Abstract
The manifestation of Lewy bodies (LB) in the brain is a hallmark of Parkinson’s disease. Here, we present a comprehensive analysis of protein elements in Lewy bodies by comparative mass spectrometry. Cortical LB inclusions were enriched by sucrose gradient centrifugation from postmortem brains, and a negative control sample was prepared from specimen without LB pathology. Whereas ~550 proteins were identified in the LB-enriched sample by mass spectrometry, quantitative comparison with the control sample revealed that ~40 proteins were co-enriched with α-synuclein, the major component in Lewy bodies. As expected, the list of proteins included previously reported constituents, such as those involved in protein folding, membrane trafficking and oxidative stress. More interestingly, we discovered in the LB-enriched sample several kinases (MAPKK1/MEK1, protein kinase C, and doublecortin-like kinase), a novel deubiquitinating enzyme (otubain 1), and numerous ubiquitin ligases (KPC and SCF). The proteomic studies provide enzyme candidates to investigate the regulation of α-synuclein and/or other LB proteins, which may contribute to the formation of Lewy bodies and the toxicity of α-synuclein in the related neurodegenerative disorders.
Keywords: Lewy body, Parkinson’s disease, dementia with Lewy bodies, LC-MS/MS, proteomics
2. INTRODUCTION
Parkinson’s disease (PD) is characterized by selective degeneration of dopaminergic neurons of the substantia nigra in the brain, and by neuronal inclusions called Lewy bodies - proteinaceous aggregates primarily composed of α-synuclein (1, 2). Lewy bodies have also been found in cases of dementia with Lewy bodies (DLB), the Lewy body variant of Alzheimer’s disease, Down’s syndrome, and several other neurological diseases, collectively under the common denomination of synucleinopathies (3). α-synuclein in Lewy bodies has been found to undergo a number of posttranslational modifications including phosphorylation (4), ubiquitination (5, 6) and oxidative nitration (7). These modifications are proposed to modulate the equilibrium among multiple forms of α-synuclein (i.e. monomer, soluble oligomers and aggregates) and the assembly of Lewy bodies, which may eventually modulate neuronal toxicity of α-synuclein, although it is arguable whether the toxicity stems from the soluble forms or the aggregates. Identification of proteins associated with Lewy bodies may offer molecular clues to underlying mechanisms.
Traditionally, LB-associated components have been identified by immunohistochemistry. This approach relies on speculation or a priori knowledge of proteins expected to be present in LB and the availability of specific antibodies to the protein of interest. Recent advances in mass spectrometry-based proteomics approaches provide an opportunity to directly sequence proteins from minute amounts of isolated Lewy bodies, as hundreds of proteins can be identified from a mixture containing a few micrograms of proteins (8, 9). Utilizing this unbiased method, we identified approximately 550 proteins in a LB-enriched fraction, in which potential LB proteins were distinguished from unrelated contaminants by quantitative comparison with a control sample containing no LBs. The statistical significance was evaluated by G-test and probability values, resulting in approximately 40 proteins thought to be enriched in LBs, including a number of novel kinases and ubiquitin enzymes. Their potential involvement in the formation of LBs and the pathogenesis of neurodegeneration are discussed.
3. MATERIALS AND METHODS
3.1. Enrichment of Lewy bodies from postmortem brains
Lewy bodies were purified from brain tissues of patients diagnosed with Lewy body variant of Alzheimer’s disease, whereas cases without LB pathology were used as negative controls, according to a previously reported protocol (10). Cortical tissues from both cases were dissected to remove white matter and then homogenized in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 M sucrose, 5 mM EDTA, 2 mM DTT and protease inhibitors, 10 ml of the buffer for 1 g of tissue). The lysates were then centrifuged at 100,000 × g for 30 min, and the pellets were resuspended in the lysis buffer and loaded on a sucrose gradient composed of three layers (2.2 M, 1.5 M and 1.2 M). After 200,000 × g centrifugation for 2.5 hours, all interfaces were collected. An aliquot of each collected sample was smeared onto a glass slide and examined by immunostaining with α-synuclein antibodies. In the LB cases, the fractions expected to contain LBs were enriched for round, α-synuclein-immunoreactive structures ranging in diameter from 5 μm to 30 μm; these structures were absent in the corresponding fractions from the control cases.
3.2. Protein identification by mass spectrometry
A pair of LB-enriched and control fractions were dissolved in a gel loading buffer containing 2% SDS at 95 °C for 5 min, and separated on an SDS gel followed by Coomassie staining. The whole gel lane of each sample was excised into 10 bands and in-gel digested. The resulting peptides were analyzed by reverse-phase liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (11). Briefly, the peptide mixtures were dissolved in Buffer A (0.4% acetic acid, 0.005% heptafluorobutyric acid, and 5% acetonitrile), loaded onto a 75-μm-inner diameter self-packed column (10 cm of 5-μm resin), eluted with a gradient of Buffer B (0.4% acetic C18 acid, 0.005% heptafluorobutyric acid, and 95% acetonitrile). The eluates were monitored in a MS survey scan followed by three data-dependent MS/MS scans on an LCQ-Deca XP-Plus ion trap mass spectrometer (Thermo Finnigan).
The acquired MS/MS spectra were searched against the human reference database of the National Center for Biotechnology Information using the SEQUEST algorithm (12), with the following parameters: parent ion mass tolerance (3 Da), oxidized methionine (+16 Da) and carboxymethylated cysteine (+57 Da). To remove false positives caused by random matching or spectra of poor quality, we adopted the target-decoy strategy (13, 14) to evaluate false discovery rate. Peptide matches were classified according to trypticity (fully, partial and non-tryptic) as well as precursor ion-charge state (1+, 2+, and 3+), and then filtered by XCorr and ΔCn values to reduce the false discovery rate to near zero. Basically, the filtering cutoffs were adjusted to discard all peptide matches from the decoy database. According to the rule of parsimony, all accepted proteins sharing peptides were grouped together, in which only the top protein with highest spectral counts was selected to represent the group.
3.3. Quantitation by spectral counts: p-value based on G-test and χ2 distribution
Among the total ~700 proteins identified in the LB and/or control fractions, numerous proteins were only identified in only one sample. A common practice is to assign the undetected proteins with an arbitrarily low number of spectral counts (i.e, 0.5) with the reasoning that those proteins might display the most dramatic changes in abundance.
The G-test (15) is a likelihood ratio test for discrete frequency data, and has been used to assess protein abundance based on spectral counts assuming a binomial distribution (16, 17). According to the null hypothesis, the expected spectral count is set to be equal to the average spectral count of the two samples:
| (1) |
where flb and f̂lb are the detected and expected spectral counts of a given protein in the LB fraction, respectively; similarly, fct and f̂ct are the corresponding values in the control sample. The G value of each protein is then calculated as
| (2) |
By combining the above two equations, the following equation is used to derive the G value.
| (3) |
Although theoretical distribution of the G values is complex, the distribution of the values approximately follows the χ2 distribution with one degree of freedom, and therefore allows the computation of p-value (15). The p-value is the probability of observing a random variable larger than G in the χ2 distribution with one degree of freedom.
4. RESULTS
4.1. Biochemical isolation and proteomics analysis of Lewy bodies from postmortem brains
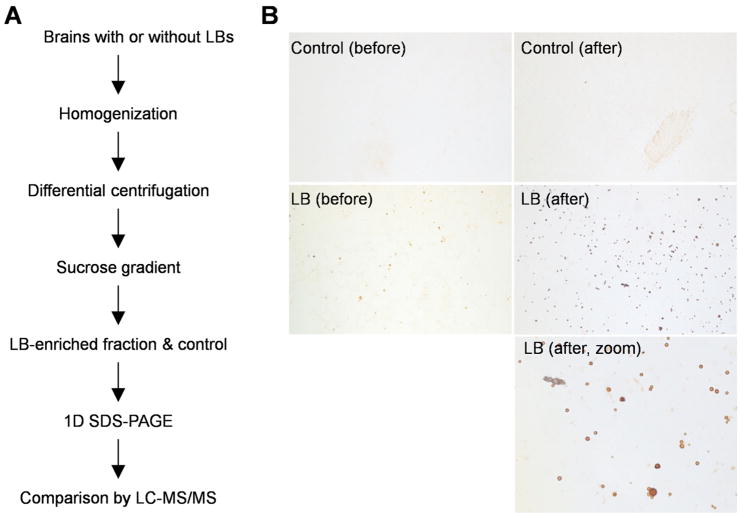
The purification and mass spectrometric analysis of Lewy bodies were achieved by sucrose density centrifugation and LC-MS/MS as illustrated in Figure 1A. Postmortem brain tissues with or without LBs were homogenized in the presence of 1M sucrose beneficial for the separation of myelin-rich layer during centrifugation. The LB-containing pellet was further resolved in a discontinuous sucrose gradient, and the resulting fractions were examined by immunostaining with α-synuclein antibodies to validate LB enrichment (Figure 1B). Judged by the number of LBs detected microscopically, the enrichment factor varied among samples in repeated analyses, and the LB inclusions were spread into different interfaces in the sucrose gradient, suggesting that the LBs are heterogeneous with respect to their densities (data not shown). For large-scale protein identification, we selected one pair of LB and control samples that were collected from the same interface of two cases, respectively. In the LB sample, LB particles were concentrated approximately 7-fold by the sucrose gradient (Figure 1B). By contrast, there were no LB inclusions detected in the control sample before and after the step of sucrose gradient.
Figure 1.
Preparation of the LB-enriched sample. A, flow chart for the experimental design. Brain tissues with or without LB were lysed, differentially centrifuged, and subsequently applied on sucrose gradient. LB-enriched fractions were collected and analyzed by geLC-MS/MS. B, immunostaining results showing the LBs before and after biochemical enrichment.
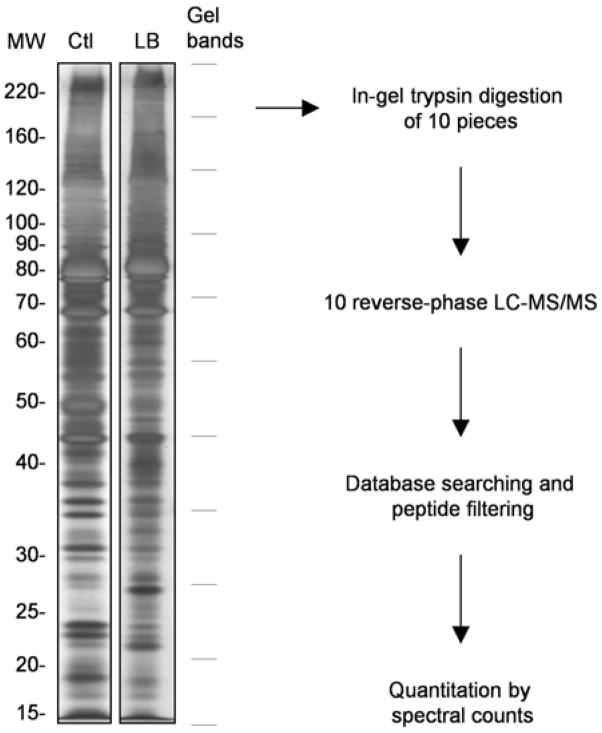
To identify novel components in the LBs, the LB fraction was subsequently analyzed by 1D SDS gel electrophoresis followed by liquid chromatography-tandem mass spectrometry. We tested several buffers (containing Triton X-100, sarkosyl, SDS, urea or formic acid) to dissolve the LB pellet and found that the SDS buffer led to the best yield of proteins. When the LB sample and the control were compared on a stained SDS gel (Figure 2), at least a few protein bands showed clear differences although the overall protein patterns in both samples were similar to each other, suggesting that the majority of proteins in the samples had comparable abundance, and therefore quantitative comparison was essential to identify LB-associated proteins in the noise of common contaminants. To reduce variations in key experimental parameters such as digestion activity, HPLC column performance, and ionization efficiency, we performed the analyses of the two samples in parallel under identical LC-MS/MS conditions. From the LC-MS/MS analysis, approximately 50,00 MS/MS spectra were acquired for each sample and searched against a human protein database. The matched peptides were further filtered rigorously, leading to the identification of ~700 proteins by combining the LB fraction and the control sample (Supplementary Table S1).
Figure 2.
Sample processing and data analysis. Proteins in the LB and control fractions were extracted by a SDS-containing buffer, run in parallel on a SDS-PAGE gel. Each sample lane was cut into 10 pieces that were subjected to trypsin digestion and LC-MS/MS analysis. The gel excision pattern is shown on the right according to the marker. Database searching was performed with SEQUEST followed by peptide filtering. Finally, quantitation was carried out with protein spectral counts.
4.2. Quantitative comparison reveals novel proteins associated with Lewy bodies
To determine which proteins are significantly more abundant in the LB fraction than in the control, we used a label-free quantitation method based on spectral count (17, 18). The spectral count of a given protein is the number of its matching MS/MS spectra. As the peptide ions are selected for MS/MS sequencing based on ion intensity, the value of spectral count of a protein is capable of reflecting its abundance in the starting materials.
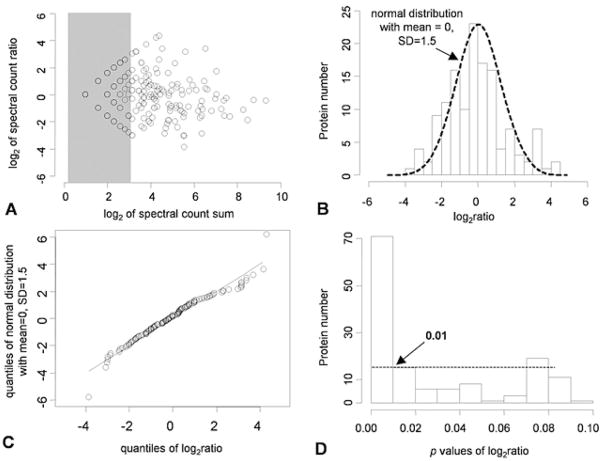
A series of statistical analyses were performed to evaluate the datasets. (i) The effect of spectral counts on the precision of the spectral count ratio was tested by plotting the sum of the spectral counts from two samples versus the ratio (Figure 3A). When the sum of spectral counts was too low (i.e. <3 at the log2 scale), the quantize effect was significant and the reliability of the ratio was greatly reduced (shown in the shaded area). Thus, these data points were excluded from further statistical analyses. (ii) The histogram of the dataset was fitted to the Gaussian distribution according to the null hypothesis (Figure 3B). The mean of zero indicated no detectable systematic error in the analysis, consistent with the gel picture that the two samples had very similar protein composites (Figure 2). Moreover, a quantile-quantile plot confirmed the Gaussian distribution of the log ratios of the spectral counts (Figure 3C). (iii) Additional statistical methods were adopted to derive p-values for each protein comparison (see Materials and Methods), which represented the statistical significance of the abundance change. The histogram of an expanded fraction of p-values from zero to 0.1 is shown in Figure 3D, and from the histogram, the selection of a p-value of 0.01 was deemed a reasonable cutoff to detect significant changes.
Figure 3.
Statistical data analyses. A, the scatter plot of log2 values of the sum versus the ratio of spectral counts in the LB and control fractions. B, the histogram of log2 transformed spectral count ratios superimposed with the density plot of a Gaussian distribution. C, quantile-quantile plot (q-q plot) of log2 of spectral count ratios with a simulated normal distribution. D, the histogram of an expanded fraction of p-values from zero to 0.1. The dashed horizontal line shows the level under which the frequency of p-values start to flatten, which indicates the closeness to uniform distribution due to randomness.
Based on the p-value criteria, a list of ~40 proteins was generated for the LB sample (Table 1). However, the p-value cutoff has the potential to exclude interesting proteins with biological relevance, especially for large-scale proteomics that are normally not repeated due to the significant effort required, thus resulting in insufficient data points for statistical evaluation. Therefore, the proteins below the cutoff were further examined manually, and some proteins were included in the table according to the involvement of the protein in neurodegeneration, the clustering of protein families, or the magnitude of protein changes (Table 1). To evaluate their potential roles in the formation of LBs, the proteins were classified into several functional groups, including chaperones, cytoskeleton components, oxidative stress, protein kinases, protein trafficking, ubiquitination, and lysosomal degradation, indicating that the cellular events governing LB formation are complex and diverse.
Table 1.
The list of proteins enriched in the Lewy body sample
| Protein names | GenBank™ accession number | Spectra counts (LB) | Spectra counts (control) | log2(LB/control)1 | p value2 |
|---|---|---|---|---|---|
| Chaperones | |||||
| Heat shock protein 90kda | NP_031381.2 | 59 | 30 | 1.0 | 1.9E−03 |
| Cytoskeleton components | |||||
| Spectrin, alpha | NP_003118.1 | 100 | 31 | 1.7 | 6.2E−10 |
| Spectrin, beta | NP_003119.1 | 84 | 8 | 3.4 | 0.0E+00 |
| Plectin 1 | NP_958786.1 | 44 | 5 | 3.1 | 2.4E−09 |
| Gelsolin | NP_000168.1 | 9 | 1 | 3.2 | 6.7E−03 |
| WD repeat-containing protein 1 | NP_059830.1 | 9 | 1 | 3.2 | 6.7E−03 |
| Oxidative stress | |||||
| Carbonyl reductase 1 | NP_001748.1 | 21 | 2 | 3.4 | 1.9E−05 |
| Peroxiredoxin 53 | NP_036226.1 | 4 | 0 | 3.0 | 7.8E−02 |
| Protein disulfide isomerase-associated 3 | NP_005304.3 | 8 | 0 | 4.0 | 4.7E−03 |
| Kinases | |||||
| Mek1, mitogen-activated protein kinase kinase 1 | NP_002746.1 | 12 | 0 | 4.6 | 2.9E−04 |
| Protein kinase c, beta 1 | NP_002729.2 | 9 | 1 | 3.2 | 6.7E−03 |
| Doublecortin-like kinase3 | NP_004725.1 | 4 | 0 | 3.0 | 7.8E−02 |
| Protein trafficking | |||||
| Adaptor-related protein complex 2, alpha 1 | NP_570603.2 | 10 | 0 | 4.3 | 1.2E−03 |
| Adaptor-related protein complex 2, beta 1 | NP_001273.1 | 8 | 0 | 4.0 | 4.7E−03 |
| Coatomer protein complex, subunit alpha | NP_004362.1 | 12 | 0 | 4.6 | 2.9E−04 |
| Calcium-dependent secretion activator | NP_003707.2 | 13 | 0 | 4.7 | 1.4E−04 |
| Clathrin heavy chain 1 | NP_004850.1 | 70 | 0 | 7.1 | 0.0E+00 |
| Dynactin 1 | NP_004073.2 | 10 | 0 | 4.3 | 1.2E−03 |
| Dynamin-like protein | NP_036193.1 | 11 | 0 | 4.5 | 5.8E−04 |
| Dynein, heavy polypeptide 1 | NP_001367.2 | 29 | 6 | 2.3 | 5.0E−05 |
| Protein ubiquitination | |||||
| Ubiquitin3 | NP_003324.1 | 11 | 3 | 1.9 | 2.7E−02 |
| Ubiquitin-activating enzyme E1 | NP_695012.1 | 26 | 0 | 5.7 | 1.7E−08 |
| E3 ubiquitin ligase kpc1, ring finger protein 1233 | NP_071347.2 | 5 | 0 | 3.3 | 3.9E−02 |
| Cullin 13 | NP_003583.2 | 2 | 0 | 2.0 | 3.3E−01 |
| F-box protein 23 | NP_036300.2 | 4 | 2 | 1.0 | 4.1E−01 |
| Ubiquitin c-terminal hydrolase l1 | NP_004172.2 | 10 | 0 | 4.3 | 1.2E−03 |
| Ubiquitin-specific protease otubain 13 | NP_060140.1 | 6 | 1 | 2.6 | 4.7E−02 |
| Proteasome 26s non-atpase subunit 23 | NP_002799.3 | 2 | 0 | 2.0 | 3.3E−01 |
| Lysosomal degradation | |||||
| Vacuolar protein sorting 35 | NP_060676.2 | 18 | 0 | 5.2 | 4.5E−06 |
| Atpase, lysosomal 70 kd, v1 subunit alpha | NP_001681.2 | 18 | 1 | 4.2 | 1.7E−05 |
| Others | |||||
| Alpha-synuclein | NP_009292.1 | 9 | 0 | 4.2 | 2.4E−03 |
| Aconitase 2 | NP_001089.1 | 18 | 3 | 2.6 | 5.7E−04 |
| Glucose phosphate isomerase | NP_000166.2 | 22 | 0 | 5.5 | 2.8E−07 |
| Glyceraldehyde-3-phosphate dehydrogenase | NP_002037.2 | 9 | 1 | 3.2 | 6.7E−03 |
| Imp cyclohydrolase | NP_004035.2 | 11 | 0 | 4.5 | 5.8E−04 |
| Calgranulin beta | NP_002956.1 | 9 | 0 | 4.2 | 2.4E−03 |
| Collapsin response mediator protein HCRMP-2 | NP_001377.1 | 20 | 1 | 4.3 | 4.4E−06 |
| Guanine nucleotide binding protein alpha | NP_620073.1 | 21 | 7 | 1.6 | 6.8E−03 |
& 2 The values were derived from spectral counts of identified proteins in the Lewy body sample and the control,
Proteins may be of interest but have p values > 0.01.
5. DISCUSSION
This work revealed ~40 proteins co-purified with LB inclusions in the background of several hundreds of proteins, primarily accomplished by quantitative comparison of the LB-enriched sample with the control fraction. As anticipated, many previously reported LB proteins were identified, such as α-synuclein (1, 2), UCH-L1 (19), and ubiquitin-activating enzyme E1 (20). More intriguing were a number of novel regulatory proteins involved in phosphorylation and ubiquitination that emerged from this large-scale proteomic analysis.
5.1. Comparison of different strategies for LB purification
As an alternative to the biochemical approach used here, the LB aggregates in brain specimens may be enriched by laser-capture microdissection (LCM) (21). The LCM strategy is capable of procuring and thereby concentrating specific microscopic structures from brain sections, and has been utilized for proteomic analysis of senile plaques in Alzheimer’s disease in our group (22). During the preparation of this manuscript, another group has reported a qualitative analysis of LBs isolated by LCM, identifying a total of ~300 proteins (23). Without a negative control sample, however, it is not clear whether those proteins are present in the LBs themselves or are derived from the surrounding neuropil tissues. Indeed, close examination of our LCM preparations (data not shown) has suggested that it is difficult to capture LBs without introducing significant contamination, as the surrounding tissues are always co-isolated with the target structures during the procedure. This becomes a major concern when the targeted structure is small and near the size limit of the LCM (~5 μm in diameter) (24). Further purification from LCM samples is, unfortunately, not practical due to the minute amount of tissues captured (~2 ng proteins from a plaque region of 60 μm in diameter and 10 μm in thickness) (25). Alternatively, LBs can be isolated by sucrose density gradient from brain homogenates (10). Using this method, we were able to enrich LBs by approximately seven-fold. To further distinguish the LB proteins from co-purified contaminants, control tissue without LB pathology was processed in parallel for quantitative comparison. This was an essential component of our analysis because less than 10% of the proteins we identified showed statistically significant enrichment in the LB sample.
5.2. Potential roles of identified kinases and ubiquitin ligases
The main LB protein, α-synuclein, has been found to be modified by a number of posttranslational events, as exemplified by phosphorylation (4, 26) and ubiquitination (5). In LBs, α-synuclein has been shown to be highly phosphorylated at Ser 129, and this phosphorylation facilitates the assembly of fibril formation in vitro. Mutation of this Ser residue causes the suppression of dopaminergic neuron loss in the fly model of Parkinsonism (27). Moreover, ubiquitination of α-synuclein occurs primarily on the phosphorylated form of α-synuclein, suggesting that the addition of ubiquitin might be dependent on prior phosphorylation of α-synuclein (26). In this study, we detected MAPKK1/MEK1, protein kinase C and doublecortin-like kinase with higher abundance in the LB fraction than in the control, suggesting that these kinases may participate in the modification of α-synuclein and/or other LB components. Indeed, both MEK1 and α-synuclein are upregulated in a human neuroblastoma cell line treated with MPP+, a toxin that can induce Parkinsonism in human, indicating that the MAP kinase pathway is activated under the toxic insult (28).
In addition to ubiquitin-activating enzyme E1, UCH-L1, and proteasome subunits, a number of novel ubiquitin enzymes were found to be co-purified with LBs (Table 1 and supplemental Table S1). Those are KPC, SCF (skip–cullin–F-box), Ube4b, and otubain 1. The KPC complex is a ring-type ubiquitin ligase composed of KPC1 and KPC2; and it functions to promote the degradation of p27 during the cell cycle (29). SCF is a family of cullin-type ligases that also regulate the timing of the cell cycle. Recently, many ubiquitin ligases that are well studied in cell cycle control have been found to play critical roles in postmitotic neurons (30). Moreover, Ube4b may directly modulate axonal degeneration, as it is overexpressed as a chimeric protein in the C57BL/WldS mouse that exhibits protection of Wallerian degeneration of injured axons (31). Otubain 1 is a member of a newly discovered ubiquitin protease family that removes ubiquitin from its modified targets (32). In those ligases, two proteins (KPC2 and Ube4b) were sequenced only once in the LB sample but not in the control, which cannot be viewed as reliable evidence, indicated by the high p value. Nevertheless, the discovery of these regulatory proteins provides molecular clues for the follow-up studies. Given that the ubiquitin enzymes that modify LB components are still elusive, the pathological involvement of those enzymes in synucleinopathies awaits future investigation.
The study described here is a pilot quantitative proteomics analysis of a LB-enriched fraction. The evolution of mass spectrometry instrumentation and the advancement of quantitative strategies will enable more extensive characterization of LBs in the future. The results of this study provide a list of candidates of interest, which are instructive for further hypothesis-driven research of the molecular mechanisms of neurodegeneration.
Supplementary Material
Acknowledgments
The first two authors contributed equally to the work. We thank the members in the lab for critical discussion of the manuscript. This work was partially supported by NIH grants ES012068, and the Emory Alzheimer’s Disease Center AG025688.
Abbreviations
- LB
Lewy body
- PD
Parkinson’s disease
- AD
Alzheimer’s disease
- DLB
dementia with Lewy bodies
- LCM
laser capture microdissection
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
Footnotes
References
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Mezey E, M Dehejia A, Harta G, Tresser N, Suchy SF, Nussbaum RL, Brownstein MJ, Polymeropoulos MH. Alpha synuclein is present in Lewy bodies in sporadic Parkinson’s disease. Mol Psychiatry. 1998;3(6):493–9. doi: 10.1038/sj.mp.4000446. [DOI] [PubMed] [Google Scholar]
- Galvin JE, Lee VM, Trojanowski JQ. Synucleinopathies: clinical and pathological implications. Arch Neurol. 2001;58(2):186–90. doi: 10.1001/archneur.58.2.186. [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, Iwatsubo T. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4(2):160–4. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujiwara H, Nonaka T, Wakabayashi K, Takahashi H, Lee VM, Trojanowski JQ, Mann D, Iwatsubo T. Phosphorylated alpha-synuclein is ubiquitinated in alpha-synucleinopathy lesions. J Biol Chem. 2002;277(50):49071–6. doi: 10.1074/jbc.M208046200. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293(5528):263–9. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290(5493):985–9. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- Yates JR., 3rd Mass spectral analysis in proteomics. Annu Rev Biophys Biomol Struct. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Yamaguchi H, Fujimuro M, Yokosawa H, Ihara Y, Trojanowski JQ, Lee VM. Purification and characterization of Lewy bodies from the brains of patients with diffuse Lewy body disease. Am J Pathol. 1996;148(5):1517–29. [PMC free article] [PubMed] [Google Scholar]
- Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36(10):1083–91. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- Eng J, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. Freeman W. H; New York: 1995. [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005 doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Xia Q, Hendrickson EL, Wang T, Lamont RJ, Leigh JA, Hackett M. Protein abundance ratios for global studies of prokaryotes. Proteomics. 2007;7(16):2904–19. doi: 10.1002/pmic.200700267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 1990;161(2):153–60. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Shashidharan P, Perl DP, Jenner P, Olanow CW. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16(11):2136–48. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14(7):272–6. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- Liao L, Cheng D, Wang J, Duong DM, Losik TG, Gearing M, Rees HD, Lah JJ, Levey AI, Peng J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279(35):37061–8. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- Leverenz JB, Umar I, Wang Q, Montine TJ, McMillan PJ, Tsuang DW, Jin J, Pan C, Shin J, Zhu D, Zhang J. Proteomic identification of novel proteins in cortical lewy bodies. Brain Pathol. 2007;17(2):139–45. doi: 10.1111/j.1750-3639.2007.00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JY, Hanfelt J, Peng J. Clinical proteomics in neurodegenerative diseases. Proteomics Clin Appl. 2007;1 doi: 10.1002/prca.200700378. in press. [DOI] [PubMed] [Google Scholar]
- Gozal YM, Cheng D, Duong DM, Lah JJ, Levey AI, Peng J. Merger of laser capture microdissection and mass spectrometry: a window into the amyloid plaque proteome. Methods Enzymol. 2006;412:77–93. doi: 10.1016/S0076-6879(06)12006-6. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, Diep L, Keim PS, Shen X, Chataway T, Schlossmacher MG, Seubert P, Schenk D, Sinha S, Gai WP, Chilcote TJ. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem. 2006;281(40):29739–52. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- Chen L, Feany MB. Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci. 2005;8(5):657–63. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- Gomez-Santos C, Ferrer I, Reiriz J, Vinals F, Barrachina M, Ambrosio S. MPP+ increases alpha-synuclein expression and ERK/MAP-kinase phosphorylation in human neuroblastoma SH-SY5Y cells. Brain Res. 2002;935(1–2):32–9. doi: 10.1016/s0006-8993(02)02422-8. [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6(12):1229–35. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47(5):629–32. doi: 10.1016/j.neuron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, Fernando FS, Tarlton A, Andressen C, Addicks K, Magni G, Ribchester RR, Perry VH, Coleman MP. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4(12):1199–206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4(5):517–22. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.