Abstract
The TOR kinases are conserved negative regulators of autophagy in response to nutrient conditions, but the signaling mechanisms are poorly understood. Here we describe a complex containing the protein kinase Atg1 and the phosphoprotein Atg13 that functions as a critical component of this regulation in Drosophila. We show that knockout of Atg1 or Atg13 results in a similar, selective defect in autophagy in response to TOR inactivation. Atg1 physically interacts with TOR and Atg13 in vivo, and both Atg1 and Atg13 are phosphorylated in a nutrient-, TOR- and Atg1 kinase-dependent manner. In contrast to yeast, phosphorylation of Atg13 is greatest under autophagic conditions and does not preclude Atg1-Atg13 association. Atg13 stimulates both the autophagic activity of Atg1 and its inhibition of cell growth and TOR signaling, in part by disrupting the normal trafficking of TOR. In contrast to the effects of normal Atg13 levels, increased expression of Atg13 inhibits autophagosome expansion and recruitment of Atg8/LC3, potentially by decreasing the stability of Atg1 and facilitating its inhibitory phosphorylation by TOR. Atg1-Atg13 complexes thus function at multiple levels to mediate and adjust nutrient-dependent autophagic signaling.
INTRODUCTION
Eukaryotic cells have evolved multiple cellular processes and intracellular signaling systems that allow them to effectively respond to stressful environmental conditions. A central response to nutrient stress is the self-digestive process known as autophagy, in which portions of cytoplasm are sequestered into specialized vesicles known as autophagosomes (Mizushima, 2007; Xie and Klionsky, 2007). Subsequent fusion of these vesicles with the endosomal/lysosomal compartment leads to hydrolytic digestion of the autophagosome cargo, providing an internal source of nutrients that can support energy production or biosynthesis. This process is inhibited by the presence of abundant nutrients largely through the activity of the target of rapamycin (TOR) family of serine/threonine protein kinases (Pattingre et al., 2007). As autophagy has been found to influence an increasing spectrum of health and disease-related functions including aging, cancer, and neurodegenerative diseases (Levine and Kroemer, 2008; Mizushima et al., 2008), understanding the signaling mechanisms by which TOR regulates autophagy is an important goal.
The process of autophagosome formation is driven by the activity of the autophagy-related (ATG) genes, which were identified through genetic screens in yeast and in several cases have been shown to promote autophagy in higher eukaryotes as well (Suzuki and Ohsumi, 2007). In Saccharomyces cerevisiae, the Ser/Thr kinase Atg1 appears to play a unique role in initiating autophagosome formation in response to nutrient- and TOR-dependent signals. Loss of TOR signaling results in the activation and assembly of Atg1 into a multiprotein complex containing Atg13, Atg17, and other multifunctional proteins (Funakoshi et al., 1997; Matsuura et al., 1997; Kamada et al., 2000; Cheong et al., 2005; Kabeya et al., 2005). Formation of this complex is regulated at least in part through TOR-dependent phosphorylation of Atg13, causing decreased affinity for Atg1 under favorable growth conditions. The Atg1 complex promotes both recruitment and subsequent release of additional Atg proteins to a poorly characterized autophagosome initiating structure known as the phagophore assembly site or preautophagosomal structure (PAS; Reggiori et al., 2004; Cheong et al., 2008; Kawamata et al., 2008).
In higher eukaryotes, the mechanisms of autophagy regulation by TOR are less well understood. Disruption of the mammalian Atg1 orthologue Ulk1 through siRNA-mediated depletion or conditional knockout has been reported to either disrupt or to have no effect on starvation-induced autophagy (Chan et al., 2007; Kundu et al., 2008). This discrepancy may reflect the presence of the related Ulk2 gene, as expression of kinase-defective alleles of either Ulk1 or Ulk2 was found to disrupt autophagy (Hara et al., 2008), whereas Ulk2 knockdown did not (Chan et al., 2007). Ulk1 has also been implicated in axon outgrowth and branching, functions that are apparently unrelated to its presumptive role in autophagy (Tomoda et al., 2004).
Putative metazoan orthologues of Atg13 and Atg17 have also been recently identified and demonstrated to function in autophagy. siRNA-mediated knockdown of a human Atg13-related gene was shown to disrupt starvation-induced autophagy and trafficking of Atg9 (Chan et al., 2008). Atg13 was found to associate with and be phosphorylated by both Ulk1 and Ulk2, independent of nutrient conditions (Chan et al., 2008). Finally, the focal adhesion kinase–interacting protein FIP200 was recently shown to be required for autophagy and has been proposed to function as a mammalian counterpart to Atg17 (Hara et al., 2008).
Here we address these issues in Drosophila, which possesses a single Atg1 gene previously shown to act downstream of TOR to promote autophagy (Scott et al., 2004, 2007). We demonstrate that a Drosophila Atg13 orthologue is required for autophagy and associates with and activates Atg1, but that the functional and regulatory interactions among TOR, Atg1, and Atg13 have diverged in significant ways from their established roles in yeast.
MATERIALS AND METHODS
Drosophila Stocks and Genetic Manipulations
Flies were raised at 25°C on standard cornmeal/molasses/agar media. The following D. melanogaster strains were used: UAS-RhebAV4 (gift of F. Tamanoi, University of California, Los Angeles, CA), UAS-RhebEP50.084, Rheb2D1 (gifts of E. Hafen, Institute for Molecular Systems Biology, Zürich, Switzerland), UAS-Tsc1 and UAS-Tsc2 (gifts of N. Tapon, Cancer Research UK, London, United Kingdom), UAS-bsk (gift of D. McEwen, University of Texas Health Science Center, San Antonio, TX), Tsc129 (gift of D. Pan, Johns Hopkins University, Baltimore, MD), UAS-d4E-BP (gift of N. Sonenberg, McGill University, Montreal, QC, Canada), UAS-LAMP-green fluorescent protein (GFP; gift of Helmut Krämer, University of Texas, Dallas, TX), Atg1[Δ3D] (Scott et al., 2004), UAS-Atg1[6B] and UAS-Atg1[GS10797] (Scott et al., 2007), UAS-TOR[F4A] (Hennig and Neufeld, 2002), and the Atg13 inverted repeat UAS line 7331R-1 (Genetic Strain Research Center, Mishima, Japan). Additional strains were obtained from the Bloomington Stock Center (Bloomington, IN).
Deletions in the CG7331/Atg13 locus were generated by transposase-mediated imprecise excision of the viable P element line GS11822 (Drosophila Genetic Resource Center, Kyoto, Japan), which is inserted 34 bases after the annotated CG7331 transcription start site. Potential deletions were screened by PCR using primers flanking the locus and confirmed by sequence analysis.
Heat shock-induced Flippase (hsFLP)/Flippase Recognition Target (FRT)-mediated loss of function clones in the larval fat body were induced in 0–6 h embryos by a 1-h heat shock at 37°C and were marked by fat body–specific activation of Upstream Activating Sequence (UAS)-green fluorescent protein (GFP) lines on FRT-linked chromosomes. Gain-of-function (flip-out) clones were generated through spontaneous hsFLP-dependent activation of Act>CD2>GAL4.
Transgenic Lines
UAS-mCherry-Atg8a.
The Drosophila Atg8a coding region from pUAST-GFP-Atg8a (Juhász et al., 2008) was ligated as a 0.4-kb BsrGI/XbaI fragment into pUAST-mCherry (gift of Amy Kiger, University of California, San Diego, CA) that had been modified to remove a second BsrGI site.
UAS-GFP-Ref(2)P.
The Ref(2)P coding region from cDNA GH06306 was PCR amplified with KpnI and XbaI sites added at the 5′ and 3′ ends, respectively. The product was ligated into the KpnI and XbaI sites of pUAST-GFP-Atg8a (Juhász et al., 2008), replacing the Atg8a open reading frame with that of Ref(2)P.
UAS-Myc-Atg1.
The Drosophila Atg1 coding region was amplified from pUAST-Atg1 (Scott et al., 2007) using primers with BamHI and EcoRI sites and then ligated with PCR-amplified 6xMyc sequences flanked by SpeI and BglII sites into SpeI/EcoRI–digested pUAST-MCS-6xMyc (gift of M. O'Connor, University of Minnesota, Minneapolis MN), replacing the 6xMyc sequences with 6xMyc-Atg1.
UAS-Myc-Atg1-KD.
A 1.8-kb fragment containing the Myc tag and 5′ end of the Atg1 gene was excised from UAS-Myc-Atg1 and used to replace the equivalent region of UAS-Atg1-K38Q (Scott et al., 2007).
UAS-Atg13.
Drosophila Atg13 cDNA LD09558 (Drosophila Genomics Resource Center, Bloomington, IN) was digested with EcoRI and XhoI and ligated into the EcoRI/XhoI sites of pUAST.
UAS-Atg13-Flag and UAS-Atg13-GFP were generated by the Gateway System (Invitrogen, Carlsbad, CA). Atg13 coding sequences were amplified from cDNA LD09558 with flanking attB sites and then recombined into pDONOR 221 to produce the Atg13 entry clone. This entry clone was subsequently recombined into pTWF and pTWG (Drosophila Genomic Resource Center) to create UAS-Atg13-Flag and UAS-Atg13-GFP, respectively.
UAS constructs were coinjected with Δ2–3 helper plasmid into yw embryos according to standard methods.
Autophagy Induction and Detection
To induce starvation, larvae were isolated 48 h after hatching, cultured in fresh fly media under uncrowded conditions for 16–24 h and then transferred to 20% sucrose solution for 4 h before dissection. For rapamycin treatment larvae were cultured under uncrowded conditions in fly media containing 2 μM rapamycin for 24 h before dissection. LysoTracker Red (Invitrogen) staining was performed as described (Juhász and Neufeld, 2008).
Histology and Imaging
For analysis of fluorescent proteins, 10–12 larvae per genotype were bisected and inverted in PBS, fixed overnight at 4°C in 4% paraformaldehyde/PBS, washed extensively in PBS + 0.1% Triton X-100 (PBT), and counterstained with DAPI. A single section of fat body from each carcass was dissected and mounted in FluoroGuard (Bio-Rad Laboratories, Hercules, CA). Samples were prepped for immunohistochemistry by fixing and washing as above, blocking in PBT + 4% normal goat serum, and then incubating overnight in blocking solution containing 1.25 μg/ml anti-Myc mouse monoclonal 9E10 (gift of M. Stewart, North Dakota State University, Fargo, ND) or 2 μg/ml anti-Flag mouse monoclonal M5 (Sigma-Aldrich, St. Louis, MO). Rabbit anti-Ref(2)P serum (Wyers et al., 1993) was used at a dilution of 1:1000. Texas Red–conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at 1.25 μg/ml. TUNEL labeling was performed as described (Scott et al., 2007). Phalloidin (Molecular Probes, Carlsbad, CA) staining was performed as described (Hennig et al., 2006).
Confocal images were captured on a Zeiss Axioplan-2 microscope (Thornwood, NY) equipped with a CARV spinning disk system and a Hamamatsu ORCA-ER digital camera (Bridgewater, NJ), using a 63× Plan-Apochromat 1.4 NA objective lens and Axiovision software. Live images of LysoTracker Red–stained samples were obtained on a Zeiss Axioscope-2 microscope equipped with a Nikon DXM1200 digital camera (Melville, NY), using a 40× Plan-Neofluar 0.75 NA objective lens and Nikon ACT-1 software. Images were further processed and assembled into figures using Adobe Photoshop CS2 (San Jose, CA).
Immunoprecipitation and Western Blot Analysis
Larvae were collected 72–96 h after egg laying. hsGAL4-mediated expression of Atg1 and Atg13 transgenes was induced by a 2-h heat shock at 37°C, followed by a 4-h recovery either in standard fly media (fed) or 20% sucrose (starved). For immunoprecipitation of Atg1/Atg13, 90 pairs of fat bodies were lysed in 600 μl ice-cold RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% DOC, 1 mM PMSF, 10 mM NaF, 1 mM NaVO3, and complete protease inhibitor cocktail; Roche, Indianapolis, IN). Cell lysate was centrifuged to clear debris and lipid and then incubated with anti-c-Myc agarose gel (Sigma-Aldrich) at 4°C for 2 h. For immunoprecipitation of Atg1/TOR, fat bodies were lysed in 0.3% CHAPS mammalian (m)TOR extraction buffer (Kim et al., 2002). After incubation, the gel was washed with ice-cold extraction buffer and then boiled in SDS sample buffer. For phosphatase assays, 15 pairs of fat bodies were lysed in RIPA buffer or 2× CIP (calf intestinal phosphatase) buffer containing 2% NP-40 and complete protease inhibitor cocktail, EDTA-free (Roche). Cell lysate was incubated at 30°C for 30 min with or without CIP (Sigma-Aldrich), and the reaction was terminated by adding SDS sample buffer. For general Western blots, fat bodies were lysed directly in SDS sample buffer. The following antibodies were used: mouse anti-Flag M5 1:2000 (Sigma-Aldrich), mouse anti-Myc 9E10 1:2000, rabbit anti-phospho-T398 dS6K 1:250 (Cell Signaling Technology, Beverly, MA), rabbit anti-dS6K 1:1000 (gift of M. Stewart), mouse anti-α-tubulin 1:2000 (Calbiochem, Darmstadt, Germany), mouse anti-β-tubulin E7 1:1000 (Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti-phospho-Thr37/46 4EBP 1:250 (Cell Signaling Technology), rabbit anti-d4EBP 1868 1: 1000 (gift of N. Sonenberg).
RESULTS
Atg13 Plays an Essential Positive Role in Autophagy
The human KIAA0652 protein was identified as a potential Atg13 orthologue through reiterative PSI-Blast analysis of human protein datasets, using fungal Atg13 orthologues as query sequences (Meijer et al., 2007). Although the sequence identity between the human and S. cerevisiae proteins is limited to ∼15% over their carboxy-terminal halves, this region is more highly conserved among other metazoan orthologues, including 43% identity (E value, 6e-41) between human KIAA0652 and the Drosophila protein encoded by the CG7331 gene (Figure 1A), which we refer to hereafter as Atg13. To initiate a genetic analysis of Drosophila Atg13, we generated targeted deletion alleles using a P excision strategy (see Materials and Methods). Two deletions were isolated (Figure 1, B and C): Δ74, which removes 246 base pairs flanking the annotated transcription start site, and Δ81, which deletes 593 base pairs including the translation start site and the first 104 codons of Atg13. Each deletion resulted in a highly penetrant lethality at the late pupal stage that could be rescued to adulthood by expression of wild-type or epitope-tagged Atg13. The molecular nature of these alleles, the similar severity of their phenotypes, and their lack of enhancement by a deficiency for this region (unpublished data) indicate these are likely null alleles of Atg13; for simplicity we refer to cells or animals homozygous for either allele as Atg13−/−.
Figure 1.
Generation of mutations in Drosophila Atg13. (A) Amino acid sequence alignment of the C-terminal region of Atg13 orthologues from yeast, human (KIAA0652) and fly (CG7331). Identical residues are boxed. Asterisks indicate the extent of the Δ81 deletion. (B) Schematic showing the extent and orientation of the Δ74 and Δ81 deletions relative to the Atg13 transcript and the P element GS11822. The dashed line indicates sequences used to generate the inverted repeat RNAi construct 7331-R1. (C) Genomic DNA from larvae homozygous or heterozygous for Atg13 deletions Δ74 and Δ81 was analyzed by PCR using the primers indicated by arrows in B. A 1720-base pair fragment is amplified from wild-type and heterozygous samples. Animals carrying the Δ74 allele generate a smaller (1474 base pairs) product, whereas those carrying the Δ81 allele generate a 3.3-kb product, reflecting both a 2.2-kb remnant of the excised P element and a 593-base pair deletion of genomic DNA, as determined by sequence analysis.
To assess the requirement for Atg13 in autophagy, we generated clones of Atg13−/−cells in the larval fat body, which acts as a nutrient store and has a robust autophagic response to starvation. This mosaic approach allowed us to compare autophagic responses in side-by-side mutant and wild-type control cells under identical experimental conditions. Cells mutant for Atg13 failed to induce autophagy in response to starvation, as indicated by their lack of punctate localization of fluorescently tagged (mCherry)-Atg8a (Figure 2A), an Atg8/LC3 orthologue that specifically labels autophagic vesicles (Scott et al., 2004). Direct inhibition of TOR signaling by rapamycin treatment also induced autophagy in control cells but not Atg13−/− cells (Figure 2D). Atg13 mutant cells also failed to label with LysoTracker Red (Figure 2B), an acidophilic dye that tracks the expansion and acidification of the lysosomal compartment resulting from autophagosome-lysosome fusion, thus serving as an in vivo measure of flux through the autophagic pathway in this system (Juhász and Neufeld, 2008). Accordingly, we observed a time-dependent colabeling of mCherry-Atg8a with the lysosomal marker LAMP-GFP in wild-type animals but not Atg13 mutants (Supplemental Figure S1A).
Figure 2.
Atg13 is required for starvation-induced autophagy downstream of TOR. (A) Atg13 mutant cells are defective for 4-h starvation-induced autophagy. Shown is a mosaic fat body tissue comprised of clones of Atg13−/− cells (GFP-negative) and heterozygous control cells (GFP-positive); all cells express mCherry-Atg8a as a marker for autophagic vesicles (autophagosomes and autolysosomes), which appear as punctate structures in control cells. (B) Generation of acidified autolysosomes in response to 4-h starvation is defective in clones of Atg13 mutant cells (GFP-negative), as indicated by lack of LysoTracker Red staining. (C) Atg13−/− cells (mCherry-negative) accumulate high levels of transgenic GFP-Ref(2)P. (D) Atg13 mutant cells (GFP-negative) fail to induce mCherry-Atg8a labeled vesicles in response to 24-h rapamycin treatment. (E) Loss of Atg13 does not disrupt induction of autophagy in response to activation of Jnk signaling. Expression of the Jun-kinase homolog Bsk (GFP-positive cells) results in high levels of punctate LysoTracker staining in both Atg13 mutant animals (shown) and wild-type control animals (unpublished data). Scale bar in A represents 10 μm in A, C, and D and 25 μm in B and E. Genotypes: (A and D) hsFLP; Cg-GAL4/+; UAS-mChAtg8a FRT82B UAS-GFPnls/FRT82B Atg13Δ74; (B) hsFLP; Cg-GAL4/+; FRT82B UAS-GFPnls/FRT82B Atg13Δ81; (C) hsFLP; Cg-GAL4/UAS-GFP-Ref(2)P; FRT82B UAS-mCherry/FRT82B Atg13Δ81; and (E) hsFLP; Act>y+>GAL4 UAS-GFPnls/+; UAS-bsk Atg13Δ81/Atg13Δ81.
As a direct measure of autophagic degradation, we assessed the levels of Ref(2)P, a P62 orthologue previously shown to be a selective autophagy target (Lindmo et al., 2008). Levels of both endogenous Ref(2)P and transgenic GFP-Ref(2)P were markedly increased in Atg13−/− clones (Figure 2C and Supplemental Figure S1B). We observed a similar autophagy defect in cells expressing an interfering RNA against Atg13, as well as in clones of Atg1 mutant cells (Supplemental Figure S2, A–C and F). Both Atg1 and Atg13 were also required for rapamycin-induced autophagy in the neuroepithelial cells of the eye imaginal disk and for developmentally induced autophagy in the larval fat body (Supplemental Figure S2, D–E and G). In contrast, these genes were not required for induction of autophagy in response to activation of Jnk signaling (Figure 2E and Supplemental Figure S2, H–J), which activates autophagy independently of nutrient signals in this system. We conclude that Atg1 and Atg13 share a similar essential role in nutrient-dependent autophagic induction and that other signals may be sufficient to bypass this requirement.
Phosphorylation of Atg1 and Atg13 Is Regulated by Nutrient Status
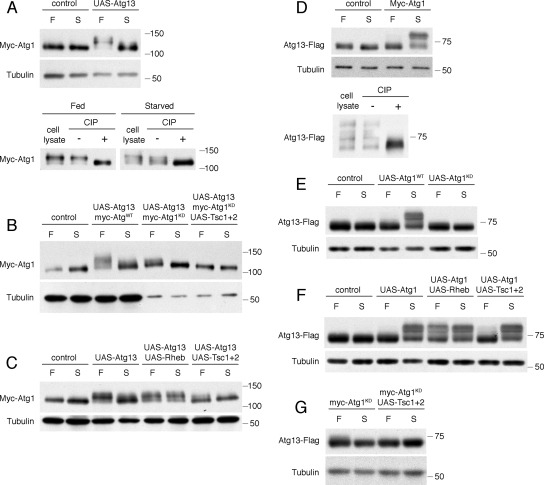
In yeast, both Atg1 and Atg13 are phosphoproteins (Matsuura et al., 1997; Kamada et al., 2000), and the level of Atg13 phosphorylation is highly sensitive to TOR signaling. We examined the effects of nutrient conditions and direct genetic manipulation of TOR activity on the phosphorylation status of epitope-tagged Atg1 and Atg13, from extracts of dissected fat body tissues. When expressed individually, Myc-Atg1 and Atg13-Flag each appeared as a single major band (Figure 3, A and D, control lanes). A faint, phosphatase-sensitive subpopulation with a slower migration rate could occasionally be observed (Supplemental Figure S3, A and D), suggesting a low level of phosphorylation of these proteins. Changes in nutrient status or TOR activity by Ras homolog enriched in brain (Rheb) or Tsc1/Tsc2 overexpression had minor effects on phosphorylation status (Supplemental Figure S3, B and E). Interestingly, starvation led to a moderate, reproducible increase in Myc-Atg1 levels (1.4-fold; p = 0.012, two-tailed Student's t test).
Figure 3.
Coexpression of Atg1 and Atg13 increases their nutrient-dependent phosphorylation. Shown are immunoblot analyses of fat body tissues dissected from Drosophila larvae expressing the indicated transgenes using the hs-GAL4 driver. (A) Expression of Atg13 increases the phosphorylation of Myc-Atg1, particularly under fed conditions. The slower form of Myc-Atg1 under either fed or starved conditions is eliminated by calf intestinal phosphatase (CIP) treatment. (B) Phosphorylation of kinase-defective Myc-Atg1 under fed conditions is less relative to wild-type Myc-Atg1. Coexpression of Tsc1 and Tsc2 further reduces Myc-Atg1KD phosphorylation. Samples expressing Myc-Atg1KD (lanes 5–8) were diluted fivefold to equalize levels of Myc-Atg1WT and Myc-Atg1KD expression. (C) Coexpression of Rheb increases the level of Myc-Atg1 phosphorylation under starvation conditions, and Tsc1/Tsc2 decreases the level of Myc-Atg1 phosphorylation under fed conditions. (D) Mobility of Atg13-Flag under starvation conditions is markedly decreased by coexpression of Myc-Atg1, because of increased phosphorylation as shown by CIP treatment. (E) Coxpression of kinase-defective Myc-Atg1 does not lead to increased Atg13-Flag phosphorylation. (F) Coexpression of Rheb increases Atg13-Flag phosphorylation under fed conditions. Overexpression of Tsc1/Tsc2 has little effect on Atg13-Flag phosphorylation. (G) Phosphorylation of Atg13-Flag is undetectable upon expression of kinase-defective Myc-Atg1 and disruption of TOR signaling by Tsc1/Tsc2 overexpression.
In contrast to the limited phosphorylation of Atg1 and Atg13 when expressed singly, we found that coexpression of these proteins had a striking effect on their migration. Under fed conditions, the majority of Myc-Atg1 migrated as a hyperphosphorylated form when coexpressed with Atg13 (Figure 3A). Under starvation conditions, Myc-Atg1 migrated at an intermediate rate. This effect of Atg13 on Myc-Atg1 phosphorylation was largely, but incompletely, reduced for the kinase-defective mutant (Figure 3B), suggesting that both autophosphorylation and exogenous phosphorylation of Myc-Atg1 is stimulated by coexpression of Atg13. Consistent with this, the phosphorylation remaining on the kinase-defective mutant was eliminated upon inhibition of TOR by overexpression of Tsc1/Tsc2 (Figure 3B). Tsc1/Tsc2 overexpression also led to a reduction in phosphorylation of wild-type Myc-Atg1, whereas activation of TOR by overexpression of Rheb increased Myc-Atg1 phosphorylation under starvation conditions, when TOR is normally less active (Figure 3C). We conclude that Atg1 is phosphorylated in a TOR- and nutrient-dependent manner. Despite its positive role in autophagy, Atg13 appears to stimulate the TOR-mediated phosphorylation of Atg1, as well as increasing Atg1 autokinase activity.
Coexpression of these proteins had an even stronger effect on the phosphorylation of Atg13-Flag, resulting in a dramatic upshift (Figure 3D). This large shift was fully reversed with phosphatase treatment and may reflect the abundance of serine and threonine residues in the Atg13 protein (18% of total). In contrast to S. cerevisiae Atg13, which is phosphorylated only under nutrient-replete conditions, hyperphosphorylation of Drosophila Atg13-Flag was strongest in starved animals, and required Atg1 kinase activity (Figure 3E). Atg13-Flag phosphorylation was also observed under fed conditions in extracts from later-stage larvae, which have higher basal levels of TOR activity (Lee et al., 2007), and this was inhibited by Tsc1/Tsc2 overexpression (Supplemental Figure S3C). Accordingly, overexpression of Rheb resulted in high levels of Atg13-Flag phosphorylation under both fed and starved conditions (Figure 3F). As observed for Atg1, Atg13-Flag phosphorylation was undetectable when both TOR and Atg1 activity was inhibited (Figure 3G). Thus, Atg13 is phosphorylated in a TOR and Atg1 kinase-dependent manner under fed conditions and in an Atg1-dependent manner under starvation conditions. Atg13 may be direct substrates for these kinases in vivo; alternatively, the activity of one kinase may indirectly facilitate increased phosphorylation of Atg13 by another.
Physical and Genetic Interactions between Atg1 and Atg13
In yeast, phosphorylation of Atg13 regulates its association with Atg1, such that loss of TOR signaling leads to dephosphorylation of Atg13 and increases its affinity for Atg1. Drosophila Atg13 was previously found to interact directly with Atg1 in a genomic-scale 2-hybrid screen, despite an apparent lack of conservation of the Atg1 interaction domain as defined in S. cerevisiae (residues 432-520 of yeast Atg13; Kamada et al., 2000). We therefore tested for in vivo interactions between Atg1 and Atg13 in response to nutrient signaling. Immunoprecipitation of Myc-Atg1 from extracts of dissected fat body tissues consistently led to coprecipitation of Atg13-Flag (Figure 4A). This association was dependent on expression of Myc-Atg1 and was observed under both fed and starved conditions. Relative to the abundance of Myc-Atg1, which was higher in starved samples, starvation resulted in a 3.9-fold increase in the amount of coprecipitated Atg13-Flag (p = 0.006, two-tailed Student's t test). Overall, the level of immunoprecipitable Atg13-Flag increased ∼10-fold under starvation conditions, presumably reflecting increases in both Myc-Atg1 protein levels and its affinity for Atg13-Flag. Interestingly, hyper- and hypo-phosphorylated forms of Atg13-Flag interacted with Myc-Atg1 to a comparable extent. Atg13-Flag also associated with kinase-defective Myc-Atg1, indicating that the lack of Atg13 phosphorylation in animals expressing Atg1KD (Figure 3E) is not due to a lack of Atg1-Atg13 association. We conclude that the physical association between Atg1 and Atg13 is conserved between yeast and fly, but the mechanisms of regulation appear to differ. While nutrient- and TOR-dependent phosphorylation of Atg13 prevents formation of the active Atg1/Atg13 complex in S. cerevisiae, Drosophila Atg13 is most highly phosphorylated during starvation and associates with Atg1 under both autophagy-conducive and -prohibitive conditions. Our results suggest that phosphorylation-dependent regulation of Atg1-Atg13 complex formation is not likely to account for autophagy induction in this system. These differences may in part reflect higher levels of basal autophagy in metazoans.
Figure 4.
Biochemical and genetic interactions between Atg1 and Atg13. (A) Atg1 and Atg13 associate under both fed and starved conditions. Fat body extracts from larvae expressing Atg13-Flag ± Myc-Atg1 using the hs-GAL4 driver were immunoprecipitated with anti-Myc beads. Both hyper- and hypo-phosphorylated forms of Atg13 are coprecipitated with Myc-Atg1, and starvation increases the interaction. Atg13 is also coprecipitated with kinase defective Myc-Atg1, which expresses at a substantially higher level than wild-type Myc-Atg1. (B–D) Atg13 stimulates the autophagic activity of Myc-Atg1. Representative images of fat body tissues expressing mCherry-Atg8a and indicated transgenes in response to hs-GAL4, 24 h after 1-h heat shock. Under fed conditions, mCherry-Atg8a is diffuse in controls (B) and forms small, perinuclear autophagosomes in cells expressing Myc-Atg1 (C). Coexpression of Myc-Atg1 and Atg13 at low levels (D) leads to a strong induction of autophagosome formation. (E–G) Myc-Atg1 and Atg13 cooperate to suppress cell growth. Expression of either Myc-Atg1 (E) or Atg13 (F) in GFP-marked fat body cell clones does not have an appreciable effect on cell size. Cells coexpressing Myc-Atg1 and Atg13 are markedly reduced in size relative to neighboring control (GFP-negative) cells (G). Cell boundaries are highlighted by Alexa Fluor 555 phalloidin staining in red. (H–K) Expression of either Myc-Atg1 or Atg13 in the developing eye using the GMR-GAL4 driver has no discernable effect on eye development (I and J), whereas coexpression leads to a disruption of ommatidial patterning (K). (L) Constitutive expression of Myc-Atg1 or Atg13 throughout the larval fat body using the Cg-GAL4 driver does not affect larval growth or developmental timing. Coexpression of these transgenes leads to a growth and developmental arrest at the first instar stage of development. (M) Atg13 is required for induction of autophagy by Atg1 overexpression under fed conditions. Cg-GAL4-driven expression of Myc-Atg1 induces punctate localization of mCherry-Atg8a in GFP-positive control cells, but not in clones of cells mutant for Atg13, marked by lack of GFP. Scale bar in B represents 10 μm in B–G and M, 100 μm in H–K, and 700 μm in L. Genotypes: (B) hs-GAL4 UAS-mCherry-Atg8a/+; (C) hs-GAL4 UAS-mCherry-Atg8a/UAS-Myc-Atg1; (D) UAS-Atg13/+; hs-GAL4 UAS-mCherry-Atg8a/UAS-Myc-Atg1; (E) hsFLP; Act>CD2>GAL4 UAS-GFP/UAS-Myc-Atg1; (F) hsFLP; UAS-Atg13/+; Act>CD2>GAL4 UAS-GFP/+; (G) hsFLP; UAS-Atg13/+; Act>CD2>GAL4 UAS-GFP/UAS-Myc-Atg1; (H) GMR-GAL4/+; (I) GMR-GAL4/+; UAS-Myc-Atg1/+; (J) GMR-GAL4/UAS-Atg13; (K) GMR-GAL4/UAS-Atg13; UAS-Myc-Atg1/+; (L) left to right: Cg-GAL4/+, Cg-GAL4/+; UAS-Myc-Atg1/+, Cg-GAL4/UAS-Atg13, Cg-GAL4/UAS-Atg13; UAS-Myc-Atg1/+; and (M) hsFLP/UAS-Myc-Atg1; Cg-GAL4/+; UAS-mCherry-Atg8a FRT82B UAS-GFPnls/FRT82B Atg13Δ74.
To ask if these biochemical interactions reflect relevant physiological effects in vivo, we tested whether Atg13 affected Atg1-dependent phenotypes. We previously showed that high levels of UAS-regulated Atg1 can override fed signals to induce autophagy (Scott et al., 2007). We found that our UAS-Myc-Atg1 lines expressed at substantially lower levels than these previously published untagged Atg1 lines (Supplemental Figure S3F). Expression of Myc-Atg1 at low levels using hs-GAL4 resulted in a weak induction of autophagy: few, small autophagosomes were observed, mostly confined to the perinuclear region (Figure 4, B and C). Coexpression of Atg13 resulted in a marked increase in the number, size, and distribution of autophagosomes (Figure 4D). Similar effects were observed using another weakly expressed, untagged Atg1 line, monitoring autophagy with LysoTracker (Supplemental Figure S2, K and L). We also found that in clones of cells mutant for Atg13 autophagy was no longer induced by high levels of Atg1 (Figure 4M). Thus, Atg13 is required for and stimulates the autophagy-promoting function of Atg1.
We next tested the effects of Atg13 on additional Atg1-dependent phenotypes. Strong overexpression of Atg1 was previously shown to reduce cell size, inhibit larval growth, and disrupt the morphology of the compound eye (Scott et al., 2007). Neither Myc-Atg1 nor Atg13 on their own had any effects in these assays, but coexpression of these proteins resulted in phenotypes similar to those caused by high levels of native Atg1 overexpression (Figure 4, E–L). Atg13 was also required for induction of apoptosis caused by overexpression of Atg1 (Supplemental Figure S4, A and B). In contrast to these effects on Atg1-driven autophagy and apoptosis, loss of Atg13 did not suppress the cell size reduction and developmental arrest caused by high Atg1 levels (Supplemental Figure S4, C and D), suggesting that sufficient levels of Atg1 overexpression may override the requirement for Atg13 in some cases or that Atg1 may possess Atg13-independent functions.
Finally, we examined genetic interactions between Atg1 and Atg13 loss-of-function mutants. Animals homozygous for either mutation survive to the late pupal stage of development; in contrast, Atg1−/− Atg13−/− double mutant animals showed a fully penetrant embryonic lethality (0/200 hatched embryos). Interestingly, hsGAL4-driven expression of Myc-Atg1 was sufficient to partially rescue the lethality, but not the autophagy defect, of Atg13−/− animals (Atg13−/− + UAS-Atg13-Flag: 63.6% survival to adulthood, n = 223; Atg13−/− + UAS-Myc-Atg1: 30.0% survival to adulthood, n = 198). This suggests that the lethality of Atg13 mutants is autophagy-independent, consistent with the viability of animals mutant for other Drosophila autophagy-related genes such as Atg7 and Atg8a (Juhász et al., 2007; Scott et al., 2007). Taken together, the extensive genetic interactions between Atg1 and Atg13 support the conclusion that Atg13 functions cooperatively with Atg1 to promote its activity.
Atg13 Possesses an Activity Inhibitory for Autophagy
GAL4 lines that express uniformly in most tissues often cause uneven, mottled expression of transgenes in the fat body (Juhász and Neufeld, 2008). We exploited this feature of hs-GAL4 to compare the effects of different levels of UAS-regulated Myc-Atg1/Atg13 expression within the same tissue. We noticed that although weak coexpression of Myc-Atg1 and Atg13 caused induction of autophagy under fed conditions, this was not observed in cells with higher levels of expression (Figure 4D). Instead, such cells were resistant to induction of autophagy by starvation (unpublished data). We found that expression of Atg13 alone was sufficient for this inhibitory effect (Figure 5, A–C). Expression of wild-type, Flag- or GFP-tagged versions of Atg13 gave similar results.
Figure 5.
Inhibition of autophagy by Atg13 overexpression. All panels depict fat body tissues from 4-h starved animals, unless otherwise indicated. (A and B) Starvation induces induction of mCherry-Atg8a–positive autophagosomes in controls (A) but not in animals overexpressing Atg13 (B). (C) Clonal expression of Atg13-GFP cell autonomously disrupts accumulation of LysoTracker Red—labeled autolysosomes. (D and E) Distribution of Atg13-GFP changes from a diffuse pattern under fed conditions (D) to small, uniform punctate structures after 4-h starvation (E). Atg13-GFP punctae (boxed area in E, enlarged in E′) are smaller than mCherry-Atg8a labeled autophagosomes and autolysosomes (boxed area in A, enlarged in A′). (F) In cells with high levels of Atg13-GFP expression, most Atg13-GFP–positive structures fail to label with mCherry-Atg8a. Yellow circles indicate examples of colocalization. (G) Atg13-GFP localizes to mCherry-Atg8a–labeled autophagosomes in cells with low levels of Myc-Atg1 and Atg13-GFP coexpression. Yellow circles indicate examples of colocalization. (H) Loss of Atg13 leads to increased levels of Myc-Atg1. A two-cell clone of Atg13 mutant cells (marked by lack of GFP) contains higher levels of Myc-Atg1 (red) than in neighboring control cells. (I and J) Starvation-induced Atg13-GFP redistribution is blocked by coexpression of Rheb (I) but is independent of Atg1 (J). Atg13-GFP is localized to punctate structures in both control cells and in cell clones homozygous mutant for Atg1 (central cell in J). Scalebar in A represents 10 μm in all panels, except A′ and E′ 5 μm, in C 38 μm, and in H 20 μm. Genotypes: (A) hsFLP/+; Act>CD2>GAL4 UAS-mCherry-Atg8a/+; (B–E) hsFLP/+; UAS-Atg13-GFP/+; Act>CD2>GAL4 UAS-mCherry-Atg8a/+; (F) UAS-Atg13-GFP/+; hsGAL4 UAS-mCherry-Atg8a/+; (G) UAS-Atg13-GFP/+; hsGAL4 UAS-mCherry-Atg8a/UAS-Myc-Atg1; (H) hsFLP/UAS-Myc-Atg1; Cg-GAL4/+; UAS-mCherry-Atg8a FRT82B UAS-GFPnls/FRT82B Atg13Δ74; (I) hsFLP/+; UAS-Atg13-GFP/+; Act>CD2>GAL4 UAS-GFPnls UAS-RhebAV4; and (J) hsFLP/+; UAS-Atg13-GFP/Cg-GAL4; FRT82B UAS-myrRFP/Atg1Δ3D FRT80B.
To further investigate these effects, we examined the localization of GFP-tagged Atg13. Despite inhibiting autophagosome and autolysosome formation, Atg13-GFP was redistributed from a diffuse localization under fed conditions to punctate structures in response to starvation (Figure 5, D–E). These structures were considerably smaller and more uniform in size than typical autophagosomes and tended to form in clusters of two to five spots (Figure 5, E′ and A′). A fraction of the Atg13-GFP punctae colabeled with mCherry-Atg8a, suggesting they are autophagy-related vesicles (Figure 5F). Consistent with this, when coexpressed at low levels with Myc-Atg1, nearly all Atg13-GFP punctae colocalized with mCherry-Atg8a to autophagosomes (Figure 5G). The finding that high levels of Atg13-GFP lead to small vesicles that are mostly devoid of mCherry-Atg8a suggests that Atg13 overexpression may suppress recruitment of Atg8, which is required for autophagosome expansion (Xie et al., 2008). This may reflect the ability of Atg13 to stimulate TOR-dependent phosphorylation of Atg1 (Figure 3C) or may indicate a dominant-negative effect of high levels of Atg13 on Atg1 complex formation. In addition, Atg13 also appears to reduce Atg1 expression, as we found that Myc-Atg1 levels were increased in Atg13 mutant clones (Figure 5H). These effects of Atg13 are likely on the stability of Atg1, as the UAS-Myc-Atg1 transgene is regulated by exogenous transcriptional and translational control sequences.
Induction of Atg13-GFP punctae by starvation was blocked by overexpression of Rheb or mutation of Tsc1 (Figure 5I and unpublished data), indicating this is a TOR-dependent response. In contrast, Atg13-GFP punctae formation did not require Atg1 (Figure 5J). These data suggest that recruitment of Atg13 to punctate structures represents an early step in autophagosome induction, perhaps analogous to the early role played by Atg13 in yeast in recruiting other Atg proteins to the PAS.
Together, these results indicate that Atg13 can play dual roles in both activating and inhibiting autophagy. The positive function of Atg13 in autophagy induction is dominant over its negative role, as loss of Atg13 results in an autophagy block.
Atg1 and Atg13 Cooperate to Inhibit TOR Signaling through Effects on Its Trafficking
In both Drosophila and mammalian cells, Atg1 has been shown to antagonize TOR signaling, resulting in a self-reinforcing inhibitory feedback loop (Lee et al., 2007; Scott et al., 2007). We find that this signal also involves Atg13, as coexpression of Atg13 increased the inhibition of TOR by Myc-Atg1 (Figure 6, A and B). TOR signaling has recently been shown to be regulated in response to nutrient levels through effects on its trafficking between vesicular, perinuclear, and endocytic compartments of the cell (Sancak et al., 2008). Furthermore, Atg1 regulates the trafficking of Atg9 and Atg23 in yeast and mammalian cells (Reggiori et al., 2004; Young et al., 2006), and interacts with components of the endocytic pathway (Tomoda et al., 2004).
Figure 6.
Reciprocal inhibitory interactions between Atg1/Atg13 and TOR. (A and B) Suppression of TOR activity by Myc-Atg1 and Atg13. Fat body extracts from late third instar animals expressing Myc-Atg1 and/or Atg13 with hs-GAL4 were assayed for phosphorylation of endogenous dS6K. (A) Phosphorylation of dS6K Thr398 is moderately inhibited by Myc-Atg1 expression, unaffected by Atg13 expression, and strongly reduced by coexpression of Myc-Atg1 and Atg13. Levels of endogenous d4EBP, which is transcriptionally inhibited by TOR signaling, rise to detectable levels in response to Myc-Atg1 and Atg13 expression (A). (B) Phosphorylation of Thr37/46 of overexpressed d4EBP is inhibited in response to Atg1 overexpression. (C–E) Disruption of TOR trafficking by Myc-Atg1 and Atg13. hs-GAL4-driven Flag-TOR is localized to a perinuclear compartment in control fat body cells 24 h after induction (C). Coexpression of low levels of Myc-Atg1 and Atg13 leads to accumulation of Flag-TOR in punctate/vesicular structures (D). Constitutively expressed Flag-TOR (E) localizes to the surface of vesicles and punctate structures in control cells (GFP-positive in E′). These structures and overall levels of Flag-TOR are reduced in clones of cells mutant for Atg13 (GFP-negative cells in E′). (F and G) TOR activity inhibits Myc-Atg1 accumulation. Myc-Atg1 was uniformly expressed in all fat body cells using Cg-GAL4, and its levels in clones of Tsc1 mutant cells (F) and Rheb mutant cells (G) were compared with that in neighboring control cells (GFP positive cells in F′ and G′). Loss of Tsc1 reduces and loss of Rheb increases Myc-Atg1 levels. Scale bars, (C–G) 10 μm. (H) Coimmunoprecipitation of Atg1 and TOR from fed and starved animals. Fat body extracts from larvae expressing hs-GAL4-driven Flag-TOR alone or Flag-TOR and Myc-Atg1 together were immunoprecipitated with Anti-Myc beads. (I) Hs-GAL4-driven coexpression of Flag-TOR stimulates phosphorylation of Myc-Atg1. Genotypes: (C) hsFLP/+; UAS-Flag-dTORF4A/+; Act>CD2>GAL4; (D) hsFLP/+; UAS-FlagTORF4A/UAS-Atg13; Act>CD2>GAL4/UAS-Myc-Atg1; (E) hsFLP/+; UAS-FlagTORF4A/+; r4-GAL4 FRT82B UAS-GFPnls/FRT82B Atg13Δ81; (F) hsFLP/UAS-MycAtg1; Cg-GAL4/+; FRT82B UAS-GFPnls/FRT82B Tsc129; (G) hsFLP/UAS-MycAtg1; Cg-GAL4/+; FRT82B UAS-GFPnls/FRT82B Rheb2D1.
We therefore asked whether Atg1/Atg13 might influence TOR signaling through effects on its intracellular trafficking. To do so, we examined the localization of Flag-tagged TOR in response to altered levels of Atg1 and Atg13. In control animals, Flag-TOR was found predominantly in a diffuse, perinuclear pattern at short time points (<24 h) after induction (Figure 6C) and in a more vesiculated pattern at later times or in constitutively expressing lines (Figure 6E), as previously described (Hennig et al., 2006). Manipulation of Atg1 and Atg13 levels had a dramatic effect on this localization. In cells coexpressing MycAtg1 and Atg13, the diffuse perinuclear expression of Flag-TOR was disrupted; Flag-TOR localized instead to the surface of discrete vesicles, still concentrated in the perinuclear region of the cell (Figure 6D). Expression of high levels of Atg1 alone had a similar effect (unpublished data). Conversely, cells mutant for Atg13 displayed a less vesicular localization of constitutively expressed Flag-TOR, as well as a reduction in overall levels of the protein (Figure 6E). Myc-Atg1 and Atg13-Flag levels were also strongly affected by TOR function, reduced in cells with high TOR activity (mutant for Tsc1) and increased in cells with low TOR activity (mutant for Rheb), consistent with the negative effects of TOR signaling on autophagy (Figure 6, F and G, and unpublished data). The control of these transgenes by exogenous transcriptional and translational regulatory elements again indicates that these effects are likely posttranslational. We conclude that TOR may regulate autophagy in part by controlling the stability of Atg1 and Atg13, in addition to its effects on Atg1 activity. In turn, Atg1-Atg13 complexes regulate TOR in part by influencing its intracellular distribution and trafficking.
Given these reciprocal effects on the activity, levels and localization between TOR and Atg1, we asked whether these proteins interact physically. Immunoprecipitation with Myc antibodies reproducibly led to coprecipitation of Flag-TOR from fat body extracts of animals coexpressing Myc-Atg1 and Flag-TOR, but not from animals expressing Flag-TOR alone (Figure 6H), indicating that these proteins can associate in vivo. This interaction was observed in extracts from both fed and starved animals and did not show a consistent response to changes in nutrient conditions. In addition, coexpression of Flag-TOR increased the phosphorylation of Myc-Atg1 (Figure 6I), consistent with a close association of these proteins. We conclude that the extensive interactions between TOR and Atg1 are mediated in part by their incorporation into a common complex.
DISCUSSION
The remarkable conservation of autophagy-related proteins and processes between yeast and higher eukaryotes has contributed to a rapid advance in our understanding of this process in multicellular animals. The data presented here extend this conservation to the mechanisms of autophagy regulation by nutrient-dependent TOR signaling. As in yeast, Drosophila Atg13 forms a complex with Atg1, stimulates its activity and is highly phosphorylated in a nutrient and TOR-dependent manner. Although the relevant substrates of Atg1 remain to be identified, these results suggest that this fundamental pathway from nutrient signal to autophagic induction has been largely retained between yeast and metazoans. This regulatory link between TOR and Atg1/Atg13 thus represents one of the most highly conserved TOR outputs described to date.
Despite this conservation, we also identify critical differences in the behaviors of these proteins, which likely stem in part from differences in the physiology of lower versus higher eukaryotes. While this manuscript was under review, Chan et al. (2008) reported an essential role in autophagy for human Atg13 using RNAi-mediated knockdown in cultured cell lines and described interactions between Atg13 and Ulk1 and Ulk2 similar to those reported here. In this issue of Molecular Biology of the Cell, Hosokawa et al. (2009) and Jung et al. (2009) also describe related interactions between mammalian Atg13 and Ulk kinases. Together, these studies point to a model whereby Atg13 is phosphorylated and interacts with Atg1 under both fed and starved conditions in metazoans, in contrast to the growth-dependent phosphorylation and dissociation of Atg13-Atg1 observed in yeast. Although the greater nutrient buffering capacity of multi- versus unicellular animals probably contributes to these differences, the requirement for significant basal rates of autophagy in the maintenance of large, long-lived metazoan cells may dictate that the Atg1-Atg13 complex remain partially active even under fed conditions. Furthermore, although Atg1 can clearly associate with phosphorylated Atg13, dephosphorylation of TOR-dependent sites on Atg13 may be masked by increased phosphorylation by Atg1, but may nonetheless contribute to regulation of Atg1-Atg13 interaction or activity. The observation that Atg1 kinase activity plays a role under both fed and starved conditions in supporting hyperphosphorylation of Atg13 (Supplemental Figure S3C) suggests that rather than switching Atg1 kinase between off and on states, nutrient signals may instead affect its substrate specificity or accessibility.
Metazoan Atg1-Atg13 complexes have also accrued additional regulatory mechanisms that have not been described in yeast, including negative feedback from Atg1 to TOR, phosphorylation of Atg13 by Atg1, and nutrient-dependent effects on the stability of these proteins. These additional layers of regulation are in keeping with the elaboration of intracellular signaling pathways in metazoans. The observation that Atg13 phosphorylation is both TOR- and Atg1-dependent under fed conditions suggests a model whereby phosphorylation by one of these kinases may serve as a priming event for the other. In addition, the role of Atg1-dependent phosphorylation of Atg13 under starvation conditions remains an important question. Although the Atg1-independent localization of Atg13 to autophagosomes and its activation of Atg1 indicate that Atg13 functions upstream of Atg1, the finding that Atg13 acts as a substrate for Atg1-dependent phosphorylation raises the possibility that Atg13 may also act to transduce signals downstream of Atg1. Identification of additional Atg1 substrates and Atg13 interacting proteins may help clarify this issue.
Given its positive role in autophagy induction, the ability of Atg13 to inhibit autophagy when overexpressed was unexpected. This may in part reflect a dominant negative effect of Atg13 overexpression causing titration of Atg1 complexes or competition with other Atg1 substrates. However, the observation that Atg13 stimulates TOR-dependent phosphorylation of Atg1 and that Atg1 levels increase in Atg13 mutant cells suggests that Atg13 has both positive and negative roles in autophagy induction. These opposing activities of Atg13 are reminiscent of the mTOR complex I component Raptor, which plays an essential role in TOR signaling yet also inhibits TOR activity under starvation conditions (Kim et al., 2002). Our results suggest that Atg13 may play an analogous role, switching between states of promoting or inhibiting autophagy, thereby sharpening the response to changes in nutrient conditions. These findings suggest that the relative ratio of Atg13 to Atg1 or to other components of the complex may play an important role in dictating its activity. In this regard, the differential regulation of Atg1 and Atg13 levels by TOR could provide an additional mechanism whereby TOR signaling can influence autophagic activity. Although the autophagy-defective phenotypes of Atg1 and Atg13 mutants reveals the dominant positive roles of these genes, disruption of other putative components of this complex leads to autophagy induction (unpublished data), suggesting some components may play primarily negative roles.
In conclusion, our results demonstrate that Atg1-Atg13 complexes play an essential, conserved role in promoting autophagy in response to TOR inactivation, with additional regulatory functions unique to metazoans. Further insight into the regulation of this complex and identification of its targets may lead to additional means of manipulating autophagy rates for therapeutic purposes.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the contribution of Charles Nelson in isolating Atg13 deletion alleles and Laura Muller for generation of UAS-mCherry-Atg8a and UAS-GFP-Ref(2)P lines. We thank Harald Stenmark (Norwegian Radium Hospital, Oslo, Norway) and Didier Contamine (Université Versailles, France) for antibodies and Noboru Mizushima and Do-Hyung Kim for communicating results before publication. This work was supported by National Institutes of Health Grant RO1 GM62509 to T.P.N.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-12-1250) on February 18, 2009.
REFERENCES
- Chan E. Y., Kir S., Tooze S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domain using an Atg13-independent mechanism. Mol. Cell. Biol. 2008;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura A., Noda T., Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig K. M., Colombani J., Neufeld T. P. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J. Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig K. M., Neufeld T. P. Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis. 2002;34:107–110. doi: 10.1002/gene.10139. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., et al. Nutrient-dependent mTORC1 association with ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Erdi B., Sass M., Neufeld T. P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J. H., Yang Y., Sass M., Baehrecke E. H., Backer J. M., Neufeld T. P. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Neufeld T. P. Experimental control and characterization of autophagy in Drosophila. Methods Mol. Biol. 2008;445:125–133. doi: 10.1007/978-1-59745-157-4_8. [DOI] [PubMed] [Google Scholar]
- Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Kabeya Y., Sekito T., Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kundu M., Lindsten T., Yang C. Y., Wu J., Zhao F., Zhang J., Selak M. A., Ney P. A., Thompson C. B. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Kim S., Lee J., Park J., Lee G., Kim Y., Kim J. M., Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–365. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K., Brech A., Finley K. D., Gaumer S., Contamine D., Rusten T. E., Stenmark H. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4:500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada M., Wada Y., Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Meijer W. H., van der Klei I. J., Veenhuis M., Kiel J. A. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Espert L., Biard-Piechaczyk M., Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Juhász G., Neufeld T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Tomoda T., Kim J. H., Zhan C., Hatten M. E. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18:541–558. doi: 10.1101/gad.1151204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Dru P., Simonet B., Contamine D. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J. Virol. 1993;67:3208–3216. doi: 10.1128/jvi.67.6.3208-3216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A. R., Chan E. Y., Hu X. W., Kochl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.