 1999), was markedly reduced or absent upon deletion of Hsl1p regions 6, 8, or 9 (Figure 1B). These same deletions markedly elevated Swe1p-mediated CDK tyrosine phosphorylation (Figure 1C and Supplemental Figure S2), suggesting that Swe1p down-regulation was impaired. In addition, these mutants displayed a G2 delay or lethal arrest when combined with deletion of MIH1, the Cdc25-related phosphatase that counteracts Swe1p action (Figure 1D, Supplemental Figure S2, and Table 3). Because active CDK promotes a depolarizing apical-isotropic switch in bud growth (Lew and Reed, 1993), these Swe1p-mediated G2 delays are associated with the development of elongated buds (Figure 1D and Supplemental Figure S2). Together, these data suggest that regions 6, 8, and 9 are critical for Hsl1p function. Similar but less severe functional deficits were noted upon deletion of region 5 (Figure 1, Supplemental Figure S2, and Table 3). Because regions 5, 8, and 9 each encompassed more than one sequence conservation peak, we generated smaller deletions (Figure 1A) that narrowed down the functionally relevant regions to 5b and 8a (Figure 1, Supplemental Figure S2, and Table 3). Deletions of 9a, 9b, or 9c all had deleterious effects, although none was as severe as deletion of the entire region 9. In summary, regions 6, 8a, and 9 are each critical for Hsl1p function, region 5b plays a less important role, and regions 1, 2, 3, 4, 5a, 7, 8b, and 8c are largely dispensable.
Table 3.
Rescue of hsl1Δ mih1Δ by HSL1 regional deletions
| HSL1 allele | Viable | Microcolony | Inviable |
|---|---|---|---|
| WT | 13 | ||
| Δ1 | 21 | ||
| Δ2 | 13 | ||
| Δ3 | 10 | 1 | |
| Δ4 | 11 | ||
| Δ5 | 12 | ||
| Δ5a | 20 | 1 | |
| Δ5b | 13 | ||
| Δ6 | 4 | 10 | |
| Δ7 | 23 | 3 | |
| Δ8 | 14 | ||
| Δ8a | 12 | 2 | |
| Δ8b | 14 | ||
| Δ8c | 14 | ||
| Δ9 | 4 | 2 | 17 |
| Δ9a | 13 | ||
| Δ9b | 3 | 1 | 16 |
| Δ9c | 11 |
Diploids containing marked mih1Δ, hsl1Δ, and regional HSL1 mutants were sporulated, tetrads were dissected, and viability of hsl1Δ mih1Δ mutants containing the indicated HSL1 regional deletions was inferred from marker segregation. The few viable colonies from Δ9 and Δ9b seem likely to contain suppressor mutations.
Region 9 Is Required for Hsl1p Localization, and Region 8a Is Required for Hsl7p Recruitment
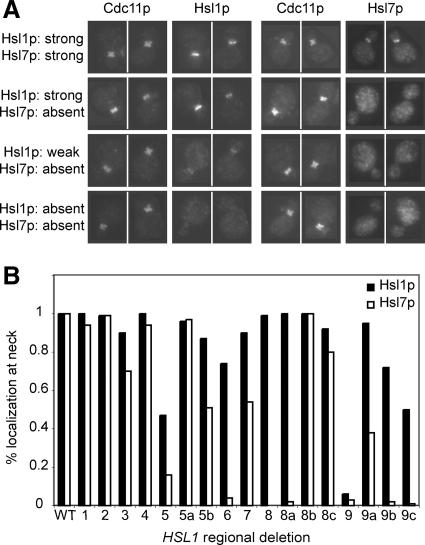
The Hsl1p mutants that exhibited normal (or near normal) function also exhibited wild-type (or near wild-type) localization of both Hsl1p and Hsl7p (Figure 2). Deletion of region 9 resulted in the complete disappearance of both Hsl1p and Hsl7p from the neck, implying that this region is critical for Hsl1p localization. Deletion of region 8 or 8a had no effect on Hsl1p localization but abolished Hsl7p localization, suggesting that 8a mediates Hsl7p recruitment. Several other deletions (5, 5b, 6, 9a, 9b, and 9c) partly reduced both Hsl1p and Hsl7p localization, with Hsl7p being more strongly affected than Hsl1p in every case. It is noteworthy that all mutations that reduced Hsl1p function also reduced localization of Hsl7p to the neck, consistent with the notion that Hsl7p localization is the major (and perhaps only) function of Hsl1p in Swe1p regulation.
Figure 2.
Effect of regional deletions on localization of Hsl1p and Hsl7p. (A) Localization of Hsl1p-myc and Hsl7p-HA was determined by immunofluorescence microscopy. The four categories of staining pattern are illustrated with pairs of cells double-stained for Cdc11p (septin) and Hsl1p (left), or Cdc11p and Hsl7p (right). Top row, wild type; second row, region 8 deleted; third row, region 9a deleted; and bottom row, region 9 deleted. (B) Localization was quantitated based on the percentage of cells with detectable neck staining for Hsl1p or Hsl7p, normalized to wild type.
Region 8a Mediates Hsl7p Interaction with Assistance from Region 9
A fusion of GST to the Hsl1p C-terminal 531 residues (987-1518, encompassing regions 7-9) was able to effectively recover Hsl7p-HA from yeast lysates (Figure 3). Hsl7p binding was abolished by deletion of region 8a and reduced by deletion of region 9 or 9c. Region 8 alone was able to bind Hsl7p, although not as robustly as the larger 987-1518 fragment (Figure 3). In vitro kinase assays performed with Hsl1p-myc immunoprecipitates revealed that all Hsl1p mutants were capable of robust autophosphorylation but that region 8a was required for Hsl7p phosphorylation, and region 9 enhanced Hsl7p phosphorylation approximately threefold (Figure 4). In sum, the localization, binding, and phosphorylation data indicate that Hsl1p-Hsl7p interaction is mediated primarily by region 8a and that region 9 increases the interaction efficacy.
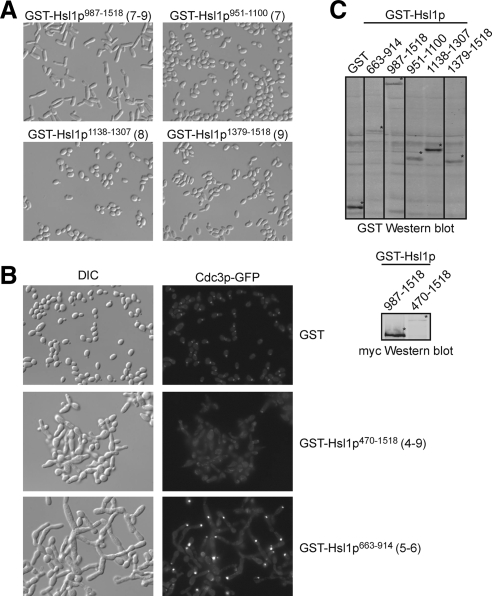
Multiple Hsl1p Regions Can Interface with Septins
Overexpression of the Hsl1p 987-1518 fragment also led to septin disorganization and the development of elongated buds (Szkotnicki et al., 2008), consistent with the presence of septin-interacting domains in regions 7–9 (Hanrahan and Snyder, 2003). However, separate overexpression of regions 7, 8, or 9 did not affect septins or bud morphology (Figure 5A), suggesting that several septin binding regions may be required for the Hsl1p 987-1518 fragment to induce septin disorganization. Unexpectedly, overexpression of an Hsl1p fragment encompassing regions 5 and 6 had an even more dramatic effect, promoting gross septin disorganization and development of large, branched, tubular cells (Figure 5B). This observation suggests that regions 5 and/or 6 also harbor septin-interaction motifs, although we have thus far been unable to detect septin interactions biochemically with any portion of Hsl1p.
Figure 5.
Effects of overexpressing Hsl1p regions on septins and cell morphology. (A) Bud elongation caused by Hsl1p987-1518 containing regions 7–9 is not recapitulated by overexpression of individual regions. Cells containing the indicated GST-Hsl1p fragments were grown in galactose containing medium, fixed, and photographed using differential interference contrast (DIC) microscopy. (B) Overexpression of GST-Hsl1p470-1518 containing regions 4–9 causes much more severe bud morphology defects, and displaces septins from the neck. Both defects are recapitulated by GST-Hsl1p663-914 containing only regions 5 and 6, although in this case septin disappearance from the neck is accompanied by aggregation in bright foci frequently at the bud tip (visualized using GFP-Cdc3p). (C) Western blot showing GST fusions. Additional lanes from the gel were removed using Photoshop, as indicated by black lines. GST-Hsl1p470-1518 was not detected by the GST antibody (due to lower expression) but could be detected using anti-myc to recognize the C-terminal 13-myc tag (bottom).
Deletion of Region 5 Allows Hsl1p to Remain Active after Septin Disassembly or Deletion of Region 9
We suspected that some of the evolutionarily conserved regions that were not required for Hsl1p-mediated Swe1p down-regulation might instead be important for regulation of Hsl1p by septins. Previous studies showed that both Hsl1p autophosphorylation and Hsl7p phosphorylation are lost when septins disassemble upon shift of temperature-sensitive septin mutants to restrictive temperature (Barral et al., 1999; Theesfeld et al., 2003). However, we found that Hsl1p lacking region 5 remained highly phosphorylated, as did Hsl7p, even in the absence of assembled septins (Figure 6A), suggesting that Hsl1p region 5 is important for turning off Hsl1p when it is not bound to septins. Similar, although smaller, effects were observed upon deletion of the smaller 5a or 5b segments (Figure 6B).
Figure 6.
Hsl1p region 5 is important for Hsl1p regulation by septins. (A) Deletion of region 5 prevents the loss of Hsl1p and Hsl7p phosphorylation after septin disassembly. Wild-type (Septin +) and cdc12-6 (Septin −) cells containing the indicated Hsl1p regional deletions were shifted from 18°C to 37°C for 20 min, lysed by TCA precipitation, and Hsl1p-myc and Hsl7p-HA were detected by Western blotting. Hsl7p panels from different gels are separated by vertical bars. (B) Deletion of 5a or 5b partly recapitulates the phenotype of the full region 5 deletion in this assay. (C) Deletion of region 5 restores autophosphorylation to Hsl1p lacking region 9. Cdc11p, loading control. (D) Deletion of region 5 does not restore function of Hsl1p lacking region 9. Western blots of whole cell lysates were probed for Swe1p-phosphorylated Cdc28p tyr19 (P-Y19) and total CDK (PSTAIR). High P-Y19 results from failure to degrade Swe1p and is similar in Hsl1pΔ9- and Hsl1pΔ5Δ9-expressing cells. (E) Deletion of region 5 does not restore neck localization of Hsl1p lacking region 9. Immunofluorescence as in Figure 2. Left, representative cells double-stained for Cdc11p (septin, top) and Hsl1p (bottom). Right, localization was quantitated based on the percentage of cells with detectable neck staining for Hsl1p, normalized to wild type. (F) Deletion of region 5 or its subregions does not prevent bud elongation following septin disassembly. Cells containing wild-type (top) or temperature-sensitive mutant (bottom) septin Cdc12p and the indicated allele of Hsl1p were grown at 24°C, shifted to 37°C for 3 h, fixed, and photographed by DIC microscopy. White boxes indicate cells from different fields.
Deletion of region 9 abolished Hsl1p localization to the neck (Figure 2) as well as catalytic activity (as judged by Hsl1p autophosphorylation) and function (Figure 1). However, we found that deleting both region 5 and region 9 restored a significant degree of Hsl1p autophosphorylation (Figure 6C), although it did not restore Hsl1p function (Figure 6D) or localization (Figure 6E), implying that region 5 prevents activation of the delocalized Hsl1pΔ9 mutant.
A previous analysis of Hsl1p concluded that residues 987-1100 (comprising most of region 7) contain an autoinhibitory domain and that removal of this domain from Hsl1p allows the “activated” mutant to down-regulate Swe1p and thereby reduce bud elongation after septin disassembly (Hanrahan and Snyder, 2003). However, we were unable to reproduce this finding (Szkotnicki et al., 2008), and unlike Hsl1p lacking region 5, the Hsl1p lacking region 7 was rapidly dephosphorylated upon septin disassembly (Figure 6A).
Because our analysis suggested that region 5 (rather than region 7) might encode an autoinhibitory domain, we also tested whether deletion of region 5 (or 5a, which is dispensable for Hsl1p function) would bypass the need for septins in Swe1p down-regulation. We found that deleting these regions did not alter the Swe1p-mediated G2 delay (as judged by bud elongation) that follows septin disassembly (Figure 6F). These findings suggest that even if Hsl1p remains active as a kinase, it must localize to the neck for effective Swe1p degradation.
Relationship between Hsl1p Region 5 and Hsl1p Activation by the Upstream Kinase Elm1p
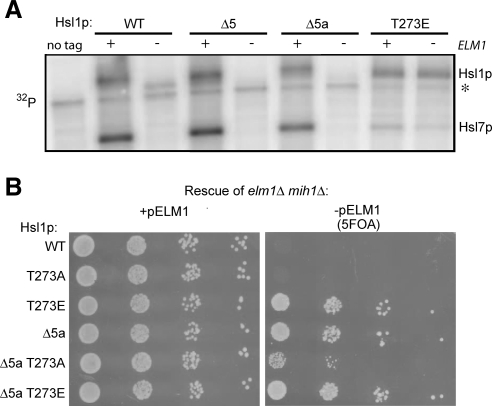
We recently reported that full Hsl1p activation requires phosphorylation of T273 in the kinase activation loop, which is promoted by the kinase Elm1p (Szkotnicki et al., 2008). Nonphosphorylatable Hsl1pT273A mutants display greatly reduced kinase activity in vitro and reduced biological potency in vivo, whereas a phospho-mimic Hsl1pT273E mutant is active (at a reduced level) and functional even in an elm1Δ mutant background. elm1Δ mih1Δ mutants are synthetically lethal, and the lethality can be rescued by Hsl1pT273E (Szkotnicki et al., 2008). Because our regional analysis suggested that region 5 was important for Hsl1p regulation, we investigated the relationship between this domain and the activation of Hsl1p via Elm1p.
The in vitro kinase activity of Hsl1p toward recombinant Hsl7p was dramatically reduced when Hsl1p was isolated from elm1Δ cells (Szkotnicki et al., 2008), and we found that deletion of region 5 did not alter this behavior (Figure 7A). Thus, deleting region 5 does not bypass the requirement for Elm1p in full Hsl1p activation.
Figure 7.
Relation between Hsl1p region 5 and Elm1p-mediated Hsl1p activation. (A) Deletion of region 5 or 5a does not bypass the need for Elm1p in Hsl1p kinase activation. Hsl1p carrying the indicated mutations was immunoprecipitated from wild-type (+) or elm1Δ (−) strains and assayed for autophosphorylation and Hsl7p phosphorylation as in Figure 4. Asterisk denotes nonspecific background band. (B) Deletion of region 5a bypasses the need for Elm1p in Hsl1p function. A mih1Δ elm1Δ strain kept alive with a URA3-marked plasmid carrying ELM1 was transformed with plasmids expressing the indicated alleles of Hsl1p. On 5-fluoroorotic acid (5FOA) media, only cells that have lost the ELM1 plasmid can live, providing an assay for whether the Hsl1p allele can function without Elm1p.
Deletion of region 5 made Hsl1p insensitive to septin disorganization (Figure 6A), but this region was also required for full Hsl1p function (Figure 1), suggesting that it can exert both positive and negative effects on Hsl1p. Deletion of region 5a partly reduced the sensitivity of Hsl1p to septin disorganization (Figure 6B) with no ill effects on Hsl1p function (Figure 1), so we used the Hsl1pΔ5a mutant to test whether this deletion allows Hsl1p function even in the absence of the upstream kinase Elm1p. Hsl1pΔ5a was able to rescue the elm1Δ mih1Δ lethality (Figure 7B), suggesting that deletion of region 5a can promote Hsl1p function even in the absence of Elm1p.
Rescue of elm1Δ mih1Δ lethality was also observed (albeit somewhat less robustly) with a double mutant Hsl1pT273A, Δ5a (Figure 7B), suggesting that deletion of 5a can promote Hsl1p function without T273 phosphorylation.
DISCUSSION
Conserved Elements in Hsl1p
Guided by evolutionary conservation, we identified several motifs important for Hsl1p regulation and function. The large nonkinase portion of Hsl1p-family kinases differs significantly between species: no homology is evident between S. cerevisiae Hsl1p and Schizosaccharomyces pombe Nim1 outside the kinase domain (Supplemental Figure S1), and a comparison of S. cerevisiae and Candida albicans Hsl1p found only one very small conserved motif N-terminal to the kinase domain (our region 1) (Wightman et al., 2004). Because of this rapid divergence, we sought to identify functional motifs by comparing the closer Saccharomyces relatives S. cerevisiae, Saccharomyces bayanus, and Saccharomyces castelli. Scoring only residues that were identical in all three species, we identified a patchwork of conserved regions separated by nonconserved linkers. Five of these regions (Figure 1, 5a, 5b, 6, 8a, and 9) proved critical for Hsl1p function and/or regulation, but six other regions (Figure 1, regions 1, 3, 4, 7, 8b, and 8c) could be deleted with little apparent effect in the assays we used. It is possible that these regions are important for regulating Hsl1p in response to specific stresses not examined here, as suggested by the identification of a phosphorylation site targeted by the osmostress MAPK Hog1p in motif 8b (Clotet et al., 2006). Alternatively, these regions could mediate distinct Hsl1p functions, such as the proposed roles for Hsl1p in targeting Cdc5p to the neck (Sakchaisri et al., 2004) or regulating microtubule interactions with the neck (Kusch et al., 2002).
Hsl1p Localization and Septin Interaction
Hsl1p localization to the neck was abolished upon deletion of region 9 and partly reduced upon deletion of regions 5b or 6. The simplest interpretation of these findings is that regions 5b, 6, and 9 each interact with septins at the neck, helping to localize Hsl1p. Consistent with that view, overexpression of regions 5 + 6 or regions 7 + 8 + 9 caused septin disorganization at the neck, and previous studies indicated that multiple fragments from regions 7, 8, and 9 could interact with septins in the yeast two-hybrid assay (Hanrahan and Snyder, 2003). We were unable to demonstrate a convincing septin interaction by coimmunoprecipitation (data not shown), although others have detected such interactions (Barral et al., 1999). We suspect that the physiological binding partner for Hsl1p is the septin filament array, rather than unassembled septin complexes. Previous findings indicated that Hsl1p activation requires septin interaction (Barral et al., 1999) and that Hsl1p activation may require a specific septin filament arrangement found only in budded cells (Theesfeld et al., 2003). In that context, the observation that several regions in Hsl1p may interact with septins supports the possibility that Hsl1p's multiple septin contacts may allow it to “sense” septin organization.
Hsl1p-Hsl7p Interaction
Deletion of region 8a abolished Hsl1p-Hsl7p coimmunoprecipitation as well as Hsl7p localization and phosphorylation, without affecting Hsl1p localization or autophosphorylation. This mutant was nonfunctional, confirming the hypothesis that Hsl7p binding by Hsl1p is a key step in Swe1p degradation (Cid et al., 2001). Region 8 alone was able to bind Hsl7p, suggesting that 8a is the major binding site for Hsl7p. However, the presence of region 9 was important for optimal binding as well as phosphorylation of Hsl7p. This is likely a separate role for region 9 from its role in Hsl1p localization to the septins at the neck, because the kinase assays were performed in vitro with recombinant Hsl7p. It is interesting to note that effective Hsl7p localization to the septin cortex requires both bud emergence and Hsl1p catalytic activity, whereas Hsl1p localization can occur efficiently even when budding is blocked or the kinase is inactivated (Theesfeld et al., 2003). Thus, although a small fragment of Hsl1p can directly bind Hsl7p in vitro, effective interaction in vivo is more complex. We speculate that Hsl7p binding site(s) in regions 8a and/or 9 are masked when Hsl1p is inactive and that a conformational change in Hsl1p, perhaps coupled to Hsl1p-mediated phosphorylation(s) (likely on Hsl1p itself or Hsl7p), increases Hsl7p affinity to promote effective recruitment once cells have budded.
Hsl1p Regulation by Septins
A previous study on Hsl1p regulation (Hanrahan and Snyder, 2003) suggested that a part of region 7 (residues 987-1100) constituted an autoinhibitory domain that bound to and inhibited the Hsl1p kinase domain. This region was also suggested to bind septins, leading to a model in which septin binding activates Hsl1p via relief of autoinhibition. Our findings, both here and in Szkotnicki et al. (2008), do not support the specifics of that model, as deletion of region 7 (or the 987-1100 piece) had little effect on Hsl1p regulation or function in any of the conditions we tested. However, the overall autoinhibition strategy may well apply, as suggested by the behavior of mutants lacking all or part of Hsl1p region 5.
Deletion of region 5 allowed Hsl1p and Hsl7p to remain phosphorylated after septin disassembly, suggesting that it is region 5 that normally turns off Hsl1p kinase activity when septins disassemble. Thus, we suggest that region 5 is autoinhibitory for Hsl1p activity and function. Region 5 contains two conserved sequence elements, one of which (5a) is highly conserved even in the more distantly related fungi Ashbya gossypii and Kluyveromyces lactis (Supplemental Figure S1) and seems to be purely autoinhibitory. The other motif (5b) contributes to autoinhibition but is also required for optimal Hsl1p localization and function. The neighboring region 6 is also important for Hsl1p localization and function, and a fragment of Hsl1p containing regions 5 and 6 has dramatic effects on septin organization when overexpressed. We speculate that these regions mediate both autoinhibition and septin interaction, thereby allowing septin conformation to regulate Hsl1p activity.
Two Parallel Pathways Activate Hsl1p
We recently described a pathway for Hsl1p regulation via phosphorylation of the activation-loop threonine (T273) in the kinase domain, which is promoted by the upstream neck-localized kinase Elm1p (Szkotnicki et al., 2008). T273 phosphorylation stimulates Hsl1p kinase activity, and a phospho-mimic Hsl1pT273E mutant can bypass the need for Elm1p in activating Hsl1p (Szkotnicki et al., 2008). In contrast, we found that deleting region 5 did not bypass the need for Elm1p in activating Hsl1p. Conversely, although deleting region 5 did bypass the need for septins in activating Hsl1p, the phospho-mimic Hsl1pT273E mutant did not (Szkotnicki et al., 2008). These findings rule out simple linear models in which T273 phosphorylation is needed only to relieve region 5-mediated autoinhibition, or alternatively region 5 is needed only to restrain T273 phosphorylation. Instead, the data suggest that two parallel inputs, T273 phosphorylation and relief of region 5-mediated autoinhibition, are required for full Hsl1p activation.
Strikingly, deletion of region 5a allowed bypass of the elm1Δ mih1Δ synthetic lethality, even though it did not restore Hsl1p kinase activity. This result suggests that the basal kinase activity of Hsl1p lacking T273 phosphorylation is sufficient to promote a significant level of Swe1p down-regulation once region 5-mediated autoinhibition is relieved.
Our data also highlight an apparent difference in Hsl1p kinase activity as measured in vitro (by 32P incorporation into Hsl1p itself or added recombinant Hsl7p) and in vivo (by monitoring phosphorylation-dependent mobility shifts in Hsl1p and Hsl7p). Both assays indicate severely reduced activity of Hsl1p from elm1Δ strains. However, in vitro both Hsl1pΔ9 (which cannot localize to the septin cortex) and wild-type Hsl1p isolated from septin-mutant strains retain robust activity, despite the lack of autophosphorylation in vivo. Region 5 is responsible for the lack of autophosphorylation in vivo, suggesting that kinase activity is restrained by an autoinhibited conformation in these cases. We speculate that this conformation is destabilized by the extraction or immunoprecipitation procedure used for the in vitro assay, explaining why the autoinhibited Hsl1p seems to be active in vitro.
An interesting conclusion emerging from these considerations is that “autoinhibited” Hsl1pΔ9 must be phosphorylated at T273 (otherwise it would not be active in vitro). The inference that an Hsl1p that does not localize to the septins is still phosphorylated at T273 suggests that the Elm1p-mediated Hsl1p activation pathway can occur efficiently in the cytoplasm, which is surprising considering that both Elm1p and Hsl1p are normally concentrated at the neck.
In summary, our findings suggest that septins regulate Hsl1p via region 5, whereas T273 phosphorylation represents a separate Hsl1p activation pathway that does not require neck localization. The physiological inputs affecting this pathway remain to be determined.
Relation between Hsl1p Kinase Activity and Biological Function
A previous report concluded that activated Hsl1p could promote Swe1p degradation even in the absence of assembled septins, suggesting that the only role for septins in Swe1p degradation is to activate Hsl1p (Hanrahan and Snyder, 2003). However, we were unable to reproduce that finding (Szkotnicki et al., 2008), and we found that none of the activated alleles discussed above were able to down-regulate Swe1p in the absence of assembled septins. Similarly, an activated mutant unable to localize to the neck (Hsl1p lacking both region 5 and region 9) was unable to down-regulate Swe1p. These observations suggest that in addition to activating Hsl1p, septins provide an essential localization platform for Swe1p degradation. This is consistent with the view that Hsl1p acts by bringing together Swe1p and its upstream kinases at the neck (Sakchaisri et al., 2004; Asano et al., 2005), allowing productive Swe1p phosphorylation and targeting Swe1p for degradation.
Implications for Other Systems
Intriguingly, recent studies showed that septins in mammalian neurons form ring structures (approximately the same size as those in yeast) at the base of dendritic spines (Barral and Mansuy, 2007; Tada et al., 2007; Xie et al., 2007). Investigation of septin roles in that system is still in its infancy, but dendritic spines are sites of intensive information processing, whose number and shape are thought to change during learning. This raises the fascinating possibility that septin “sensors” analogous to those in yeast may contribute to cognitive function in mammals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Audrey Howell, Steve Haase, Sally Kornbluth, and Lew laboratory members for comments on the manuscript. This work was supported by National Institutes of Health grant GM-53050 (to D.J.L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-08-0848) on February 18, 2009.
REFERENCES
- Asano S., Park J. E., Sakchaisri K., Yu L. R., Song S., Supavilai P., Veenstra T. D., Lee K. S. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M. N. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 2003;116:4119–4130. doi: 10.1242/jcs.00706. [DOI] [PubMed] [Google Scholar]
- Barral Y., Mansuy I. M. Septins: cellular and functional barriers of neuronal activity. Curr. Biol. 2007;17:R961–R963. doi: 10.1016/j.cub.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Barral Y., Parra M., Bidlingmaier S., Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Caviston J. P., Longtine M., Pringle J. R., Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Smith K. W., Gustin M. C. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 1992;118:561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid V. J., Shulewitz M. J., McDonald K. L., Thorner J. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol. Biol. Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J., Escote X., Adrover M. A., Yaakov G., Gari E., Aldea M., de Nadal E., Posas F. Phosphorylation of Hsl1 by Hog1 leads to a G(2) arrest essential for cell survival at high osmolarity. EMBO J. 2006;25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delley P. A., Hall M. N. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 1999;147:163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Gladfelter A. S., Pringle J. R., Lew D. J. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Hanrahan J., Snyder M. Cytoskeletal activation of a checkpoint kinase. Mol. Cell. 2003;12:663–673. doi: 10.1016/j.molcel.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Keaton M. A., Bardes E. S., Marquitz A. R., Freel C. D., Zyla T. R., Rudolph J., Lew D. J. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr. Biol. 2007;17:1181–1189. doi: 10.1016/j.cub.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaton M. A., Szkotniki L., Marquitz A. R., Harrison J., Zyla T. R., Lew D. J. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol. Biol. Cell. 2008;19:4006–4018. doi: 10.1091/mbc.E08-03-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch J., Meyer A., Snyder M. P., Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 2003;15:648–653. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., Theesfeld C. L., McMillan J. N., Weaver E., Pringle J. R., Lew D. J. Septin-dependent assembly of a cell-cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.-J., Lu Q., Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- McMillan J. N., Longtine M. S., Sia R.A.L., Theesfeld C. L., Bardes E.S.G., Pringle J. R., Lew D. J. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan J. N., Theesfeld C. L., Harrison J. C., Bardes E. S., Lew D. J. Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:3560–3575. doi: 10.1091/mbc.E02-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H. E., Wittenberg C., Cross F., Reed S. I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I. Conservation of mitotic controls in fission and budding yeast. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- Sakchaisri K., Asano S., Yu L. R., Shulewitz M. J., Park C. J., Park J. E., Cho Y. W., Veenstra T. D., Thorner J., Lee K. S. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. USA. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz M. J., Inouye C. J., Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R.A.L., Bardes E.S.G., Lew D. J. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia R.A.L., Herald H. A., Lew D. J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkotnicki L., Crutchley J. M., Zyla T. R., Bardes E. S., Lew D. J. The checkpoint kinase Hsl1p is activated by Elm1p-dependent phosphorylation. Mol. Biol. Cell. 2008;19:4675–4686. doi: 10.1091/mbc.E08-06-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Simonetta A., Batterton M., Kinoshita M., Edbauer D., Sheng M. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld C. L., Zyla T. R., Bardes E. G., Lew D. J. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell. 2003;14:3280–3291. doi: 10.1091/mbc.E03-03-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Barral Y. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Wightman R., Bates S., Amornrrattanapan P., Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Vessey J. P., Konecna A., Dahm R., Macchi P., Kiebler M. A. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.