Summary
Mammalian T lymphocytes are a prototype for development from adult pluripotent stem cells. While T-cell specification is driven by Notch signaling, T-lineage commitment is only finalized after prolonged Notch activation. However, no T-lineage specific regulatory factor has been reported that mediates commitment. We used a gene-discovery approach to identify additional candidate T-lineage transcription factors and characterized expression of >100 regulatory genes in early T-cell precursors using realtime RT-PCR. These regulatory genes were also monitored in multilineage precursors as they entered T-cell or non-T-cell pathways in vitro; in non-T cells ex vivo; and in later T-cell developmental stages after lineage commitment. At least three major expression patterns were observed. Transcription factors in the largest group are expressed at relatively stable levels throughout T-lineage specification as a legacy from prethymic precursors, with some continuing while others are downregulated after commitment. Another group is highly expressed in the earliest stages only, and is downregulated before or during commitment. Genes in a third group undergo upregulation at one of three distinct transitions, suggesting a positive regulatory cascade. However, the transcription factors induced during commitment are not T-lineage specific. Different members of the same transcription factor family can follow opposite trajectories during specification and commitment, while factors co-expressed early can be expressed in divergent patterns in later T-cell development. Some factors reveal new regulatory distinctions between αβ and γδ T-lineage differentiation. These results show that T-cell identity has an essentially complex regulatory basis and provide a detailed framework for regulatory network modeling of T-cell specification.
INTRODUCTION
T-cell development is an exceptionally favorable system in which to examine the mechanism of differentiation from stem cells in general. It offers the most completely identified sequence of developmental intermediates known for any post-embryonic stem-cell derivatives. Cells with T-cell precursor activity become physically segregated from other blood-cell precursors by migrating to the thymus at a very early stage of their development (Shortman and Wu, 1996; Ceredig and Rolink, 2002; Petrie and Zuniga-Pflucker, 2007; Rothenberg et al., 2008). This allows these intermediates to be purified for analysis with minimal interference from irrelevant cell populations. Also, because individual T-cell precursors can be isolated relatively easily, it has been possible to define the series of distinct steps at which particular developmental alternatives are lost during the stages leading up to lineage commitment (Rothenberg, 2007). T-lineage specification is both triggered and sustained by Notch signaling. Yet the way lineage-specific gene expression actually starts and how T-lineage identity is cemented at commitment are both questions still to be resolved.
T lymphocyte development from hematopoietic stem cells is not driven by a single “master gene”, but rather depends upon a large number of distinct regulatory factors, as shown by genetic evidence in targeted mutant mice (Anderson, 2006; Rothenberg and Taghon, 2005). Notch pathway signaling, triggered by ligands in the thymic microenvironment, is a central element, needed both to begin T-cell specification and to complete lineage commitment. However, to promote T-lineage development this signal must act on cells that also have an appropriate constellation of intrinsic regulatory activities. Among the transcription factors already known to have critical roles are GATA-3, Myb, Runx1+CBFβ, Ikaros, TCF-1 activated by Wnt, RBP-J activated by Notch, basic helix-loop-helix “E proteins”, Gfi1, and in an early hit-and-run role, hematopoietic transcription factor PU.1. Most of these regulatory inputs are needed to allow recognizable T-lineage precursors to emerge in the thymus from the earliest stages, long before the cells express T-cell receptors for antigen and undergo selection events that depend on those receptors. Most of the same factors are then used repeatedly over many cell cycles for days or weeks to define T-cell subtypes (Rothenberg et al., 2008; Ho and Pai, 2007; Tanigaki and Honjo, 2007; Anderson, 2006; Rothenberg and Taghon, 2005; Staal et al., 2001).
Although identities of these factors are clear, the transcriptional mechanisms that result in T-cell specification have remained more obscure than those involved in specification of many other hematopoietic cell types, especially as contrasted with another lymphoid cell type, namely B lymphocytes (Rothenberg, 2007; Rothenberg and Taghon, 2005). B cell identity is now known to be created through the action of a cascade of transcription factors in which key roles are played by EBF and Pax5, two dedicated factors that are B-lineage specific in expression as well as function within the hematopoietic system (Cobaleda et al., 2007; Singh et al., 2007; Nutt and Kee, 2007; Hagman and Lukin, 2006). Because of their specific expression patterns, mutations in these factors also yield selective defects in B-cell development that can be staged and characterized in molecular terms. These factors also act effectively as positive B-lineage regulators in both gain of function and loss of function experiments. The result has been a definitive elucidation of the B-lineage specification process (Cobaleda et al., 2007; Singh et al., 2007; Nutt and Kee, 2007; Hagman and Lukin, 2006). There are indeed two well-studied factors that are T-lineage specific in expression, namely GATA-3 and TCF-1 (product of the Tcf7 gene), and it has been known for years that these are both required specifically for T-cell development (Verbeek et al., 1995; Ting et al., 1996; Hattori et al., 1996). However, in marked contrast to the B-cell factors, these do not appear to upregulate T-lineage target genes specifically and do not enhance T-cell development in gain of function experiments (Taghon et al., 2007; Kirstetter et al., 2006; Scheller et al., 2006; Baba et al., 2005; Staal et al., 2004; Anderson et al., 2002a). One possible interpretation of existing data is that T-lineage choice is fundamentally different from B lineage choice; but another possibility has been that the EBF and Pax5 equivalents for T cells simply have yet to be recognized. Therefore, a crucial question is whether other transcription factors may exist in early T-lineage cells that could play more cell-type specific roles.
This study and its predecessors (Tydell et al., 2007; Anderson et al., 1999) were undertaken to identify any “missing” regulators of T-cell development that may be T-cell lineage specific, to track the major changes in transcription factors generally as cells progress toward T-lineage commitment, and to clarify how their roles and expression are coordinated with the observed landmarks in T-cell development (Rothenberg et al., 2008). In this study, we have relied on cDNA macroarray screening for gene discovery and on sensitive, quantitative realtime RT-PCR (qPCR) measurements to identify a large group of regulatory gene candidates and track the way that they and their family members are expressed in early T-lineage precursors. The results show that the T-cell specification process entails discontinuous upregulation of one group of transcription factor genes, surprisingly stable expression of a larger group of stem cell-inherited factors until after commitment, and downregulation of another substantial group of previously expressed transcription factor genes. However, few regulatory genes overall have highly specific T-cell expression, and while T-lineage commitment also causes upregulation of certain transcription factor genes, most of these are not T-lineage specific. These results add substantial detail to the transcription factor dynamics that underlie T-cell specification and strengthen the probability that there is a fundamental divergence between the overall regulatory strategies involved in T-cell and B-cell lineage choice.
MATERIALS AND METHODS
Animals
Cells were obtained from C57BL/6 mice and from mice of TCRβ−/− and Rag2−/− genotypes on a C57BL/6 background, all maintained in our breeding colony at the California Institute of Technology (Caltech). “Adult” samples were from 4–6 week old mice for thymocyte isolation and from 8–12 week old mice for bone marrow isolation. Fetal liver was obtained from timed pregnancies of C57BL/6 or (C57BL/6 × DBA/2) F1 mice at E14.5 (day of plug = E0.5). Animals were kept in microisolator cages in an AAALAC-accredited facility with the immune-deficient mutants maintained with sterilized cages, water, food, and bedding. Euthanasia and animal care followed NIH guidelines, under protocols approved by the Institute Animal Care and Use Committee.
Gene discovery by macroarray screening
New cDNA clones of interest were isolated from a C.B17-Prkdcscid/scid (SCID) mouse thymocyte cDNA library that was constructed and arrayed onto four large nylon filters as previously described (Anderson et al., 1999; Tydell et al., 2007). SCID thymocytes were used as starting material because their mutant phenotype prevents T-cell development from continuing through β-selection. Thus, >90% of these populations naturally consist of T-cell precursors in the DN1-DN3 stages. To select for cDNAs that might encode gene regulatory factors, four “macroarray” filters containing approx 73,000 clones were screened as previously described (Tydell et al., 2007) with probes designed to identify conserved domains associated with transcription factors.
To make these initial probes, gene sequences of representatives from various transcription factor families were aligned using ClustalX and primers were designed to their DNA binding domains or protein interaction domains. Primers used are tabulated in Table 1. The putative functional domain coding regions were then amplified from bulk SCID thymus cDNA and gel-purified. Domain-specific DNA probes were generated by random priming in the presence of α-32P dCTP, using the gel-purified PCR amplification products as templates. Probe was hybridized to the macro-array filters overnight and washed with 3–4 low stringency washes to enable fully and partially matched hybrids to be detected. Macroarray filters were exposed to film and clones identified by positively hybridizing spot pairs that were picked from the archival library for further analysis as previously described (Tydell et al., 2007). A total of 1490 clones were selected into 96-well plates for sequencing and re-arraying onto smaller nylon filters.
TABLE 1.
Conserved Domain Primers for Macroarray Screen
| A. AT-Hook | L. Homoeodomain | ||
| AT-Hook.For | 5′-ctccgaaagagcccagtgaa-3′ | HOXB4.For | 5′-cggagctcggccaagtctatga-3′ |
| AT-Hook.Rev | 5′-ggtttcctccctggagctgt-3′ | HOXB4.Rev | 5′-ccggttatggaacgagatcttga-3′ |
| B. ARID | M. Leucine Zipper | ||
| ARID.For | 5′-ggacctttgaggagcagttcaa-3′ | TSC.For | 5′-gtgggaccgagccgctgcagat-3′ |
| ARID.Rev | 5′-ctcataggggtaaagatacttcat-3′ | TSC.Rev | 5′-gtcttcagcagattgttctcct-3′ |
| C. bHLH | N. LIM Domain | ||
| MyoD.For | 5′-ccaccaacgctgatcgccgcaa-3′ | Lim-1.For | 5′-cggagattaccagagtgagta-3′ |
| MyoD.Rev | 5′-cgcagcagagcctgcagacctt-3′ | Lim-1.Rev | 5′-gcaagctgggctcaggacta-3′ |
| D. Bromodomain | 4.5Lim.For | 5′-gtcaggaatgccacaagcccat-3′ | |
| Bromo.For | 5′-catcattaaacaccccatggacctcagta-3′ | 4.5Lim.Rev | 5′-gcacaggatattcttgtccca-3′ |
| Bromo.Rev | 5′-gctcacctggcatcttggcaaatctcat-3′ | O. NFAT | |
| E. Chromodomain | NFAT.p-For | 5′-ctcctctgccagcttcatt-3′ | |
| Chromo.Chd1-For | 5′-caaccatttatgctgtcgaa-3′ | NFAT.p-Rev | 5′-ggcagaggagcaaccaaa-3′ |
| Chromo.Chd1-Rev | 5′-ctgctgcttcagggtctctt-3′ | NFAT.x.For | 5′-cctttgagtgcccaagtatccaa-3′ |
| Chromo.Cbx/Mpc2-For | 5′-cttcgcggtggagagcat-3′ | NFAT.x.Rev | 5′-gagatgctgctggcaggacta-3′ |
| Chromo.Cbx/Mpc2-Rev | 5′-cctttccctgttctggaa-3′ | P. PAS | |
| F. Cut-Homoeodomain | AHR.PAS-A.For | 5′-caggcgctgaatggctttgt-3′ | |
| Cux.For | 5′-gtcaaggaggtgctcaccgacaa-3′ | AHR.PAS-A.Rev | 5′-cttctgtatggatgagctcat-3′ |
| Cux.Rev | 5′-cagccacagctgcatacgcacaa-3′ | AHR.PAS-B.For | 5′-cgaaccaaaaacttcatcttca-3′ |
| G. Ezrin-Radixin-Moesin (Protein Interaction) | AHR.PAS-B.Rev | 5′-gtctgcagcatggatgaact-3′ | |
| ERM.For | 5′-gacctgaagaccacatacgatgaa-3′ | PER.PAS-A.For | 5′-ggaacccggataccttcgctgt-3′ |
| ERM.Rev | 5′-ctgcagggtctgcctggccagtt-3′ | PER.PAS-A.Rev | 5′-acgtcatgaggagccaggagct-3′ |
| H. Forkhead Box | PER.PAS-B.For | 5′-gatccctcctgagaagaggat-3′ | |
| Fox.For | 5′-tcacctatgccacccttat-3′ | PER.PAS-B.Rev | 5′-cagatcctgaggtaggtaa-3′ |
| Fox.Rev | 5′ctcctcttcttgcgaaactca-3′ | Q. PDZ | |
| I. GCM Box | PDZ.For | 5′-cctctgacaatctctagtctgaa-3′ | |
| GCM.For | 5′-gcgcaacaccaacaaccacaa-3′ | PDZ.Rev | 5′-ctttgtagagtcatattcaa-3′ |
| GCM.Rev | 5′-cttctgccttctgtctctgactt-3′ | PDZk1.For | 5′-gcagaggcagctggcttgaa-3′ |
| J. HLH | PDZk1.Rev | 5′-ccagcaccaacagagtagt-3′ | |
| Myf5.For | 5′-caggctggccactgcctcat-3′ | LAP.For | 5′-gttgcgtttgctcagacgaga-3′ |
| Myf5.Rev | 5′-gttggtggtggtgcacctctt-3′ | LAP.Rev | 5′-caagcttgatgcaccgcgacgct-3′ |
| K. HMG Box | WEGbox.For | 5′acagataatgggggttgggct-3′ | |
| SOX.For | 5′-gcaagccccatgctaagct-3′ | WEG/PHD.Rev | 5′-gtattgaacattgtctgcgccatt-3′ |
| SOX.Rev | 5′-ggtgattggcatggcggct-3 | W. ZINC FINGERS | |
| R. PHD | Btb/Poz | ||
| PHD.For | 5′-gtgcttgcatgacatgtaataaa-3′ | BTB/POZ.For | 5′-catttgctgcagtgcctgaacgagca-3′ |
| WEG/PHD.Rev | 5′-gtattgaacattgtctgcgccatt-3′ | BTB/POZ.Rev | 5′-catctggagatagttggcagctgcaa-3′ |
| ZIP/PHDf.For | 5′-cagcagcattgaacagaagga-3′ | ROG.For | 5′-ggcaggagcaactaggatgat-3′ |
| ZIP/PHDf.Rev | 5′-ggtagtcccctttcatgttat-3′ | ROG.Rev | 5′-ggtcagtgggatcctctgat-3′ |
| S. REL | C2H2 | ||
| NFkB.For | 5′-gctattcggtgagtaaagaa-3′ | Multizf.For | 5′-gcctcacagatcaccgaga-3′ |
| NFkB.Rev | 5′-gtttttgtagccctattttcat-3′ | Multizf.Rev | 5′-ggaagaggcttaaattgtt-3′ |
| T. RNA Binding Domains | Fog | ||
| hnRNP.A2/B1-For | 5′-ggctgcaaggcctcattccatt-3′ | FOG.For | 5′-gccccaggatgaagagaaa-3′ |
| hnRNP.A2/B1-Rev | 5′-gcctatcggtaattatttcaatagt-3′ | FOG.Rev | 5′-gcagtctttgcaggggaa-3′ |
| hnRNP.U-For | 5′-caggggcgaggcagggccaa-3′ | Gli | |
| hnRNP.U-Rev | 5′-gacacaccgtaggaagctctt-3′ | GLI.spfc-For | 5′-gagacaaactgccactgggat-3′ |
| hnRNP.x-For | 5′-ggagagtggtgcacgtatcaa-3′ | GLI.spfc-Rev | 5′-gctcacagatgtaaggcttct-3′ |
| hnRNP.x-Rev | 5′-gcaaccaccttttccaataaga-3′ | Groucho | |
| Rbm.For | 5′-gaaccagaggtgcttcaagaa-3′ | Grg.For | 5′-gaaatgcacaaacagactgaaat-3′ |
| Rbm.Rev | 5′-cccttggtaatggatactcat-3′ | Grg.Rev | 5′-ggtgcttcttgtcatcttttatt-3′ |
| U. STAT | Ring Finger | ||
| STAT.For | 5′-gaccctgtccctccctgtggt-3′ | Ring/B-box.For | 5′-gatcgaggatcttctgta-3′ |
| STAT.Rev | 5′-cccatgatagccccatcattcca-3′ | Ring/B-box.Rev | 5′-ggcctcatgattgctgta-3′ |
| V. T-BOX | ZIC | ||
| T-box.For | 5′-gagatgatcatcactaagcaa-3′ | ZICspfc.GLI-For | 5′-ctgttccgcaaccgtggctt-3′ |
| T-box.Rev | 5′-ggcagcctctggctctccat-3′ | ZICspfc.GLI-Rev | 5′-gggcagcatagtgctcggat-3′ |
Sequencing was performed by the modified Sanger’s method at the Institute for Systems Biology, Seattle, WA. The sequences were identified by BLASTn, BLASTx (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and BLAT alignments against the Genbank “nr” databases and the mouse genome (up to builds 35 & 36; http://genome.ucsc.edu). New sequences that appeared to encode transcription factors or potential transcription-modifying factors, as well as sequences that defined new splice isoforms of known transcription factor genes, were submitted to Genbank as Expressed Sequence Tags (accession numbers listed in Table 2).
TABLE 2.
EST Sequences from SCID Thymocyte Library
| Gene | Alt Name | EST ID# | GenBank Accession # | Date Submitted |
|---|---|---|---|---|
| ARHGEF2 | 39175456 | EB739685 | 5/10/2006 | |
| 39175457 | EB739686 | |||
| ARID1A | 39175458 | EB739687 | 5/10/2006 | |
| 39175459 | EB739688 | |||
| BAHD1 | 39175460 | EB739689 | 5/10/2006 | |
| 39175461 | EB739690 | |||
| BRD3 | 38305921 | EB359574 | 4/6/2006 | |
| 38305922 | EB359575 | |||
| 38305923 | EB359576 | |||
| BTF3 | 39175462 | EB739691 | 5/10/2006 | |
| 39175463 | EB739692 | |||
| Cbx4 | 39175464 | EB739693 | 5/10/2006 | |
| CEBPd | 39175465 | EB739694 | 5/10/2006 | |
| 39175466 | EB739695 | |||
| CENTb1 | 39175467 | EB739696 | 5/10/2006 | |
| CIZ | Zfp384 | 39175468 | EB739697 | 5/10/2006 |
| 39175469 | EB739698 | |||
| 39175470 | EB739699 | |||
| 39175471 | EB739700 | |||
| 39175472 | EB739701 | |||
| 39175473 | EB739702 | |||
| CRABP1 | 39781094 | EC277794 | 6/8/2006 | |
| 39781095 | EC277795 | |||
| CSF2Ra | 39781134 | EC277834 | 6/8/2006 | |
| 39781096 | EC277796 | |||
| CTBP2 | 39781097 | EC277797 | 6/8/2006 | |
| 39781098 | EC277798 | |||
| CUTL1 | 39781099 | EC277799 | 6/8/2006 | |
| 39781100 | EC277800 | |||
| DAPLE | CCdc88c | 39781101 | EC277801 | 6/8/2006 |
| 39781102 | EC277802 | |||
| 39781103 | EC277803 | |||
| 39781104 | EC277804 | |||
| DBP | 39781105 | EC277805 | 6/8/2006 | |
| 39781106 | EC277806 | |||
| DIMP | Dmtf1 | 39781107 | EC277807 | 6/8/2006 |
| 39781108 | EC277808 | |||
| DSC43 | 39781109 | EC277809 | 6/8/2006 | |
| 39781110 | EC277810 | |||
| 39781111 | EC277811 | |||
| 39781112 | EC277812 | |||
| 39781113 | EC277813 | |||
| 39781114 | EC277814 | |||
| 39781115 | EC277815 | |||
| 39781116 | EC277816 | |||
| 39781117 | EC277817 | |||
| ELK3 | 39781118 | EC277818 | 6/8/2006 | |
| 39781119 | EC277819 | |||
| 39781120 | EC277820 | |||
| 39781121 | EC277821 | |||
| 39781122 | EC277822 | |||
| ELK4 | 39781123 | EC277823 | 6/8/2006 | |
| 39781124 | EC277824 | |||
| 39781125 | EC277825 | |||
| ESR1 | 39781126 | EC277826 | 6/8/2006 | |
| 39781127 | EC277827 | |||
| 39781128 | EC277828 | |||
| 39781129 | EC277829 | |||
| 39781130 | EC277830 | |||
| 39781131 | EC277831 | |||
| 39781132 | EC277832 | |||
| 39781133 | EC277833 | |||
| FLI1 | 39877808 | EC365110 | 6/13/2006 | |
| 39877809 | EC365111 | |||
| 39877810 | EC365112 | |||
| 39877811 | EC365113 | |||
| 39877812 | EC365114 | |||
| GATA3 | 39877813 | EC365115 | 6/13/2006 | |
| 39877814 | EC365116 | |||
| GIMAP6 | 39877815 | EC365117 | 6/13/2006 | |
| GNAI2 | 39877816 | EC365118 | 6/13/2006 | |
| 39877817 | EC365119 | |||
| GSE1 | 39877818 | EC365120 | 6/13/2006 | |
| 39877819 | EC365121 | |||
| HKR2 | Zscan22 | 39877820 | EC365122 | 6/13/2006 |
| HKR3 | 39877821 | EC365123 | 6/13/2006 | |
| HMGA1 | 38305924 | EB359577 | 4/6/2006 | |
| 38305925 | EB359578 | |||
| 38305926 | EB359579 | |||
| 38305927 | EB359580 | |||
| 38305928 | EB359581 | |||
| 38305929 | EB359582 | |||
| 38305930 | EB359583 | |||
| 38305931 | EB359584 | |||
| 38305932 | EB359585 | |||
| 38305933 | EB359586 | |||
| IARS2 | 39877822 | EC365124 | 6/13/2006 | |
| IKAROS | 39877823 | EC365125 | 6/13/2006 | |
| 39877824 | EC365126 | |||
| 39877825 | EC365127 | |||
| 39877826 | EC365128 | |||
| 39877827 | EC365129 | |||
| 39877828 | EC365130 | |||
| 39877829 | EC365131 | |||
| 39877830 | EC365132 | |||
| JMJD3 | 39877864 | EC365166 | 6/13/2006 | |
| 39877831 | EC365133 | |||
| 39877832 | EC365134 | |||
| 39877833 | EC365135 | |||
| KLF2 | 39877834 | EC365136 | 6/13/2006 | |
| 39877835 | EC365137 | |||
| 39877836 | EC365138 | |||
| 39877837 | EC365139 | |||
| 39877838 | EC365140 | |||
| 39877839 | EC365141 | |||
| 39877840 | EC365142 | |||
| 39877841 | EC365143 | |||
| 39877842 | EC365144 | |||
| 39877843 | EC365145 | |||
| KLF13 | 39877844 | EC365146 | 6/13/2006 | |
| 39877845 | EC365147 | |||
| 39877846 | EC365148 | |||
| 39877847 | EC365149 | |||
| 39877848 | EC365150 | |||
| 39877849 | EC365151 | |||
| 39877850 | EC365152 | |||
| 39877851 | EC365153 | |||
| 39877852 | EC365154 | |||
| 39877853 | EC365155 | |||
| KLF15 | 39877854 | EC365156 | 6/13/2006 | |
| 39877855 | EC365157 | |||
| 39877856 | EC365158 | |||
| 39877857 | EC365159 | |||
| 39877858 | EC365160 | |||
| 39877859 | EC365161 | |||
| 39877860 | EC365162 | |||
| 39877861 | EC365163 | |||
| 39877862 | EC365164 | |||
| 39877863 | EC365155 | |||
| MTA2 | 40881949 | EE265166 | 8/3/2006 | |
| MTA3 | 40881950 | EE265167 | 8/3/2006 | |
| 40881951 | EE265168 | |||
| NCOR2 | 40881952 | EE265169 | 8/3/2006 | |
| PAX1 | 40881953 | EE265170 | 8/3/2006 | |
| 40881954 | EE265171 | |||
| PCBP2 | 40881955 | EE265172 | 8/3/2006 | |
| 40881956 | EE265173 | |||
| Per1 | 40881976 | EE265193 | 8/3/2006 | |
| 40881957 | EE265174 | |||
| POU2F1 | 40881958 | EE265175 | 8/3/2006 | |
| 40881959 | EE265176 | |||
| 40881960 | EE265177 | |||
| 40881961 | EE265178 | |||
| POU6F1 | 40881962 | EE265179 | 8/3/2006 | |
| 40881963 | EE265180 | |||
| 40881964 | EE265181 | |||
| PPARd | 40881965 | EE265182 | 8/3/2006 | |
| 40881966 | EE265183 | |||
| PPRC1 | 40881967 | EE265184 | 8/3/2006 | |
| PurB | 40881968 | EE265185 | 8/3/2006 | |
| RARa | 40881969 | EE265186 | 8/3/2006 | |
| 40881970 | EE265187 | |||
| 40881971 | EE265188 | |||
| RBAK | 40881972 | EE265189 | 8/3/2006 | |
| 40881973 | EE265190 | |||
| 40881974 | EE265191 | |||
| 40881975 | EE265192 | |||
| SBF1 | 41528217 | EE663535 | 8/24/2006 | |
| SFPI1 | 41528238 | EE663556 | 8/24/2006 | |
| 41528249 | EE663567 | |||
| SOX4 | 41528260 | EE663578 | 8/24/2006 | |
| 41528271 | EE663589 | |||
| 41528176 | EE663494 | |||
| 41528187 | EE663505 | |||
| 41528189 | EE663507 | |||
| 41528190 | EE663508 | |||
| 41528191 | EE663509 | |||
| 41528192 | EE663510 | |||
| SPI-B | 41528193 | EE663511 | 8/24/2006 | |
| SUPT5h | 41528194 | EE663512 | 8/24/2006 | |
| 41528195 | EE663513 | |||
| TBX21 | 41528196 | EE663514 | 8/24/2006 | |
| 41528197 | EE663515 | |||
| 41528198 | EE663516 | |||
| TCF3 | E2A | 41528199 | EE663517 | 8/24/2006 |
| 41528200 | EE663518 | |||
| 41528201 | EE663519 | |||
| 41528202 | EE663520 | |||
| 41528203 | EE663521 | |||
| 41528204 | EE663522 | |||
| TCF7 | 38305934 | EB359587 | 4/6/2006 | |
| 38305935 | EB359588 | |||
| 38305936 | EB359589 | |||
| 38305937 | EB359590 | |||
| 38305938 | EB359591 | |||
| 38305939 | EB359592 | |||
| 38305940 | EB359593 | |||
| 38305941 | EB359594 | |||
| 38305942 | EB359595 | |||
| 38305943 | EB359596 | |||
| TCF12 | HEB | 41528205 | EE663523 | 8/24/2006 |
| 41528206 | EE663524 | |||
| 41528207 | EE663525 | |||
| 41528208 | EE663526 | |||
| 41528209 | EE663527 | |||
| THRAP3 | 41528210 | EE663528 | 8/24/2006 | |
| 41528211 | EE663529 | |||
| TLE3 | 41528212 | EE663530 | 8/24/2006 | |
| 41528213 | EE663531 | |||
| 41528214 | EE663532 | |||
| Tmsb4x | 41528215 | EE663533 | 8/24/2006 | |
| 41528216 | EE663534 | |||
| TRIM24 | 41528218 | EE663536 | 8/24/2006 | |
| TRIM25 | EFP | 41528219 | EE663537 | 8/24/2006 |
| 41528220 | EE663538 | |||
| TRIM28 | 41528221 | EE663539 | 8/24/2006 | |
| TSC-22d1 | 41528222 | EE663540 | 8/24/2006 | |
| 41528223 | EE663541 | |||
| 41528224 | EE663542 | |||
| YY1 | 38305944 | EB359597 | 4/6/2006 | |
| 38305945 | EB359598 | |||
| 38305946 | EB359599 | |||
| ZBTB20 | 39175474 | EB739703 | 5/10/2006 | |
| 39175475 | EB739704 | |||
| ZBTB7a | 41528225 | EE663543 | 8/25/2006 | |
| 41528226 | EE663544 | |||
| 41528227 | EE663545 | |||
| 41528228 | EE663546 | |||
| 41528229 | EE663547 | |||
| 41528230 | EE663548 | |||
| 41528231 | EE663549 | |||
| 41528232 | EE663550 | |||
| 41528233 | EE663551 | |||
| 41528234 | EE663552 | |||
| 41528235 | EE663553 | |||
| ZC3H5 | 41528236 | EE663554 | 8/25/2006 | |
| 41528237 | EE663555 | |||
| ZFP3 | 41528239 | EE663557 | 8/25/2006 | |
| 41528240 | EE663558 | |||
| ZFP28 | 41528241 | EE663559 | 8/25/2006 | |
| ZFP32 | ZFP637 | 41528242 | EE663560 | 8/25/2006 |
| 41528243 | EE663561 | |||
| ZFP36 l2 | 41528244 | EE663562 | 8/25/2006 | |
| 41528245 | EE663563 | |||
| 41528246 | EE663564 | |||
| 41528247 | EE663565 | |||
| 41528248 | EE663566 | |||
| ZFP41 | 41528250 | EE663568 | 8/25/2006 | |
| 41528251 | EE663569 | |||
| ZFP90 | 41528252 | EE663570 | 8/25/2006 | |
| 41528253 | EE663571 | |||
| ZFP95 | Zkscan5 | 41528254 | EE663572 | 8/25/2006 |
| 41528255 | EE663573 | |||
| ZFP110 | 41528256 | EE663574 | 8/25/2006 | |
| 41528257 | EE663575 | |||
| ZFP238 | 41528258 | EE663576 | 8/25/2006 | |
| 41528259 | EE663577 | |||
| ZFP286 | 41528261 | EE663579 | 8/25/2006 | |
| 41528262 | EE663580 | |||
| ZFP287 | 41528263 | EE663581 | 8/25/2006 | |
| 41528264 | EE663582 | |||
| ZFP316 | 41528265 | EE663583 | 8/25/2006 | |
| 41528266 | EE663584 | |||
| ZFP358 | 41528267 | EE663585 | 8/25/2006 | |
| 41528268 | EE663586 | |||
| ZFP422 | 41528269 | EE663587 | 8/25/2006 | |
| 41528270 | EE663588 | |||
| ZFP598 | 41528272 | EE663590 | 8/25/2006 | |
| 41528273 | EE663591 | |||
| ZFP672 | 41528274 | EE663592 | 8/25/2006 | |
| 41528275 | EE663593 | |||
| ZFP691 | 41528276 | EE663594 | 8/25/2006 | |
| 41528277 | EE663595 | |||
| ZFPN1A4 | Eos | 41528278 | EE663596 | 8/25/2006 |
| 41528279 | EE663597 | |||
| 41528280 | EE663598 | |||
| 41528281 | EE663599 | |||
| Zip67 | 41528177 | EE663495 | 8/25/2006 | |
| 41528178 | EE663496 | |||
| 41528179 | EE663497 | |||
| 41528180 | EE663498 | |||
| 41528181 | EE663499 | |||
| 41528182 | EE663500 | |||
| 41528183 | EE663501 | |||
| Zipro1 | 41528184 | EE663502 | 8/25/2006 | |
| 41528185 | EE663503 | |||
| ZKSCAN1 | 41528186 | EE663504 | 8/25/2006 | |
| 41528188 | EE663505 |
Primary T-lineage cell samples
Cell populations of interest were collected by fluorescence-activated cell sorting (FACS), and cells were processed in Qiazol (Qiagen, USA) for RNA. RNA was purified using Qiagen RNAeasy kits (Qiagen), and treated with RNase-free DNaseI (Ambion) to remove genomic DNA contamination before conversion into first strand cDNA using Superscript Reverse Transcriptase (Invitrogen).
To isolate intermediates in T-cell development, antibody-mediated depletion with magnetic beads and multiparameter fluorescence-activated cell sorting (FACS) were used as previously described (Taghon et al., 2006; Yui and Rothenberg, 2004; Anderson et al., 1999). DN subsets were sorted from wild type C57BL/6 mouse thymi that were first depleted for CD3, CD8, TCRβ, TCRγδ, NK1.1, Gr-1, F4/80, B220, and Ter119. For “Series A”, cells were sorted into DN1 (or ETP, early T-cell precursor)(CD117+ CD44+ CD25−), DN2 (CD117+ CD44+ CD25+), DN3 (CD117− CD44− CD25+), and DN4 (CD117− CD44− CD25−). For “Series B”, cells were sorted into DN1, DN2, DN3a, and DN3b subsets using CD27 to split the DN3 subsets as described previously (Taghon et al., 2006), and isolating DN4 cells as CD44− CD25− CD24+. To examine DN subsets from Rag2−/− mutant thymus, first 10% of dissociated thymocytes were reserved to sort for DN2 = Thy1.2+ CD44+ CD25+ and DN3 = Thy1.2+ CD44− CD25+ stages. The remaining 90% cells were depleted of CD24, Gr-1, F4/80, and Ter119 and then sorted to separate Sca-1+Thy1− (precursor-enriched) and “pre-NK” = Sca1− Thy1+/− subsets (David-Fung et al., 2006). These “pre-NK” cells are strongly enriched for expression of NK cell genes such as perforin and Id2 (Wang et al., 2006; David-Fung et al., 2006; Wang et al., 1998).
To analyze the development of fetal hematopoietic precursors developing in OP9-control or OP9-DL1 culture, Lin- c-Kit+ CD27+ multilineage precursors (Lin = Ter-119, CD19, F4/80, Gr-1) were isolated by FACS and cultures were established exactly as described previously (Taghon et al., 2005). In these experiments, OP9-control cultures generated B cells predominantly while OP9-DL1 cultures generated T cells. Samples were generated from two independent large-scale time course experiments, one of which was a 10-day time-course also used for previous analyses (Taghon et al., 2005; Tydell et al., 2007), and another set in which cells retaining precursor phenotypes were re-sorted after the first two days, returned to OP9 coculture, and harvested after just 4 days overall. Some samples from these time-courses were also sorted again at day 4 to re-isolate cells retaining the Lin− c-Kit+ CD27+ precursor phenotype and to separate these from CD27− Mac-1+/Gr-1+ cells that had also been generated in the cultures.
To assess gene expression in later T-cell development, thymic cells from wild type C57BL/6 mice were sorted by FACS directly to isolate double positive (DP) and single positive CD4+ cells, while another aliquot of the thymocytes was first depleted of CD4, B220 and NK1.1 expressing cells and then sorted to isolate double negative (DN) and single positive CD8+ cells. The markers used were: DN = CD24+ CD4− CD8−, DP = CD4+ CD8+, CD4 SP4 = CD4+ CD8−, CD8 SP = TCRβ + CD24− CD4− CD8+. TCRγδ cells were obtained from TCRβ−/− thymocyte populations that were depleted for CD4 and CD8 prior to sorting for expression of CD3ε and TCRγδ. To assess non-T lineage expression in adult tissues, pro-B and pre-myeloid cells were prepared from Rag2−/− mutant bone marrow as described previously (Tydell et al., 2007).
Quantitative real-time reverse transcriptase-dependent PCR
Quantitative reverse transcriptase-dependent PCR (qPCR) was performed to measure gene expression in different samples of cDNA from the cells described above. Primers were designed to genes of interest using the PrimerExpress™1.5 (ABI) or Primer3 (http://fokker.wi.mit.edu/primer3/input.htm) programs. They were designed as far as possible to span introns of > 200 bp length, to avoid amplification of genomic DNA. Intron-exon information was verified from Ensembl (http://www.ensembl.org) and UCSC mouse genome assemblies. Primers utilized are shown in Table 3. Many primer sets used in this study have also been reported previously (Anderson et al., 2002b; Anderson et al., 2002a; Hernández-Hoyos et al., 2003; Yun and Bevan, 2003; Yui and Rothenberg, 2004; Anderson et al., 2004; Dionne et al., 2005; Taghon et al., 2005; David-Fung et al., 2006; Taghon et al., 2006; Franco et al., 2006; Taghon et al., 2007; Tydell et al., 2007). Standard curves were run for new primers to select sets with dilution curves giving close to the theoretical value of ΔCT 3.3 per tenfold dilution. Note the anomalous titration behavior of two primer sets, Stat5b and Thrap3 (Table 3), which should be treated with caution, but the great majority showed amplification consistent with 1.8–2.0 fold amplification per cycle. Samples were run in triplicate on an ABI Prism 7700 or 7900HT realtime PCR machine, and thresholds were set uniformly across all genes analyzed with a particular reference standard to calculate ΔCT = CT(gene) − CT(standard). Levels of expression were converted to units of β-actin or GAPDH expression using the formula: expression = (1.9)−ΔCt.
TABLE 3.
Primer sequences used for Quantitative PCR
| New1 primers | Gene tested | Alternative name | Primer Sequences (Forward; Reverse) | Comments Titration quality2 |
|---|---|---|---|---|
| + | 4632433K11Rik | AK014606 |
5′-ACAAGACTGACAGAGAAGACTTACAGAAA-3′ 5′-GGGCTAAAAGGACTTGGGAGTT-3′ |
|
| Actb | β-actin |
5′-ACACCCGCCACCAGTTC-3′ 5′-TACAGCCCGGGGAGCAT-3′ |
||
| Aff3 | LAF4 |
5′-TGACACCTCCCACCATGGA-3′ 5′-TTTCCTCCGTAACGCATTCC-3′ |
*(−) | |
| + | ARID1a | Smarcf1 |
5′-ACGTGGGCGTTAGACACCAT-3′ 5′-AATATTCCACAAGGAGCTCTAGCAA-3′ |
*(−) |
| + | Atxn2l |
5′-CTCTCCCTGACCCCTACAGA-3′ 5′-CAGGGACTTTCCCAGCTAT-3′ |
||
| + | B2m | β2-microglobulin |
5′-TCACTGACCGGCTTGTATGCT-3′ 5′-TGAGGCGGGTGGAACTGT-3′ |
Used in preliminary analysis only |
| + | Bahd1 | mKIAA0945 |
5′-TCCCGCTTGGAGGGATTC-3′ 5′-AGTCCCCTGCTCCATATTGCT-3′ |
|
| Bcl11a | CTIP1 |
5′-GTCTGCACACGGAGCTCTAA-3′ 5′-CACTGGTGAATGGCTGTTTG-3′ |
Tydell et al., 2007 | |
| Bcl11b | CTIP2 |
5′–GGGCGATGCCAGAATAGAT–3′ 5′–GGTAGCCTCCACATGGTCAG–3′ |
Primers from Tydell et al., 2007 | |
| + | Bcl6 |
5′-GTGGAGATGAAGCCTGTAGCA-3′ 5′-CACTTGAATCGTGCAGTGGTA-3′ |
**(−); Not expressed in thymocytes | |
| + | Brd3 |
5′-AAATGCAGGTTCCCAACAAGTG-3′ 5′-CGGCAATGACGGGTGTCT-3′ |
*(−) | |
| + | Brd4 |
5′-CCCTGAAGCCATCTACACTACGA-3′ 5′-ACCAGCAATCACGTCAACTTT-3′ |
||
| + | Cbx4 |
5′-AGCTGATGGGATATCGCAAGAG-3′ 5′-GTCTTGAAGCCCAGTCAGAACAT-3′ |
||
| + | Ccdc88c | Daple |
5′-CCTGGGTGAAAACTTTTGGA-3′ 5′-CTGGGGTCTATTTGCAGCAT-3′ |
|
| Cd3e |
5′-CGTCCGCCATCTTGGTAGAG-3′ 5′-ATTCAATGTTCTCGGCATCGT-3′ |
|||
| Cd3g |
5′-TGGAGAAGCAAAGAGACTGACA-3′ 5′-GCCATCCACTTGTACCAAATTC-3′. |
|||
| Cebpa | C/EBPα |
5′–CGGTCATTGTCACTGGTCAACT–3′ 5′-GGACAAGAACAGCAACGAGTACC-3′ |
*(++) Only one intron | |
| Cebpb | C/EBPb |
5′-GTTTCGGGACTTGATGCAATC-3′ 5′-CGCAGGAACATCTTTAAGGTGAT-3′ |
||
| Cebpd | C/EBPd |
5′-TCCACGACTCCTGCCATGTA-3′ 5′-TGAAGAGGTCGGCGAAGAGTT-3′ |
||
| Cebpg | C/EBPg |
5′-GCGCAGGTACATGTGAAGATT-3′ 5′-CTGCGACAGCTTGCTCATT-3′ |
||
| + | Cep170 | KAB1 |
5′-GCCGAGCATCCTGATCACTT-3′ 5′-AGAGAACTGAAAGACTGATGACAATAGC-3′ |
First thought to interact with KRAB domains |
| + | Csf2ra1 | GM-CSFRα |
5′-CACCGCGTCCTGTAACTCTT-3′ 5′-GCGCTCTGCCACTGGACCTCAAACT-3′ |
Data not shown; exp. declines DN1-4 |
| + | Ctbp1 |
5′-TGAGTCAGAGCCCTTCAGCTTT-3′ 5′-CAATGGACGCCTGCTCACT-3′ |
*(−) | |
| + | Cutl1 |
5′-CCATGGAAACTGTCTCTGTCTTCA- 5′-TGTCAGACTAGCTGGGTTGTTTG-3′ |
*(+) | |
| + | Dmtf1 | Dimp |
5′-ACACGGACGGGAATCTCATTC-3′ 5′-AATGCTTTGATCATCCTCAGAGG-3′ |
*(+) |
| Dtx1 | Deltex1 |
5′-GAGGATGTGGTTCGGAGGTA-3′ 5′-CCCTCATAGCCAGATGCTGT-3′ |
*(−) | |
| Elf4 | MEF (myeloid Ets) |
5′-CAGGCTCACCAAAACTGTGA-3′ 5′-TTGGTCAGCACCGTAGTCAG-3′ |
||
| + | Elk3 | ERP |
5′-CTGAGCAGAGCGCTGAGATAC-3 5′-TTCAGGATATCCGGGAAAGAG-3′ |
**(+) |
| + | Elk4 |
5′-GCTTTTCCAGTTTCCCTCTGT-3′ 5′-TCTGCAGGTCTGGAGAAAATG-3′ |
||
| + | Erg |
5′-GCCTCCCAATATGACCACAA-3′ 5′-TATTCTTTCACCGCCCACTC-3′ |
||
| + | ESR1 | Estrogen receptor 1 |
5′-GCTCCTGGACAGGAATCAAG-3′ 5′-CTTCTCCCTGCAGGTTCATC-3′ |
|
| Ets1 |
5′-AAAAGTGGATCTCGAGCTTTTCC-3′ 5′-CTTTCAAGGCTTGGGACATCA-3′ |
|||
| Ets2 |
5′-GCAAGGCAAACCAGTTATTCCT-3′ 5′-ACTTGTCAGAGAGTAGCTCCAGAAGAA-3′ |
|||
| + | Etv6 | TEL |
5′-TCCTGCTGCTGACCAAAGAG-3′ 5′-CTGGCTTGGTGTGGATAGAG-3′ |
|
| + | Fli-1 |
5′-ACAGACCAGTCCTCACGACTGA-3′ 5′-CTTTTGTTGAGGCCAGAGTTCAT-3′ |
||
| + | FoxJ3 |
5′-TGATCCCGGAAAGGGTTCTT-3′ 5′-AGGCCCGTTCTACGGATCTT-3′ |
*(−) | |
| + | Foxp1 |
5′–CTGGAAAACAGCCGAAAGAG–3′ 5′–GGTTGGAGGGGAAGGGCAGG–3′ |
||
| + | Foxp4 |
5′-CCCACAACCCTACCTTCTTCATC-3′ 5′-GGGACCGGAGAACTAATTTTCA-3′ |
*(−) | |
| Fus |
5′-CAGCAACGAGCTGGAGACTG-3′ 5′-TCTGGCTTAGGTGCCTTACACTG-3′ |
|||
| + | Gabpa |
5′-TGCACTGGAAGGCTACAGAAAA-3′ 5′-TTACCCAAACCACCCAATGC-3′ |
*(−) | |
| Gapdh |
5′-ACTCCACTCACGGCAAATTCA-3′ 5′-GCCTCACCCCATTTGATGTT-3′ |
|||
| Gata2 |
5′-ACCACAAGATGAATGGACAGAA-3′ 5′-GTCGTCTGACAATTTGCACAAC-3′ |
*(−) | ||
| Gata3 |
5′-GAGGTGGTGTCTGCATTCCAA-3′ 5′-TTTCACAGCACTAGAGACCCTGTTA-3′ |
Called “Gata3-endog” in Taghon et al., 2007 | ||
| Gata3 (alternate) |
5′-CCTGCGGACTCTACCATAAAA-3′ 5′-GTGGTGGTGGTCTGACAGTTC-3′ |
In coding region; Taghon et al 2007 | ||
| Gfi1 |
5′-AGCGTCGGAGAAGTCACTGT-3′ 5′-CAGGTCAGACCCAGCAAGAC-3′ |
|||
| Gfi1b |
5′-CCTTTGCCTGTGATGTCTGT-3′ 5′-ATGAACGCTTGAAGGCTTTG-3′ |
|||
| + | Gse1 |
5′-GGGATTGAGGCAATCTTTGA-3′ 5′-GGCTGAGGCTGTAGTTCTGC-3′ |
||
| HEBAlt |
5′-GTGCTTATCCTGTCCCTGGAATG-3′ 5′-TGGCTTGGGAGATGGGTAAC-3′ |
|||
| Hes1 | (Yun & Bevan) |
5′-TACCCCAGCCAGTGTCAACA-3′ 5′-TTCTTGCCCTTCGCCTCTT-3′ |
||
| Hes1 (alternate) |
5′-CCAAGCTAGAGAAGGCAGACA-3′ 5′-CGGTATTTCCCCAACACG-3′ |
Alternate primers, Yui & E.V.R. 2004; data not shown but similar | ||
| + | Hmga1 | HMG-I(Y) |
5′-AAAGTATCACCGGGTGGGTTTC-3′ 5′-CATTAAAGTGGGTGGAGCCAAC-3′ |
|
| + | Hprt |
5′-GAGCTACTGTAATGATCAGTCA-3′ 5′-ACCAGCAAGCTTGCAACCTT-3′ |
||
| Id1 |
5′-GGCGAGATCAGTGCCTTG-3′ 5′-AAGGGCTGGAGTCCATCTG-3′ |
**(+) | ||
| Id2 |
5′-GTCCTTGCAGGCATCTGAAT-3′ 5′-CTCCTGGTGAAATGGCTGAT-3′ |
*(+) Possible detection of genomic DNA? | ||
| Id3 |
5′-AGAGGAGCTTTTGCCACTGA-3′ 5′-TGGAGAGAGGGTCCCAGAGT-3′ |
**(−) | ||
| Ikzf1 | Ikaros |
5′-TCCCAAGTTTCAGGAAAGGA-3′ 5′-TCTGCTGTGCTCCAGAGGTA-3′ |
||
| Ikzf2 | Helios |
5′-CACCTCAGGACCCATTCTGT-3′ 5′-TGACAGCGTTCCTTGTGTTC-3′ |
||
| Ikzf3 | Aiolos |
5′-CCGAGATGGGAAGTGAGAGA-3′ 5′-CGCTTCTCACCGATGAATTT-3′ |
*(−) | |
| + | Ikzf4 | Eos; Zfpn1a4 |
5′-CCCAAACAGCCAACACTCTT-3′ 5′-TTATCCAGGAGCCGTTCATC-3′ |
|
| Il2rb | IL-2/15Rβ |
5′-ACGTCCATGCCAAGTCGAA-3′ 5′-GGAACGACCCGAGGATCAG-3′ |
||
| IL2-UP | Il2 flanking seq. |
5′-ACCTTGGGAGCTGAAATCCT-3′ 5′-TTTTGAGGGATCGCTAATGG-3′ |
Detects genomic DNA; Adachi & E.V.R., 2005 | |
| Il7ra | IL-7Rα |
5′-AGTCCGATCCATTCCCCATAA-3′ 5′-ATTCTTGGGTTCTGGAGTTTCG-3′ |
*(−) | |
| + | Jmjd3 |
5′-CCCTACCCCCAGCATCTATT-3′ 5′-GCCTAAGTTGAGCCGAAGTG-3′ |
||
| + | Klf13 |
5′-CGGGCTGCGAGAAAGTTTAC-3′ 5′-GAGCGTGCGAACTTCTTGTT-3′ |
||
| + | Klf15 |
5′-TTTTCCCGCTCAGATGAGTT-3′ 5′-GCGATGCACTTTGATGTGTT-3′ |
*(+) No expression in thymocytes | |
| + | Klf2 |
5′-ATGCACCTGAGCCTGCTAGT-3′ 5′-ATAGCACTGCCCTCTCCCTCT-3′ |
*(+) crosses tiny intron | |
| Lat |
5′-CTGTTGTCTCCTCTGCTCCTGT-3′ 5′-CTCACTCTCAGGAACATTCACG-3′ |
|||
| Lck |
5′-CTAGTCCGGCTTTATGCAGTG-3′ 5′-CCGAGGGAGTCTTGAGAAAAT-3′ |
|||
| Lef1 |
5′-ACCTACAGCGACGAGCACTT-3′ 5′-GGGTAGAAGGTGGGGATTTC-3′ |
|||
| + + |
Lima1 | Eplin |
a: 5′-AGGAGAGCTGCGTGGAGTGT -3′ 5′-GCAGTAGGAGCATCGGAAACAG-3′ b: 5′-AGGAGAGCTGCGTGGAGTGT-3′ 5′-TGCAGTAGGAGCATCGGAAAC-3′ |
**(−) *(−) |
| Mitf |
5′-CCCCAAGTCAAATGATCCAG-3′ 5′-CCTTAGCTCGTTGCTGTTCC-3′ |
Taghon et al., 2007 | ||
| + | Mlf2 |
5′-ACCAGGAGACGTCGGAGATG-3′ 5′-GCCCTATCCCGAATGTGATG-3′ |
||
| MLL1 | ALL-1 |
5′-TGCCCATAGCCCATCATCA-3′ 5′-TCTGTGAATGAGGCCGATCTG-3′ |
||
| MLL2 |
5′-CCCGGTCCCGAATCAAAC-3′ 5′-GAAGAGAACCACGGTATTTTGCAT-3′ |
|||
| + | Mta2 | Mta1l1 |
5′-CTGAAGACCCCTACCCAACTTG-3′ 5′-CCCGGTTAGCACTGGTTGTATAG-3′ |
|
| + | Mta3 |
5′-CGTGCCGTGGGAACATTT-3′ 5′-GTGTGTCCATGGCATGAAACA-3′ |
*(+) | |
| Myb | c-Myb |
5′-GAGCAGAAGAAGTTTCCCGATTT-3′ 5′-AGCGGGAATCGGATGAATCT-3′ |
||
| Mybl2 | B-myb |
5′-AAGGAGGTGCTCCGTTCTGA-3′ 5′-CCAGAGACTTGCGGACCTTCT-3′ |
||
| + | Myst3 | MOZ |
5′-CCAGTGCTGAAGGGAAAGAG-3′ 5′-ACAGATGGGGATTGGTTCAG-3′ |
*(+) |
| + | Nasp |
5′-TGTAGCCGAACTGGCACTGA-3′ 5′-TTCTACTGGCAATCATGGATACTGA-3′ |
||
| + | Ncor1 |
5′-AATCTCATCCCCTATTAAACCAAACC-3′ 5′-GTATATTACATGGAGTGCAAGAAACCA-3′ |
||
| + | Ncor2 |
5′-CGCACACCGGATCCTAGAAG-3′ 5′-TCCGCTTAAAGTACAAGATCAGCTT-3′ |
||
| Nfe2 |
5′-TATGCAGCTTTTGGCAGAGA-3′ 5′-GAGGGGCAGTGAAGACTGAA-3′ |
|||
| + | Pax1 |
5′-GGCAATGACCTTCAAACACC-3′ 5′-CTGTGAGAGGACAGCCCCTA-3′ |
No expression in thymocytes | |
| Pax5 |
5′-ACAGGACATGGAGGAGTGAATCA- 3′ 5′-CCTTGATGGGCAAGTTCCACTA-3′ |
Anderson et al., 2004; no expression in T | ||
| + | Per1 | Period |
5′-CCGAATACACACTTCGAAACCA-3′ 5′-CGAAACACATCCCGTTTGC-3′ |
|
| + | Pou2f1 | Oct1 |
5′-AACCACCAACCTGCAACCA-3′ 5′-TGCTGAGGTAGTTGCGTTAAAAGAT-3′ |
**(−) |
| + | Pou5f1 | Oct4 |
5′-GGAAAGGTGTTCAGCCAGAC-3′ 5′-CTCATTGTTGTCGGCTTCCT-3′ |
No expression in thymocytes |
| Pou6f1 | Brn-5 |
5′-CTGCAACTCCCATCCCAATC-3′ 5′-CGCAAACTCCCGGATCTCTTCT-3′ |
*(−) | |
| Ptcra | Pre-Tα, pTα |
5′-CTGGCTCCACCCATCACACT-3′ 5′-TGCCATTGCCAGCTGAGA-3′ |
||
| + | Ptma | Prothymosin α |
5′-GGCGTGCCCCACCAT-3′ 5′-CATTCTCTGCCTCCTCCACAAC-3′ |
|
| + | Rara |
5′-CTCCCAGGACCGCTTAACC-3′ 5′-GGAGCTGCAAGTCCCAAGT-3′ |
||
| + + |
Rbak |
5′-TAAGTTGGAGCAGGGAGAAGAG-3′ 5′-CAATTCCTTCTCTGGGGTCATA-3′ 5′-TCGTCATGCTACACTGTGCACTAC-3′ 5′-CCGGTGTGGACCCTTTGA-3′ |
Library-cloned new isoform Conventional isoform |
|
| + | Rnf149 |
5′-GGAATTGATGTCGATGCTGA-3′ 5′-GCCATGGGTCAATGCATATT-3′ |
||
| + | Rreb1 |
5′-TCACCACCAATGGGAACATG-3′ 5′-AACCGCCTGCGCTTCAG-3′ |
*(−) | |
| Runx1 |
5′-CTCGGCAGAACTGAGAAATG-3′ 5′-GGTGATGGTCAGAGTGA-3′ |
|||
| + | Runx2 |
5′-GCCTCCGCTGTTATGAAAAA-3′ 5′-TGGGGAGGATTTGTGAAGAC-3′ |
*(+) | |
| Runx3 |
5′-GGTTCAACGACCTTCGATTC-3′ 5′-GGTCCATCCACAGTGACCTT-3′ |
|||
| + | Saps1 | mKIAA1115 |
5′-TTCATTGGCTCAACGAAGAGA-3′ 5′-GGCTTAGGCGGATGATGTCA-3′ |
*(+) |
| Satb1 |
5′-CCACAAACACGGAGGTCTCT-3′ 5′-GCAATCCCTGAGTTCGGTTA-3′ |
|||
| + | Sbf1 |
5′-AGGAGCTGCTGGATGTGATT-3′ 5′-CCATGCTCAGAACATTGTGG-3′ |
*(+) | |
| Sfpi1 | PU.1 |
5′-CCCGGATGTGCTTCCCTTAT-3′ 5′-TCCAAGCCATCAGCTTCTCC-3′ |
||
| Sox4 |
5′-TCAAGGACAGCGACAAGATTC-3′ 5′-GCCGGTACTTGTAGTCAGGGTA-3′ |
|||
| SpiB-1 | SpiB, ex3-4 |
5′-CTGCAAGCCCTTCAGTTACC-3′ 5′-AAAGGCAGCAGTAGCAGGAT-3′ |
See Dionne et al. 2005; David-Fung et al 2006 | |
| SpiB-2 | SpiB, ex1-2 |
5′-CTTGCTCTGGAGGCTGCAC-3′ 5′-CCCCCATCTGAATCTGGGTA-3′ |
*(−); used in Yui & Rothenberg, 2004 | |
| + | Stat5b |
5′-GCTGTATCCGGCACATTCTGT-3′ 5′-GTTTGGTTGATCTGAAGGTGCTT-3′ |
**(+) Serious titration anomaly | |
| Tal1 | SCL |
5′-CTCACTAGGCAGTGGGTTCTTT-3′ 5′-GGACCATCAGAAATCTCCATCT-3′ |
*(−) | |
| + | Tbx21 | T-bet |
5′-AGGTGTCTGGGAAGCTGAGA-3′ 5′-ATTCGCCGTCCTTGCTTAGT-3′ |
**(+) |
| Tcf12 | HEB (canonical) |
5′-GAGAAGAAGACCGCTCCATGAT-3′ 5′-TGGCTTGGGAGATGGGTAAC-3′ |
||
| Tcf7 | TCF-1 |
5′-CAAGGCAGAGAAGGAGGCTAAG-3′ 5′-GGCAGCGCTCTCCTTGAG-3′ |
Intra-exon: detects N-term half of the HMG domain: “Tcf7.8” | |
| Tcf7 (alt primers) | TCF-1 |
5′-TGCTGTCTATATCCGCAGGAAG-3′ 5′-CGATCTCTCTGGATTTTATTCTCT-3 |
*(+) Crosses from exon III to IV: “Tcf7.3”; Taghon et al., 2005 | |
| Tcfe2a | E2A, Tcf3 |
5′-CAGATGGTGGCCTGGATACT-3′ 5′-CATCCCTGCTGTAGCTGTCA-3′ |
||
| + | Tfdp1 |
5′-GATAGGTGAATGGATCAGAGGTTAAGA-3′ 5′-CCTTGGAACTGGAGTCACAGACTT-3′ |
**(−) | |
| + | Thrap3 |
5′-GTTGTGCCGTTGCGAGAT-3′ 5′-CTTGGGCGTGTACTTTGGAT-3′ |
**(+) Serious titration anomaly | |
| + | Tle3 |
5′-TTGCGAAGAGACTGAACACAAT-3′ 5′-GCGTTCAACTCCGTCATGGT-3′ |
||
| Tox |
5′-CCATGGACCTGCCAGAGATC-3′ 5′-TTCTGCGTTCCCAATCTCTTG-3′ |
|||
| + | Trim25 | Efp |
5′-CGGAAAATTCGACACCATCT-3′ 5′-CCTTGCAATTTTGCAGCTTT-3′ |
|
| Trim44 |
5′-TCTGTGTCCTGTGTCCAGTCATT-3′ 5′-CAGTCCACCGGAATCTTTGC-3′ |
|||
| + | Tsc22d1 |
5′-AAACGCTTCCGTGAGACTTG-3′ 5′-TCACCGCATACATCAAATGG-3′ |
*(+) | |
| + | Unk |
5′-GTTGGGGCAGAGTACCTGAA-3′ 5′-ACGTCCTGTTTGGGTTGAAG-3′ |
||
| + | Yy1 |
5′-GCCAGAATGAAGCCAAGAAA-3′ 5′-GGTGTGCAGATGCTTTCTCA-3′ |
||
| + | Zbtb20 |
5′-CAGCCAAACAGAACTACGTCAA-3′ 5′-TGCTACACTGGTACGCTCTCA-3′ |
**(−) | |
| + | Zbtb48 |
5′-TGTCCCACATGTCACAAAAAGTT-3′ 5′-TCTCCTTGCGGAAGTAACACTTC-3′ |
**(+) | |
| + | Zbtb7a | LRF, Pokemon |
5′-CCCTACGAGTGTAACATCTGTAAAGTTC-3′ 5′-GTGGTTCTTCAGGTCGTAGTTGTG-3′ |
*(+) More sensitive due to shorter amplicon than for primers in Maeda et al. 2007 |
| + | Zfp1 | mKr1 |
5′-TGGAAGTGTGGAAGGCTGATG-3′ 5′-TTTCTTCAGTTGGTGGTTGGTTC-3′ |
*(−) |
| Zfp109 |
5′-GCTGCTCAGAGGAAGCTGTA-3′ 5′-CCCCAGTGAAAGGCATCTTA-3′ |
|||
| + | Zfp110 |
5′-ATGATGGAGAACTACAGCAACATGA-3′ 5′-TGCATTGGCCAGCAGTCTT-3′ |
*(−) | |
| + | Zfp238 |
5′-CGTCCTGTGATGAGAGTGATGT-3′ 5′-TGGCTCATACTGTACCCTGTTG-3′ |
*(+) | |
| + | Zfp28 | mKr5 |
5′-TGTACTCAACATCGGAGACTTCACA-3′ 5′-CTCCGTGACATCTACGATGACAG-3′ |
|
| + | Zfp287 |
5′-CCCAAGAAGACTGGGAACTG-3′ 5′-ACCATCCACGGCTCTTCTAA-3′ |
||
| + | Zfp316 |
5′-CCCGGGTATCTCCCACTACT-3′ 5′-TACACGGCCACATCTTCAAA-3′ |
||
| + | Zfp358 |
5′-CATCGGGTCCTGTTCCAG-3′ 5′-GGTCTCCATAAAGGGGCCTAA-3′ |
*(+) | |
| + | Zfp382 | KS1 |
5′-AAACCTGATATGATCCGCAAGT-3′ 5′-TCACCAAAACTTCCTCCTCTTC-3′ |
|
| + | Zfp384 | CIZ |
5′-GATTCCGCATGGTGAAGAGT-3′ 5′-GTTACCGCTACCTGCTTCCA-3′ |
|
| + | Zfp407 |
5′-TTCAGGGAGTCCTGCAGTTT-3′ 5′-TGATGAACTGGGAGCCTTCT-3′ |
||
| + | Zfp422 |
5′-CGAAGGGAATTCCGAGTTG-3′ 5′-AAATACCCCCTTTGGGCTTT-3′ |
*(+) | |
| + + |
Zfp426 |
5′-AAGCCAGACCTGAAATAAACCAGT-3′ 5′ ′-AGAGGCAATCTCCATTCCTCTTAA-3′ 5′-TCCTAGTCTGACCCCCTGGTT-3′ 5′-GCTTAGAGTCACATAGGTCCTCTTCA-3′ |
**(−); Known mRNA *(−); Novel mRNA |
|
| + | Zfp598 |
5′-CAGGTGGTCTTTGGGAAGAA-3 5′-TGCAAACACCTTTCCATCTG-3′ |
||
| + | Zfp748 | 2610014M12Rik |
5′-ACACCCGCCACCAGTTC-3′ 5′-TACAGCCCGGGGAGCAT-3′ |
|
| + | Zfp771 | DSC43 |
5′-GGGCAGCGAGGGGCACTT-3′ 5′-CACTTGGGACTGGCACCTGAATTGTTCC-3′ |
|
| Zfpm1 | FOG-1 |
5′-GCCTCACTGAGGGCCTATCC-3′ 5′-TCCTTAGCCAGCAGCTCTCATC-3′ |
||
| + | Zkscan1 |
5′-CAGGGCTCTGAGAACAGGAATGG-3′ 5′-CTGTTCCCGTTTCTCTCCAA-3′ |
||
| + | Zkscan5 | Zfp95 |
5′-CGTGGCTGTGATAGAGAGTATCCA-3′ 5′-GTGGCTCCGGGTGGTACC-3′ |
**(−) |
| + | Zkscan21 | Zipro |
5′-CCGTCACTCTCCTGGAAGAC-3′ 5′-GCCGTTCCTGAAGTTGACAT-3′ |
|
| + | Zscan22 |
5′-CCGTTCTGGTGGAGGATATG-3′ 5′-GGTGGCTCCAATTCATCTGT-3′ |
For original references to primer pairs that are not marked by “+”, see Anderson et al., 2002a, 2002b; Hernandez-Hoyos et al., 2003; Yui and Rothenberg, 2004; Anderson et al., 2004; Dionne et al., 2005; Taghon et al., 2005; Taghon et al., 2006; Franco et al., 2006; David-Fung et al., 2006; Tydell et al., 2007; and Taghon et al., 2007.
Notations about titration curves: all slopes fall within the range of -3.0 to -3.7 ΔCT per tenfold dilution except as indicated:
(−) slopes −2.6 to −2.99;
(+) slopes −3.71 to −4.1;
(−) slopes shallower than −2.6;
(+) slopes steeper than 4.1.
Each gene was interrogated in at least two independently sorted sets of samples (two sets = 1 “series”) for verification of gene expression patterns, and key genes were analyzed in four or more independently sorted series of samples. Sorted cell cDNA inputs were routinely normalized by expression of GAPDH and β-actin; in early analyses, expression of Hprt was also assessed (not shown). To test for genomic DNA contamination in cDNA samples, primers IL2-UP, from the upstream non-transcribed region flanking the mouse IL2 gene were used (Adachi and Rothenberg, 2005). In general, patterns of expression defined by changes in level >3 fold were highly reproducible among independent experiments [also see (David-Fung et al., 2006; Rothenberg et al., 2008)]. However, “absolute” levels of expression measured relative to standards could vary over time according to detailed conditions of measurement, with the worst differences (5–10 fold) associated with certain primer sets. Thus all analyses shown are based on simultaneous measurements across a full series of samples, and patterns with little variance cannot be accurately distinguished.
Clustering Algorithms
Correlation values between the expression profiles of all genes in a data set were calculated for each Series, and they were post-processed by sorting on Hierarchical (tree)(Baldi and Brunak, 2001) or Minimum Spanning Tree (MST) algorithms as described in Supplementary Methods. The values from qPCR analyses used were log transformations of averages of the two independent sets of data. Data from Series A and from Series B were analyzed separately.
A method of visual representation of the correlation data was developed as a matrix of the Pearson correlation values between expression patterns of each gene with those of each other gene. In this matrix, each value of entry ‘ij’ (row i, column j) reports the correlation between expression of gene ‘i’ and gene ‘j’, while ‘ji’ would show the correlation between the same two genes across a 45 angle of reflection. For 60 genes processed, e.g., there are 60 entries per gene with a value of 1 for each self-correlation, with these self-correlations marking the diagonal of the matrix. A grid of values relating to colors from blue to red were assigned: blue representing a maximum correlation of +1, yellow a lack of correlation, and red a complete anticorrelation at −1. This representation was applied to the genes in lists ordered either by hierarchically clustered sorting (shown) or by minimum spanning tree sorting (not shown). In the hierarchically sorted coldmap clusters, the red numbers above the dendrogram denoted the chronological order of the links drawn, with low numbers for links between genes or groups of genes with very similar expression patterns and high numbers between groups of genes with more distinct expression patterns. Cluster or branch-cutting was based on these link-orders.
The minimum spanning tree (MST) algorithm comes from the cluster analyses of Xu and Olman (Olman et al., 2003; Xu et al., 2002); where points are ordered so that the sum of their linkages is minimized. This provides a way to view the data as a fully-connected graph with the data points as the vertexes and the distances as the edges. A tree can be obtained from this graph, such that each vertex is visited once and the sum of the edges is minimal. Details of the algorithms used for MST and hierarchical clustering are presented in the Supplementary Material.
To visualize the patterns represented by each cluster as well as their relationships, Self-Organizing Matrix (SOM) analysis (Tamayo et al., 1999) was also applied to the data for each Series separately. Log-transformed expression measurements for each gene across the DN subsets were normalized to a row mean of 0 and then organized into 10–12 clusters by GeneCluster2 software (http://www.broad.mit.edu/cancer/software/genecluster2/), using two-row and three-row organizations. The configurations presented explain >84% of the gene expression variance in each Series.
RESULTS
A global search for transcription factor genes expressed in early T-lineage cells
Most factors currently known to play a role in early T-cell development have been discovered indirectly, either by serendipity or by cis-regulatory analysis of genes expressed by mature T cells. To search more broadly for transcription factors or transcription factor variants that act during early T-cell development, we took a gene discovery approach based on de novo cloning.
As a source enriched for cDNAs expressed during the choice of a T-lineage fate, we used a macroarray of two cDNA libraries prepared from immature thymic cells whose development beyond the point of commitment was blocked by a “severe combined immune deficiency” (scid) mutation in the Prkdc gene (Anderson, 1999). These populations are ~102 fold enriched, relative to normal thymus, for immature T-lineage cells in stages between entry into the thymus and commitment (Fig. 1a). This master arrayed library has been a valuable source of stage-specific cDNA clones for previous studies (Anderson et al., 1999; Chen et al., 2001; Tydell et al., 2007). To recover candidate transcription factor coding sequences, we screened filters spotted with the arrayed library iteratively by hybridization with specific probes for diverse transcription factor domains. Probes were made to detect canonical DNA binding domain types or protein-protein interaction domain types (Table 1), as described in Materials and Methods. Clones significantly reactive with these probes were recovered from the arrayed archive and sequenced. Sequences predicted to encode regulatory factors, chromatin modifying proteins, and potentially relevant signaling molecules were submitted to GenBank as summarized in Table 2. Diverse splice isoforms of known genes (e.g. Tfdp1, Rbak, and Zfp426) and many genes predicted to encode transcriptional repressors (KRAB and SCAN domain Krüppel-type zinc finger proteins) were identified from the SCID thymus cDNA library.
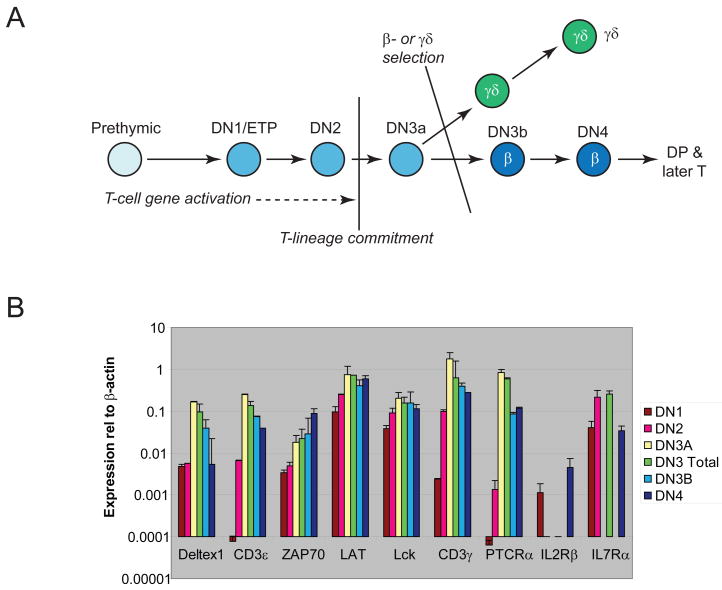
Figure 1.
Landmarks for early T-cell lineage development. (A) Diagram of stages of early T-cell development. (B) Expression of T-cell differentiation genes at successive stages of T-cell development. Samples are from “Series B”, except that Il2rb and Il7ra are shown for samples from Series A. For primer sequences, see Table 3 and (Taghon et al., 2007; Franco et al., 2006). Gene expression levels are expressed in units relative to β-actin measured in the same samples.
Expression of the newly identified transcripts was monitored in purified immature thymocytes from wildtype mice by qPCR, together with expression of known T-lineage differentiation genes and previously identified T-lineage transcription factors, as described in the following sections.
Stages of developmental activation of T-cell genes
Cells in the first stages of T-cell development that enter the thymus are diagrammed in Fig. 1A. These stages are defined as “double negative” (DN), as they lack mature T-cell markers CD4 and CD8. In order of maturity, these stages are termed DN1 (or early T-cell precursor, ETP), DN2, DN3a, DN3b, and DN4. Progression through these stages is profoundly dependent on Notch/Delta signaling. Yet the maturation process is independent of the T-cell receptor (TCR) until after the DN3a stage. At that point the cells arrest at a developmental checkpoint and cannot proceed further until successful rearrangement of TCR gene segments occurs. This conditional arrest point is known as the “β-selection checkpoint”. During the initial stages of intrathymic development, T-cell precursors retain certain alternative developmental potentials. Precursors may develop along natural-killer and/or dendritic cell lineage pathways. However, by the DN3a stage, the cells are committed to some form of T-cell fate.
Because T-lineage differentiation occurs continuously in mammals throughout life beginning at mid-gestation, the normal thymus is in a dynamic steady state in vivo. To obtain a virtual timecourse of early T-cell development for gene expression analysis, we sorted different subsets of immature mouse thymocytes from postnatal, weanling mice, defining developmental stages based on expression of the growth factor receptor c-Kit and markers CD44, CD25 (Ceredig and Rolink, 2002), and where appropriate, heat-stable antigen (CD24) and CD27 (Taghon et al., 2006). Four independent series of samples were prepared from normal, young adult mice. DN1, DN2, total DN3, and DN4 cells were collected in each of the first two sets (“sets 1 & 2”, Series A), whereas in the second two sets (“sets 4 & 5”, Series B) the DN3 cells were further subdivided into DN3a (pre-selection) and DN3b (the first stage past β-selection).
The timing of major positive regulatory changes that confer T-lineage identity was revealed by expression of known T-cell differentiation genes, such as those encoding TCR/pre-TCR complex components CD3ε and CD3γ, kinases Lck and ZAP70, the specific linker LAT, and the surrogate TCR chain Ptcra, as shown in Fig. 1B. All these genes undergo substantial increases in expression from DN1 to DN2 to DN3 or DN3a stage, with only ZAP70 expression continuing to increase further beyond the DN3a stage. The most strictly T-lineage-specific of these genes, CD3ε, CD3γ, and Ptcra, show a particularly steep upregulation between the DN1 and DN2 stages. In corresponding subsets of fetal thymocytes, the major increase in expression of each T-lineage gene occurred similarly from the DN1 to the DN3 stage. One minor difference was that the DN1 populations in those rapidly-developing cohorts appeared to show slightly higher T-lineage gene expression than adult DN1 stage cells (David-Fung et al., 2006)(M.A. Yui, data not shown). Because Notch signaling is critical for T-cell specification, the samples were also assayed for expression of the Notch target gene Deltex1 (Dtx1). Deltex1 is clearly upregulated in DN1 stage cells as compared to prethymic precursors (Tydell et al., 2007) and is further upregulated during the DN2 to DN3 transition. Its pattern of upregulation, however, fails to parallel the T-lineage-specific genes in their initial increase from DN1 to DN2 (Fig. 1B). Thus additional regulatory inputs besides Notch signaling are likely to be needed to set the timing for upregulation of T-lineage genes.
Definition of the regulatory gene set
The regulatory factor set that we assembled for expression analysis included the new candidate genes and about 20 additional transcription factor genes already implicated in T-cell specification by direct genetic or biochemical evidence (Rothenberg et al., 2008; Anderson, 2006; Murre, 2005; Rothenberg and Taghon, 2005; Staal et al., 2001). The total set of genes assayed is listed in Table 3. Genes encoding factors with a close structural relationship to known T-cell transcription factors were included in the set as well, because they might contribute partially redundant or competitive activities.
Many of the newly identified candidates encoded zinc finger factors, often for which little annotation is available. These included both Krüppel-type C2H2 zinc finger factors, likely to bind DNA directly, and RING finger factors, likely to mediate ubiquitin E3 ligase functions and/or other protein-protein interaction functions. Several Krüppel-type zinc finger factors were predicted to be obligate repressors, based on the presence of KRAB and/or SCAN domains in their sequences (Collins et al., 2001), e.g. Rbak, Zfp1, Zfp28, Zfp109, Zfp110, Zfp287, Zfp316, Zfp382, Zfp426, Zkscan1, Zkscan5, Zscan21, and Zscan22. Four transcripts encoded Zn finger factors with BTB domains, i.e. Lrf (Zbtb7a), Zbtb20, Zbtb48, and Zfp238.
Other genes were expected to encode transcriptional modifiers that work without direct DNA binding (prothymosin α = Ptma), or contribute to chromatin remodeling (Jmjd3, Myst3, Mta2, Mta3, NCoR1, NCoR2, SBF1) either to activate or to repress gene expression. Some genes encoded members of known transcription factor families that had not individually been implicated in T-cell development before, such as the POU-homeodomain factor Pou6f1 (Brn-5) and the Ikaros relative Eos (Ikzf4). Transcripts encoding novel isoforms of Rbak, Zfp426, and the winged-helix transcription factor TfDp1, were also identified and later specifically assayed, although these did not reveal a different pattern from the expression of the canonical forms.
These novel regulatory candidates were monitored in parallel with the known essential factors (Rothenberg & Taghon 2005) E2A (Tcfe2a) and its partially redundant relative HEB (Tcf12), GATA-3 (Gata3), Gfi-1, TCF-1 (Tcf7), Ikaros (Ikzf1), Myb, Runx1, and PU.1 (Sfpi1), and their relatives GATA-2, Gfi1b, LEF-1, Mybl2, Runx2, Runx3, and Spi-B. We monitored Ets1 and Ets2, which may be implicated in TCR gene expression, and other Ets family members associated with stem-cell, T-cell, or NK cell function. Also analyzed were other members of the Ikaros family and members of the helix-loop-helix inhibitor (Id) family. As landmarks for differentiation in the same samples, we assayed genes encoding the T-cell receptor signaling component CD3ε; the Notch target gene Deltex1; and the growth factor receptors IL-7Rα (CD127), IL-2Rβ (CD122), and GM-CSF-Rα (Csf2ra) to track changes in the responsiveness of the cells to different environmental signals (Fig. 1B and data not shown).. In all, 80 different candidate regulatory genes were assayed in sets 1 & 2 (series A), and 56 candidate regulatory genes were assayed in sets 4 & 5 (series B). In a review (David-Fung et al., 2006) we previously described a subset of the data from series B, for 19 genes, which is also included in this analysis for context.
The great majority of the regulatory genes were detectably expressed in at least one developmental stage at levels that were calculated to be above 10−2 times β-actin copy number. For cells with the overall RNA content of immature thymocytes (~1 pg/cell), this is consistent with a lower boundary of about 3 × 10−3 times β-actin level as the threshold for expression of at least one copy per cell in the population. The comparability of series A and series B was checked by assaying 13 transcription factor genes with important functions or well-marked, distinctive expression patterns in both series: these genes encoded Ets1, Ets2, FOG-1 (Zfpm1), GATA-3 (Gata3), HEB (Tcf12), Id2, LEF-1, Pou6f1, PU.1 (Sfpi1), Runx1, Runx3, SpiB, and TCF-1 (Tcf7). In general, there was excellent agreement between the overall patterns of expression measured across the developmental stages. Also, for more than half of these genes there was also excellent agreement with the absolute measured expression levels (within a factor of 2 or less).
Expression of landmark transcription factor genes and new candidates in normal early T-cell development
Figures 2–4 present analyses of the expression patterns of the regulatory genes we monitored in the immature DN T-lineage cells of series A and series B. In Fig. 2, actual qPCR results for the genes are shown with genes in alphabetical order. Data from Series A samples are joined by solid curves, data from Series B samples by broken-line curves. All the expression levels measured in Fig. 2 are presented in units relative to β-actin in the same samples. Several methods were used to group these regulatory genes based on their patterns of expression during T-cell specification. Fig 3 shows the correlations between expression patterns of different genes within series A and series B. Here, genes are listed in an order determined by hierarchical clustering (shown left of y axis), and the plot uses a color scale (“cold map”) to show pairwise correlations between expression patterns of each gene (y axis) with those of every other gene (x axis). Gene expression patterns were also organized by the Minimum Spanning Tree method of Xu et al. (Xu et al., 2002), which discerned similar groups (G. Buzi, data not shown). Finally, Figs. 4A, B show representative self-organizing matrix (SOM) clusterings of the normalized expression patterns for genes analyzed in each series (membership of clusters listed in Supplementary Table 1). The boundaries between clusters are somewhat flexible, but Fig. 4C–F show actual expression levels of genes that typify common patterns or features of regulatory factor expression: “rising”, “declining”, “DN3 peak”, and “legacy and loss”.
Figure 2.
Regulatory gene expression patterns through early T cell development. Gene expression levels are shown for the putative regulatory genes in this study, arranged alphabetically in order of gene names. Gene expression levels are measured by qPCR using primers shown in Table 3, and the geometric means of values determined from the two independent sets within each series are plotted on a log10 scale relative to the expression of β-actin in the same samples (0 = log101 = level of β-actin). Data from both “Series A” and “Series B” are combined in these graphs, with Series A measurements joined by continuous line spline curves and Series B measurements joined by broken-line spline curves. One group of Series B measurements that were determined separately from all the others is indicated by dotted curves (Mitf, Sox4, Bcl11a, Cebpa, and one set of Zfpm1). Where the same genes were analyzed in both series, both sets of values are shown. In some cases (SpiB-1, SpiB-2; LRF, LRF*), the same genes are assayed with different primer sets. LRF* = primers from Maeda et al. (Maeda et al., 2007). GATA-3-r = GATA-3 reassayed on the same Series B samples with the same primers, but by a different investigator > 1 yr after initial Series B measurement (David-Fung et al., 2006); included to show reproducibility. To align the Series A samples (DN1, 2, 3, and 4) with the Series B samples (DN1, 2, 3a, 3b, and 4), values were plotted on an x/y plot in which DN1 was considered “1”; DN2 was considered “2”; DN3a was considered “3.0”; DN3b was considered “3.7”; DN3 (unseparated), which is mostly DN3a, was considered “3.2”; and DN4 was considered to be “4.5”. Thus, the β-selection checkpoint is represented by “3.5”. The same convention is followed in Figs. 4C–F and 6 as well.
Figure 4.
Clusters of genes defined by expression patterns and pattern elements. (A, B) Self-organizing matrices of expression patterns of genes assayed in Series A (A) and Series B (B). Numbers of clusters and 2-row orientations were chosen to account for at least 84% of the overall variance of gene expression in each Series. Cluster number (c) and number of genes in the cluster are indicated at the top of each panel. Names of members of each of the clusters are listed in Supplementary Table 1; examples are shown in (C–F). (C) Genes in this study showing an increase from DN1 to DN3 of >3 fold. Data extracted from Fig. 2. (D) Genes in this study showing a decrease from DN1 to DN3 of >3 fold. (E) Genes with patterns featuring a major peak in expression at DN3 (DN3a) followed by a substantial decline. (F) Examples of genes showing steady “legacy” expression from DN1 to DN3, including all those with substantial decreases at β-selection. Note that distinctions between the groups in (C)–(F) are not clearcut. Certain genes, e.g. SpiB and Id2, have a “legacy” element of expression but may appear more appropriate for different categories when different primers are used (SpiB-1 vs. SpiB-2, DN3 peak) or in different sample series (Id2, declining). In some of these ambiguous cases the same gene data are shown in two panels to illustrate different pattern features, e.g. Id2, Zfp110 and Zfp287). Additional evidence for these pattern groupings comes from other independent measurements (Rothenberg et al., 2008; Taghon et al., 2006; Yui and Rothenberg, 2004).
Figure 3.
Hierarchical clustering of putative regulatory genes based on pairwise correlations. (A) Genes tracked in Series A. (B) Genes tracked in Series B. “Cold maps” are shown to depict the correlation among gene expression patterns within each series, with blue representing full correlation and red representing full anticorrelation as described in Materials and Methods. All genes analyzed in each series are listed in the same order along both the y axis and the x axis of each plot, with the origin at the upper left, so that the diagonal represents the perfect correlations of each gene’s expression pattern with itself. The order of the genes on each axis is set by hierarchical clustering of expression patterns as shown at the left of each matrix, with red numbers indicating the closeness of relationship (low numbers: close; high numbers: remote; see Supplementary Methods). Note that in this hierarchical clustering order, the clusters drawn should be considered to have rotational symmetry. Blocks of color help to visualize genes that are regulated in parallel (blue blocks) or in significantly opposing ways (orange/red blocks) through early T-cell development. Three major groups of genes with decreasing, sustained (“legacy”), and increasing expression are identified with lines over particular blocks in Panel A. Series B was used in a targeted way to assay many genes known from sources other than this gene discovery study, and so panel B is more biased toward genes with diverse, highly inflected expression patterns.
Genes with minimally changing RNA expression: legacy genes
Each clustering method showed that the majority of genes we monitored were expressed in a sustained, minimally changing (<3 fold) pattern from DN1 to DN3 stage. This inter-correlated group is the most dominant feature shown in Fig. 3 (especially 3A), and encompasses not only the genes with “legacy” patterns shown in Fig. 4F but also many genes not shown in Fig. 4 and genes with very low amplitude DN3 peaks in Fig. 4E. Although many of these stably expressed genes are poorly characterized, it must be emphasized that this group also includes many of the factors known to be essential for T-lineage specification from genetic evidence. It includes Myb, MLL1, Ikaros (Ikzf1), Gfi-1, GABPα, Stat5b, Oct1 (Pou2f1), and TOX, all of which have critical roles in T-cell development and/or in T-cell differentiation gene expression [rev. in (Rothenberg et al., 2008; Rothenberg and Taghon, 2005)(Aliahmad and Kaye, 2008; Yao et al., 2006; Ernst et al., 2004; Xue et al., 2004; Wilkinson et al., 2002; Chen and Kuo, 2001; Ullman et al., 1990)]. The stably expressed group contains at least as many known contributors to T-cell development as the group of genes that are sharply upregulated (next section). Furthermore, in terms of the provision of regulatory inputs for other genes in the T-lineage program, genes such as FOG-1 (Zfpm1), Gse1, Yy1, and Zfp598 contribute stably as well, which show slightly different expression trends from most “legacy genes” (Supplementary Table 1) but with very small changes in amplitude. The prevalence of regulatory or candidate regulatory genes showing stable expression in early T-cell development contrasted remarkably with the massive changes in T-lineage gene expression and developmental potential that occur across this interval (cf. Fig. 1B).
Upregulated genes
A surprisingly small group of genes showed definite upregulation (Fig. 4C) during DN1 to DN3 progression. Most of the regulatory genes in this group were previously known to be involved in T-cell specification, i.e., Gata3, Ets1 and Ets2, Tcf12, Tcf7, Lef1, Satb1, Ikzf3 (Aiolos), and Id3. Of the less-studied factors added to this group, Cutl1, Hmga1, Jmjd3, Mlf2, Mybl2, Pou6f1, Ptma, and Zfp382 also appeared to increase in expression with the onset of lineage-specific gene expression. However, most of these upregulated factors increased relatively little across the DN1 to DN3 progression, with gross increases of <10-fold. Some of them peaked at DN3 and decreased again after β-selection (DN3b and DN4), a pattern feature discussed further below, for an even weaker net increase.
Within the “rising” group, the timing of upregulation of different sets of regulatory genes defined at least three different positive regulatory events. First, HEBalt (a promoter-use variant of Tcf12 (Wang et al., 2006)) and Bcl11b underwent the most dramatic portion of their upregulation at the DN1 to DN2 transition, as described elsewhere (Tydell et al., 2007). At the next transition, from DN2 to DN3, Ets2, Lef1, and Pou6f1 were sharply upregulated, apparently in parallel. Then, from DN3 to DN4 stage, Ikzf3 and Tbx21 were upregulated. Id3 appeared unique in its induction in early DN3 stage (DN3a) to a peak in newly-selected DN3 cells (DN3b), followed by a striking decline in the DN4 stage. The distinctive pattern seen for Id3 could reflect its preferential use in the TCRγδ lineage (Taghon et al., 2006). These results, in accord with some previously reported data (David-Fung et al., 2006; Rothenberg et al., 2008; Taghon et al., 2006; Tabrizifard et al., 2004; Morgan et al., 1997), emphasize that T-lineage specification occurs in an ordered cascade.
Genes downregulated during specification
A prominent feature of the DN1 to DN3 progression was the downregulation of hematopoietic transcription factor genes Gfi1b, Gata2, Bcl11a, and of genes encoding several members of the Ets, Helix-loop-helix, C/EBP, and Runx transcription factor families. Many of these downregulated genes (Sfpi1 (PU.1), Tal1 (SCL), Id2, Cebpa, Bcl11a) are functionally implicated in non-T cell hematopoietic differentiation programs (Herblot et al., 2000; Ikawa et al., 2001; Anderson et al., 2002b; Liu et al., 2003; Lefebvre et al., 2005; Laiosa et al., 2006). Another “declining” gene, Erg, was previously identified (Anderson, 2006; Anderson et al., 1999) and has recently been determined to be crucial for stem-cell maintenance (Loughran et al., 2008). Genes encoding many factors not previously associated with early T-cell precursors also fell into this group, e.g. Aff3, Elk3, Etv6, Klf2, Lima1, Rnf149, Tsc22d1, and Zfp316 (possibly also Thrap3). Downregulation of these factors again occurred in distinct stages. Some transcripts diminished mostly between the DN1 and DN2 stage, including Tsc22d1, Lima1, Gata2, Cebpa and Cebpb, while others fell most abruptly only after the DN2 stage (e.g., Tal1, Sfpi1, Gfi1b, Runx2 and Runx3, and Elk3). The most dramatic shutoffs of gene expression were seen for PU.1 (Sfpi1) and SCL (Tal1), which fell at least two orders of magnitude by the DN3 stage.
Multigene downregulation after the DN3 stage
The DN3 stage is when T-lineage commitment first appears to be complete, when proliferation stops, and when survival first becomes dependent on TCR signaling (β-selection). We expected most discontinuities to occur between DN2 and DN3, with the T-cell identity stabilizing thereafter. However, a more commonly observed pattern element was a rapid decline in expression after the DN3 stage. This was seen both for genes that had been stably expressed from the DN1 stage (Fig. 4F) and for some genes that were newly upregulated at the DN2 to DN3 transition (Fig. 4E). Downregulation ranged from a slight decrease for some genes to virtual extinction of others. The Notch-induced gene Deltex1 and the PU.1 relative SpiB showed abrupt, transient peaks of expression at the DN3/3a stage followed by a sharp decline (Fig. 4E). Other genes were downregulated sharply after more gradually reaching the high point in DN3/DN3a cells. These included Runx1, Zfp287, and the Ikaros family member Ikzf4 (Eos), as well as Hes1 and HEBalt, which were also reported previously (Wang et al., 2006; Tan et al., 2005; Yui and Rothenberg, 2004). Even among genes that were also downregulated between the DN1 and DN3 stages, Zbtb20, Klf2, Runx2, and even Id2 all showed their steepest downregulation after β-selection. The shutoff of expression after the DN3a stage was the most conspicuous feature of the expression pattern for the Ets-family gene Erg. In the cases of SpiB, HEBalt, and Erg, at least, this downregulation appears to be permanent (see below) (Rothenberg et al., 2008; Anderson et al., 1999). These results suggest that although developmental commitment appears to occur at the DN3 stage before selection, many “legacy” regulatory genes are shut off only afterwards, apparently triggered by β- or γδ-selection.
Genes specifically expressed in thymic natural killer cells
One regulatory gene, Tbx21 (T-bet), showed an apparently anomalous pattern of expression in these analyses. Although it was recovered from our macroarrayed SCID thymus library in multiple clones, qPCR showed this gene to be expressed at very low levels in the DN1-DN3 T-cell precursor stages, with most of its expression beginning only as the cells progressed to DN4. Since there is no β-selection in SCID thymocytes, this appears inconsistent with the apparent high prevalence of Tbx21 transcripts in the SCID thymus cDNA library. Fig. 5 shows that this is because Tbx21 is actually expressed selectively in thymic natural killer (NK) cells, a population that is highly enriched in SCID thymocytes (Telfer and Rothenberg, 2001; Anderson et al., 1999; Wang et al., 1998). As in normal DN thymocytes, the immunodeficient cells showed increasing Hes1 expression in DN2 and DN3 cells, a sharp rise in Ets2 expression in the DN3 stage. As expected (Anderson et al., 1999), PU.1 (Sfpi1) was expressed highly in a Rag-deficient population that contains the earliest T-cell precursors (Sca-1+ Thy-1low), decreasing in DN2 and DN3 cells and in differentiating NK cells. By contrast, Tbx21 expression was highly enriched in the pre-NK population, a subset which is also marked by its very high expression of IL2rb (Fig. 5). High expression of Tbx21 was also confirmed in mature TCRγδ cells (see below), in agreement with others (Yin et al., 2002).
Figure 5.
Gene expression in thymocyte subpopulations with a genetic block to β-selection: early T vs. NK lineage gene expression. Realtime qPCR measurements are shown for subpopulations of thymocytes from B6. Rag2−/− mice, sorted as described previously (David-Fung et al., 2006; Anderson et al., 2002b; Wang et al., 1998). True DN1 cells are very rare in these thymus populations, but the Sca-1+ Thy-1low subset contains T-lineage precursor activity while pre-NK cells (Sca-1− Thy-1+/− CD24−) are marked by strong perforin expression (David-Fung et al., 2006). Note distinctively high Tbx21 and Il2rb expression in the pre-NK cells.
Discordant or compensatory expression within transcription factor families
Ets, Runx, Helix-loop-helix, and Ikaros family transcription factors are all implicated in stage-specific T-cell development and gene regulation. Our results, however, showed that these families are continuously well-represented at all stages in early T-cell development. It is the balance among family members that undergoes stage-specific developmental change. Fig. 6 (extracted from the data in Fig. 2) shows detailed patterns of expression for transcription factors of some of these families. Some of the strongest changes in expression occur, in opposite directions, among different members of the same structural families. Most dramatic are the expression patterns in the Ets factor family. While PU.1 (Sfpi1) plunges, SpiB spikes and then falls; and while Elk3, Etv6 (TEL), and Erg decrease, Ets1 and Ets2 increase (Fig. 6A). Concomitantly, as Runx2 and Runx3 decrease, Runx1 increases (Fig. 6B). Ikaros family members Ikaros (Ikzf1) and Helios (Ikzf2) are expressed fairly stably, but Aiolos (Ikzf3) dips and then rises while Eos (Ikzf4) peaks and falls (Fig. 6C). Note also that even though Lef1 and Pou6f1 are among the genes most sharply induced at the DN2 to DN3 transition, their respective relatives Tcf7 and Pou2f1 (Oct1) are also being expressed in a more sustained pattern, at apparently higher levels (cf. Fig. 2). The bHLH transcription factor families including SCL and the E proteins are also highly dynamic, as are their inhibitors (Wang et al., 2006)(cf. Fig. 2).
Figure 6.
Differential regulation of different members of the same transcription factor families. Gene expression data, extracted from Fig. 2, are shown for: (A) Ets, (B) Runx, and (C) Ikaros family members, and (D) for genes predicted to encode KRAB and SCAN domain-containing zinc finger factors.
As many “new” KRAB and SCAN domain zinc finger coding genes were identified in our screen, and are possible candidates for negative regulatory components of the commitment mechanism, it was also of interest to determine how these predicted repressors might be regulated. As shown in Fig. 6D), these genes also showed a mixture of expression patterns: many were expressed stably and at relatively high levels, while others, including the SCAN domain factor Gfi1b, were downregulated at the DN2-DN3 or later transitions. None of those identified were substantially upregulated during specification, however.
Similar or identical target sequences are thought to be bound by multiple members of the Ets, bHLH, Ikaros, Tcf/Lef, and Runx families. If nuclear concentrations of these transcription factor proteins follow the patterns of their RNAs, then these data imply that Ets, Runx, bHLH, and Ikaros binding sites in target genes may be continuously occupied throughout early T-cell development, but by shifting combinations of factors within these families.
Lineage specificity and kinetics of induction in vitro
The evidence presented so far suggests that stage-specific positive and negative regulatory changes during the DN1 to DN3 progression are superimposed on a rich background of gene expression continuing from the earliest stage through commitment. In this context, we revisited the question of which regulatory genes in our study might be T lineage-specific. Not only genes upregulated during commitment but also some of the genes with seemingly stable expression could be candidates. Most cells in the earliest stage of adult mouse T-cell development, the DN1 stage, have already been acted upon by the thymic microenvironment and are already distinct from multilineage prethymic precursors, e.g. with respect to expression of Dtx1, Gata3, Hes1, and Tcf7 (Heinzel et al., 2007; Sambandam et al., 2005; Tan et al., 2005; Tydell et al., 2007). Thus we investigated whether the stable expression of regulatory genes from DN1 to DN3 stage was actually continuing from prethymic precursors or was an immediate response to thymic signals. For this, we made use of an in vitro differentiation system in which a cohort of prethymic precursors can be traced over time as they enter the T-cell pathway (Schmitt et al., 2004b; Schmitt and Zuniga-Pflucker, 2006; Taghon et al., 2005). The T-cell program is triggered in hematopoietic progenitor cells by co-cultivation with stromal cells that have been forced to express a Delta-type Notch ligand. In the absence of the Notch ligand, or at reduced Notch signaling intensity, these same stromal cells support differentiation into B (and some NK) cells but not T cells (Schmitt et al., 2004a). Thus a comparison of the differentiation of the same precursor cells on stromal cells with and without Delta reveals not only discontinuities from prethymic input cells but also differences between the T and B-cell programs.
In two independent series of time course samples, we tracked genes representative of each expression pattern. The gene expression measurements from one well-characterized OP9 culture time course (Taghon et al., 2005; Tydell et al., 2007) are shown in Fig. 7. Similar results were obtained in a second timecourse of four days overall, and in samples re-sorted from the two timecourses at two or four days of culture (data not shown). B-cell specification was tracked by upregulation of the Pax5 gene. In these samples, the Notch target gene Dtx1 and previously characterized T-cell specific regulatory genes Gata3, Tcf7, and Bcl11b as well as the T lineage-specific differentiation gene Cd3e were upregulated only under T-cell conditions as previously reported (Taghon et al., 2005; Tydell et al., 2007). Those data are included here again as a standard for T-lineage specificity. None of the other genes in this study showed comparably strong T-lineage specificity. A group of other genes did show a weaker but detectable preference for T-cell conditions, with either accelerated or more extensive upregulation. These genes included Ets2, Foxp4, Myb, Rbak, Pou6f1, Satb1, Tcf12, and Tle3 (Fig. 7). In addition, “legacy pattern” representatives Mta3 and Stat5b appeared to be sustained better from stem-cell levels in the T-lineage conditions than in the B-lineage conditions.
Figure 7.
Cell type specificity of gene expression induced in hematopoietic progenitors on OP9 stroma. Gene expression levels are shown in units relative to β-actin at successive two-day time points, starting from multipotent c-Kit+ CD27+ Lin− hematopoietic precursors from fetal liver. Cells were cultured for up to ten days on OP9-control stroma, which does not permit T-cell development (blue shaded bars), and on OP9-DL1 stroma, which promotes T-cell development (brown to yellow shaded bars). Black bars show gene expression in input cells. Cultures were harvested for RNA preparation every two days. At day 4, cells from the cultures were collected and those retaining a precursor phenotype (c-Kit+ CD27+ Lin−) were repurified before being placed in fresh cultures for the remaining 2–6 days (Taghon et al., 2005). Similar results were obtained in experiments where the cells were not repurified at an intermediate timepoint or were repurified at day 2 instead (data not shown). By 6 d, ~50% of cells in OP9-DL1 culture are at least DN2 stage, and ~30% of cells in OP9-control culture are CD19+ (Taghon et al., 2005). Genes transiently induced on either stroma (e.g. Il2rb, Cebpa) reflect abortive emergence of NK and myeloid cells (data not shown), which are then suppressed during sustained culture under OP9-control or OP9-DL1 conditions.
Even so, the great majority of the genes analyzed were not induced preferentially during T-lineage differentiation under T-cell conditions. Although genes such as HEBalt and Pou6f1 are markedly upregulated in vivo at particular DN stages, they also were substantially induced in the B-cell conditions. Of particular interest were the four regulatory genes, Ets1, Ets2, Lef1, and Pou6f1, which are specifically induced during commitment to the T-cell lineage at the DN3 (DN3a) stage and maintained thereafter. If this event represents the definitive activation of T-lineage specific regulators, then these genes should show clear T-lineage specificity. Indeed, Ets2 showed an increasing expression specifically in T-lineage promoting conditions. But as already noted, Pou6f1 was clearly upregulated in B-cell precursors as well, and Ets1 and Lef1 showed no sustained T-lineage preference at all. SpiB, also specifically upregulated during the DN3 stage, was expressed considerably better in the B lineage cells.
These results are consistent with a separate assessment of the T-lineage specificity of >70 of these genes, in which we compared expression in adult DN pro-T cells with expression in freshly isolated bone marrow myeloid cells and pro-B cells (Table 4B, columns 1–3). This survey also included Cd3e, as a marker for T lineage specificity, the B-lineage specification gene Pax5, and Bcl6, which encodes a factor crucial for mature B-cells and is reported to provide physiological restraint on the expression of GATA-3 in peripheral T cells (Kusam et al., 2003). The results confirmed that the overwhelming majority of the transcription factor genes assessed were not T lineage-specific in expression. Of the genes analyzed in this series, only Gata3, Tcf7, Pou6f1, Ets2, and TOX (not monitored in the OP9 culture samples) showed an early T-lineage preference, i.e. with T-cell levels >3x greater than their levels in myeloid and pro-B cells. SATB1 also nearly met this criterion (Bcl11b was not assayed in these samples). Tbx21 was expressed more strongly in thymocytes than in myeloid or pro-B cells, but on closer examination, it was strongly expressed only in TCRγδ cells and in unsorted mutant thymocyte populations with a strong NK component (sorted wildtype DN cell value <0.15 of pooled “pan-DN” reference value). In this comparison as well as in Fig. 7, Table 4 confirms that the levels of Ets1 and Lef1 were no higher in pro-T cells than in pro-B cells, in agreement with their known roles in B cell development (Wang et al., 2005; Reya et al., 2000).
TABLE 4.
Expression patterns beyond the pro-T cell stages. (A) Definition of pattern types for genes analyzed in the study. Genes were classified according to their patterns of expression in the DN1-DN4 stages, based on their expression in cluster analyses such as those in Fig. 4, with adjustment “by hand” to take into account whether the dynamic ranges of their profiles exceeded the range of inter-experiment error, or whether they were essentially flat in expression across these stages. Patterns are described at the foot of Table 4A. Dynamic range indications are as follows: 10 = greater than tenfold range from peak to trough in DN stages; 3 = between threefold and tenfold range; 1 = less than threefold range. (B) Expression of genes in non-T developmental contexts and in stages after β- or γδ-selection. Gene expression levels were assayed in 2 samples each of the following: bone marrow derived pre-myeloid cells; pro-B cells, as CD19+ cells from Rag2−/− mouse bone marrow; Rag2−/− total thymocytes; TCRβ−/− total thymocytes; TCRγδ+ cells sorted from TCRβ−/− thymus; and sorted subsets from wildtype thymocytes as follows: DN (mostly DN3 & DN4); DP (CD4+ CD8+); CD4 SP (TCR+ CD4+ CD8−); and CD8 SP (TCR+ CD4−CD8+). Data from individual samples were first normalized according to β-actin levels, then the geometric mean of independent samples was calculated. To compare patterns of use for genes that are expressed at very different levels, all values were then normalized to a reference “pan-DN” level for each gene, that is the geometric mean of the values in Rag2−/−, TCRβ−/−, and sorted DN wildtype thymocytes. The Table shows the resulting relative expression values. For genes that increase in expression after β-selection, the sorted DN sample value will be greater than 1; for genes that decrease in expression from the DN1 stage through β-selection, the sorted DN sample value will be less than 1. Cells are shaded to help discern different patterns of expression: orange shaded cells show expression that is more than 3x higher than the reference pan-DN value; green shaded cells show expression that is less than 30% of the level of the reference pan-DN value; purple shaded cells show expression that is less than 1/10 of the level of the reference pan-DN value. Gray shaded cells have no data.

|
 |
 |
Taken together, these results show that the regulatory genes with strongly T-lineage specific expression are a very select subset of pro-T cell transcriptional regulators, and that within our sample they are represented only by Gata3, Tcf7, TOX, and Bcl11b. These are all factors that undergo most of their upregulation before the DN3 stage, i.e. before T lineage commitment. But the notable result is that most of the regulatory factors in our set that are specifically induced during the transition to T lineage commitment appear to be factors with substantial expression in the B-cell or myeloid lineages as well.
Roles in later T-cell development for factors used in pro-T cell specification
The downregulation during β-selection that was observed for many of the new transcription factor genes raised the question of whether factors sharing particular expression profiles during the pro-T cell stages might also be used in parallel patterns in later stages of T-cell development. Therefore, we also surveyed gene expression in a collection of samples representing later, TCR+ stages of T-cell development, from CD4+ CD8+ (double positive, DP) to mature CD4+ or CD8+ TCRαβ+ cells, and TCRγδ+ cells (Table 4B). In Table 4A, each qualitative expression pattern as defined in the DN cells (from Figs. 2–4) is coded; then within each pattern, the genes are further classified according to the overall magnitudes of their expression changes from lowest to highest level, to distinguish weak and questionable expression changes from strong ones. As for the non-T cell samples in Table 4B, expression of each gene in TCR+ subsets is presented in units relative to its expression in a pool of DN cell samples, and color coded to help discern similar and different overall patterns of cell type specificity. A number of the gene expression patterns shown here have been corroborated in a separate analysis with independent sets of samples (Rothenberg et al., 2008).
While this survey was too small to establish definitive expression levels, Table 4B shows that genes representative of each DN pattern could go on after β-selection to be expressed in diverse combinations in each of these cell types. Not only genes with sharp ascending or descending patterns, but also genes with “legacy” or “flat” expression patterns (Table 4, patterns 5 & 8) showed notably divergent patterns of expression in TCR+ stages represented by DP, CD4 SP, CD8 SP, or TCRγδ cells. As expected, progenitor-cell genes such as Sfpi1 that were repressed during commitment were never re-expressed, but also an additional group of genes (e.g. Erg, SpiB, Hes1, FUS, TfDp1, and Hmga1) remained profoundly downregulated in all TCR+ cells, even though they were shut off only at β-selection, considerably later than Sfpi1 (PU.1), Lima1, Tal1, and Gata2. Thus, a limited hit-and-run role during the pro-T cell stages is suggested for at least two distinct sets of regulatory genes. Other genes were downregulated selectively in CD4+ CD8+ DP cells, in all TCRαβ+ mature cells, or in CD4+ or CD8+ cells specifically (Table 4B). Another, unanticipated finding was a group of transcription factors that appear to distinguish sharply between TCRγδ cells and any of the TCRαβ+ subsets. Period1, Stat5b, and Zbtb48 appeared to be downregulated in TCRγδ cells as compared to TCRαβ+ cells; Dmtf1 and NCoR2 were specifically elevated in TCRγδ thymocytes, along with known TCRγδ-enriched factors Runx3 and Tbx21 (Taghon et al., 2006; Yin et al., 2002). Thus, transcription factors inherited from prethymic precursors may still help to generate T-cell functional diversity after the pro-T cell stages.
DISCUSSION
T cell lineage choice appears to encompass two major regulatory events: an asynchronous upregulation of increasing numbers of T-cell genes from the DN1 to the DN3 stage, and a sharp discontinuity in developmental potential when the cells undergo lineage commitment during transition to the DN3 stage. These two processes are followed by a TCR signal-dependent cascade of selective and developmental changes through the β-selection transition. If T-cell development followed the model of B-cell development, then the onset of T-lineage gene expression should reflect the activity of T-lineage specific transcription factors, and commitment to the T-cell lineage would reflect the advent of the definitive T-cell specific regulatory state. In this study, however, we see that the T-lineage commitment process departs from expectation in several respects. First, the essential regulatory genes that are turned on in a truly T lineage-specific way remain rare and do not seem to be upregulated during commitment. Second, many of the regulatory changes that do occur in early T-cells are to repress a subset of prethymically inherited genes rather than to upregulate new T-cell specific factors, as noted by others (Kawazu et al., 2007). Third, at the DN3 stage when commitment is thought to occur, the regulatory genes turned on are mostly not T-lineage specific. This suggests that the detectable evidence of commitment at this stage may be a readout of a more general cell biological function rather than a cell type identity function as such. Finally, we show that the loss of factors inherited from the prethymic, multipotent progenitor state may only be completed during β-selection. The β-selection (or γδ-selection) process may therefore contribute a larger aspect of the commitment mechanism than has previously been recognized.
A few caveats need to be stated. First, our analysis is not a complete accounting for all transcription factor genes. Although the gene discovery methods we have used here and in related studies (Anderson et al., 1999; Tydell et al., 2007) have expanded the pool of known candidate regulatory molecules (and their splice isoforms) in early pro-T cells, our gene lists should be considered as a complement to those in microarray studies as in work by the groups of Petrie, Staal, Ogawa, and Melchers (Tabrizifard et al., 2004; Dik et al., 2005; Kawazu et al., 2007; Hoffmann et al., 2003). In contrast to this study, most of the recent microarray studies cited have not addressed how different patterns of expression might be related to lineage specificity. Where our data overlap with these published reports, they are in general agreement, but each kind of study includes genes that are not highlighted in the other. A second caveat is that the RNA-centered analyses used here almost certainly miss some levels of regulation that alter protein levels between stages at which RNA levels are constant. Known examples relevant to T-cell development are the cases of E2A (Engel et al., 2001) and GATA-3 (Yamashita et al., 2005; Kusam et al., 2003; Hernández-Hoyos et al., 2003), both of which could be more sharply regulated between certain cellular states than their transcript levels reveal. Nevertheless, even if we miss some changes in active factor levels when RNA expression appears “flat”, when we do see major changes in RNA level, it is very likely that protein activities are also changing.
This regulatory gene expression analysis sheds light both on the cellular states that make up the stages of T-cell development and on the status of the regulatory molecules that may promote transitions between these stages. Considering these regulatory genes simply as indicators, the gene expression patterns shown here increase greatly the number of “dimensions” of data we have to distinguish DN1 from DN2 from DN3 pro-T cells. Instead of relying only on c-Kit, CD44, and CD25 to track normal developmental progress, the distinctively timed downregulation patterns of PU.1 (Sfpi1), SCL (Tal1), Gfi1b, Erg, Zfp316, Runx2 and Runx3, and Elk3 can also be used together with the distinctively timed upregulation patterns of Bcl11b, Ets2, Lef1, Id3, and Pou6f1 to provide a much finer-grained picture of normal or abnormal developmental progression. The downregulation at β-selection that normally affects Hes1, SpiB, Erg, HEBalt, and genes like Zbtb20 and Zfp287 can also be used to diagnose abnormalities of passage through this checkpoint (Yui and Rothenberg, 2004). Consideration of this expanded set of stage markers reveals reproducible variations of normal T cell development, as between the normal adult pathway and the accelerated development in the fetal thymus (David-Fung et al., 2006). In this case, because the “stage markers” actually encode regulatory factors, the differences may eventually shed light on the mechanisms controlling the differing rates of fetal and adult development.
The products of these regulatory genes also need to be considered as “drivers”. Changes in gene expression from stage to stage ultimately arise from changes in transcription factor activity. However, the often-subtle changes in expression of known essential regulators that we report raise important questions about the amount and type of change that is needed to be significant. Transcription factors work combinatorially in order to regulate target genes, and even factors showing minimal change can be functionally important participants in the T-cell developmental process through interaction with different partners. Thus, not everything that is functionally important for activation of a particular target gene needs to change in level. Among the factors known to be critical for T-cell specification (Rothenberg and Taghon, 2005; Anderson, 2006), some like Myb and Gfi1 may increase less than three-to five-fold between the stem cell state and the committed DN3 stage (Tydell et al., 2007; Rothenberg et al., 2008). These precedents suggest that “legacy” factors generally could also be potential participants in T-lineage specification, changing their functions through interaction with different partners (Sieweke and Graf, 1998).
Our results also suggest that what turns on different genes at different stages of the T-cell development process is sometimes not so much the appearance of a new transcription factor DNA-binding activity, but rather, the change in the identity of the dominant transcription factor that can bind to a given DNA regulatory site. As we show here, the overall level of DNA binding activity by Ets, Runx, and Ikaros family factors may be virtually constant from DN1 to DN3 stage. It is mainly the balance among different members of the family that undergoes dramatic developmental change. Clearly some of these families can participate in causing these changes through their own cross-regulatory relationships: Gfi1b repressing Gfi1, Runx3 repressing Runx1, and PU.1 repressing SpiB, Ets1, and Ets2 (Doan et al., 2004; Vassen et al., 2005; Spender et al., 2005; Franco et al., 2006). But for the change among family members to make a difference, it is likely that protein-protein interactions with other transcription factors are affected by the swap of one family member for another, and not just simple affinity for a DNA target site: there are clear precedents for this among Ikaros family factors (Thompson et al., 2007). By extension, each of the members of these families may compete with the others functionally. For example, as the cells progress from DN1 to DN3, downregulation of Runx2, Runx3 and Gfi1b could greatly enhance the impacts of the slight upregulation of Runx1 and Gfi1, on target genes for which these family members are not functionally equivalent.
There are two crucial watersheds in the T-cell specification process that need to be explained in regulatory terms, and our results shed light on what they actually entail. These are, first, the onset of T-lineage gene expression from DN1 to DN2, while the cells are apparently still multipotent, and second, the loss of multipotency as the cells reach the DN3 stage. This work and previous data (Tabrizifard et al., 2004; Dik et al., 2005; Tydell et al., 2007) show that the DN1 to DN2 transition involves only modest increases in expression of two T-lineage specific regulators (GATA-3 and TCF-1) and the dramatic upregulation of a third, Bcl11b. But although our gene discovery strategy has yielded many “new” potential transcriptional regulators in early T-lineage cells, no new candidates are both T-cell specific and significantly upregulated at this time. This conclusion is also supported by published microarray data (Dik et al., 2005; Tabrizifard et al., 2004; Hoffmann et al., 2003). Could GATA-3 or TCF-1 be the main driver of T-cell gene activation at the DN2 stage? This seems questionable because the major upregulation of these factors occurs much earlier, in entry to the DN1 stage (Tydell et al., 2007), which can be multiple rounds of proliferation before the DN2 stage (Petrie and Zuniga-Pflucker, 2007). Notch inputs seem to be needed throughout, and there is little indication from Hes1 or Dtx1 expression that there is any special intensification of Notch signaling at this point [rev. in (Rothenberg et al., 2008)]. Bcl11b is of great interest, but seemingly is not required for development even to the DN3 stage (Inoue et al., 2006). While Gfi1 and E protein genes are somewhat upregulated, they are not T-lineage specific. Therefore, a major timing element in this initial phase of specification may be an increased permissiveness for the activity of T-lineage specific genes through the downregulation of multiple DN1 and prethymic regulatory genes. It may indeed be the downregulation of antagonists that enables stably transcribed factors like E2A to undergo sudden gains in potency, e.g. by post-transcriptional mechanisms, during the DN1 to DN2 transition (Engel et al., 2001).
The results also imply that the commitment process itself has a compound nature that is masked in conventional comparisons between DN2 and DN3 cell potential. Negative regulation of Gfi1b, SCL, PU.1, and other factors should play a role. But the surprise is that the factors specifically upregulated during commitment are almost all shared between the T lineage and other cell types, with little if any preferential expression in T cells as opposed to B cells. Ets2 upregulation and the transient hyperactivation of Notch pathway target genes are the two main positive events that are T lineage-biased; but the Notch activation is not a stable change and the Ets2 activity appears to remain low compared to its pan-lymphoid relative Ets1. Ets2 may have a later specific role in T cell development (Zaldumbide et al., 2002), as our OP9 time course captures only the beginning of its upregulation (M.A. Y., data not shown), but its expression is not actually T-lineage specific, as it is expressed in myeloid cells as well (Ross et al., 1998). Thus the transition from multipotent to committed may not use an exclusively T-lineage specific mechanism. Note that the DN3 stage in adult mouse thymocytes is associated with a proliferative halt (Petrie and Zuniga-Pflucker, 2007), with growth resuming only if β- or γδ-selection is successful. Conceivably the change assayed as “commitment” is primarily a loss of proliferation, before new functions are conferred during the β-selection process. A permanent lockdown of commitment might only be required to preserve lineage fidelity if and when proliferation might resume. Thus it is also notable that our results show another major phase of repression during β-selection, when a large subset of the “legacy” genes, which had been stably expressed through the DN3 stage are finally turned off. In this altered regulatory environment, the proliferation associated with β- and γδ-selection would not only dilute out pre-existing transcription factor proteins but also provide opportunities to remodel chromatin, which would not be available in G1 arrest in the DN3 stage. Thus it may be only through β- or γδ-selection that commitment can actually be converted into a stable and heritable condition.
In perspective, the induction of Tcf7 and Gata3 and then Bcl11b, all in the earliest steps of T-cell development, remain the most T-lineage specific of all the regulatory features studied here. Their initial upregulation occurs in the stages when cells are still multipotent, and our results show that few if any other T-lineage specific regulators are likely to be added as the cells undergo lineage commitment. What occurs instead appears to be the stripping away of other alternative pathway regulators and neutralization of inhibitory influences, so that a latent identity already instilled in the earliest intrathymic precursors can be stabilized and revealed.
Supplementary Material
Acknowledgments
We are deeply indebted to Dr. Howard Petrie (Scripps Florida) for valuable discussions during the gestation of this work and for generous sharing of data prior to publication. We are also grateful to Dr. Hamid Bolouri (Caltech and Institute for Systems Biology) and Dr. Eric Davidson (Caltech) for insightful critiques, encouragement, and advice. We also thank Brian Birditt and Scott Bloom (Institute for Systems Biology) for their excellent work sequencing and identifying the cDNA clones from our gene discovery; Marissa Morales and Dr. Rashmi Pant, for key contributions to the gene expression measurements and their validation; Dr. Tom Taghon, for generous collaboration on making the samples for the OP9 experiments; Gillian Giorgio, for early help curating the sequence files; Stephanie Adams of the Caltech Flow Cytometry Facility, for outstanding help with the sorting; and Robin Condie and Ruben Bayon for excellent care of the animals. This work was undertaken with support from the Stowers Institute for Medical Research and then supported by grants from the NSF (MCB9983129) and the NIH (R01 CA90233 and R33 HL089123), and by the Louis A. Garfinkle Memorial Laboratory Fund, the Albert Billings Ruddock Professorship, and the DNA Sequencer Royalty Fund at Caltech.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi S, Rothenberg EV. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in competent, nontranscribing T-cells. Nucleic Acids Res. 2005;33:3200–3210. doi: 10.1093/nar/gki637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias A, Chen D, Rothenberg EV. Definition of regulatory network elements for T-cell development by perturbation analysis with PU.1 and GATA-3. Devel Biol. 2002a;246:103–121. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Pant R, Miracle AL, Sun X, Luer CA, Walsh CJ, Telfer JC, Litman GW, Rothenberg EV. Evolutionary origins of lymphocytes: ensembles of T cell and B cell transcriptional regulators in a cartilaginous fish. J Immunol. 2004;172:5851–5860. doi: 10.4049/jimmunol.172.10.5851. [DOI] [PubMed] [Google Scholar]
- Anderson MK, Weiss A, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T-cell development at the pro-T stage. Immunity. 2002b;16:285–296. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- Baba Y, Garrett KP, Kincade PW. Constitutively active β-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 2005;23:599–609. doi: 10.1016/j.immuni.2005.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Brunak S. Bioinformatics: the Machine Learning Approach. Cambridge, MA, USA: MIT Press; 2001. [Google Scholar]
- Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nat Rev Immunol. 2002;2:888–897. doi: 10.1038/nri937. [DOI] [PubMed] [Google Scholar]
- Chen F, Rowen L, Hood L, Rothenberg EV. Differential transcriptional regulation of individual TCR Vβ segments before gene rearrangement. J Immunol. 2001;166:1771–1780. doi: 10.4049/jimmunol.166.3.1771. [DOI] [PubMed] [Google Scholar]
- Chen ML, Kuo CL. A conserved sequence block in the murine and human T cell receptor Jα loci interacts with developmentally regulated nucleoprotein complexes in vitro and associates with GATA-3 and octamer-binding factors in vivo. Eur J Immunol. 2001;31:1696–1705. doi: 10.1002/1521-4141(200106)31:6<1696::aid-immu1696>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Yui MA, Morales M, Wang H, Taghon T, Diamond RA, Rothenberg EV. Progression of regulatory gene expression states in fetal and adult pro-T cell development. Immunol Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik WA, Pike-Overzet K, Weerkamp F, de Ridder D, de Haas EF, Baert MR, van der SP, Koster EE, Reinders MJ, van Dongen JJ, Langerak AW, Staal FJT. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–2519. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–746. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–1147. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Tα chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell Mol Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- Hoffmann R, Bruno L, Seidl T, Rolink A, Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J Immunol. 2003;170:1339–1353. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc Natl Acad Sci U S A. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J, Kanefuji T, Okazuka K, Watanabe H, Mishima Y, Kominami R. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in Bcl11b−/− mice. J Immunol. 2006;176:5871–5879. doi: 10.4049/jimmunol.176.10.5871. [DOI] [PubMed] [Google Scholar]
- Kawazu M, Yamamoto G, Yoshimi M, Yamamoto K, Asai T, Ichikawa M, Seo S, Nakagawa M, Chiba S, Kurokawa M, Ogawa S. Expression profiling of immature thymocytes revealed a novel homeobox gene that regulates double-negative thymocyte development. J Immunol. 2007;179:5335–5345. doi: 10.4049/jimmunol.179.8.5335. [DOI] [PubMed] [Google Scholar]
- Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- Kusam S, Toney LM, Sato H, Dent AL. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J Immunol. 2003;170:2435–2441. doi: 10.4049/jimmunol.170.5.2435. [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Xie H, Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4:525–532. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, Hilton DJ, Alexander WS, Kile BT. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Olman V, Xu D, Xu Y. Identification of regulatory binding sites using minimum spanning trees. Pac Symp Biocomput. 2003:327–338. [PubMed] [Google Scholar]
- Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- Reya T, O’Riordan M, Okamura R, Devaney E, Willert K, Nusse R, Grosschedl R. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- Ross IL, Yue X, Ostrowski MC, Hume DA. Interaction between PU.1 and another Ets family transcription factor promotes macrophage-specific Basal transcription initiation. J Biol Chem. 1998;273:6662–6669. doi: 10.1074/jbc.273.12.6662. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. Molecular Genetics of T Cell Development. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, Leutz A. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat Immunol. 2006;7:1037–1047. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004a;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, De Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004b;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. T-cell development, doing it in a dish. Immunol Rev. 2006;209:95–102. doi: 10.1111/j.0105-2896.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- Sieweke MH, Graf T. A transcription factor party during blood cell differentiation. Curr Opin Genet Devel. 1998;8:545–551. doi: 10.1016/s0959-437x(98)80009-9. [DOI] [PubMed] [Google Scholar]
- Singh H, Pongubala JM, Medina KL. Gene regulatory networks that orchestrate the development of B lymphocyte precursors. Adv Exp Med Biol. 2007;596:57–62. doi: 10.1007/0-387-46530-8_5. [DOI] [PubMed] [Google Scholar]
- Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene. 2005;24:1873–1881. doi: 10.1038/sj.onc.1208404. [DOI] [PubMed] [Google Scholar]
- Staal FJT, Weerkamp F, Baert MR, van den Burg CM, van Noort M, de Haas EF, van Dongen JJ. Wnt target genes identified by DNA microarrays in immature CD34+ thymocytes regulate proliferation and cell adhesion. J Immunol. 2004;172:1099–1108. doi: 10.4049/jimmunol.172.2.1099. [DOI] [PubMed] [Google Scholar]
- Staal FJT, Weerkamp F, Langerak AW, Hendriks RW, Clevers HC. Transcriptional control of T lymphocyte differentiation. Stem Cells. 2001;19:165–179. doi: 10.1634/stemcells.19-3-165. [DOI] [PubMed] [Google Scholar]
- Tabrizifard S, Olaru A, Plotkin J, Fallahi-Sichani M, Livak F, Petrie HT. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nat Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon TN, David ES, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- Telfer JC, Rothenberg EV. Expression and function of a stem-cell promoter for the murine CBFα2 gene: distinct roles and regulation in natural killer and T cell development. Devel Biol. 2001;229:363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, Smale ST, Fisher AG, Merkenschlager M. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- Ullman KS, Northrop JP, Verweij CL, Crabtree GR. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Ann Rev Immunol. 1990;8:421–452. doi: 10.1146/annurev.iy.08.040190.002225. [DOI] [PubMed] [Google Scholar]
- Vassen L, Fiolka K, Mahlmann S, Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33:987–998. doi: 10.1093/nar/gki243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int Immunol. 2005;17:1179–1191. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- Wang H, Diamond RA, Yang-Snyder JA, Rothenberg EV. Precocious expression of T-cell functional response genes in vivo in primitive thymocytes before T-lineage commitment. Int Immunol. 1998;10:1623–1635. doi: 10.1093/intimm/10.11.1623. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- Xu Y, Olman V, Xu D. Clustering gene expression data using a graph-theoretic approach: an application of minimum spanning trees. Bioinformatics. 2002;18:536–545. doi: 10.1093/bioinformatics/18.4.536. [DOI] [PubMed] [Google Scholar]
- Xue HH, Bollenbacher J, Rovella V, Tripuraneni R, Du YB, Liu CY, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nat Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Shinnakasu R, Asou H, Kimura M, Hasegawa A, Hashimoto K, Hatano N, Ogata M, Nakayama T. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J Biol Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O’Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Chen C, Szabo SJ, Glimcher LH, Ray A, Craft J. T-Bet expression and failure of GATA-3 cross-regulation lead to default production of IFN-γ by γδ T cells. J Immunol. 2002;168:1566–1571. doi: 10.4049/jimmunol.168.4.1566. [DOI] [PubMed] [Google Scholar]
- Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- Yun TJ, Bevan MJ. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- Zaldumbide A, Carlotti F, Pognonec P, Boulukos KE. The role of the Ets2 transcription factor in the proliferation, maturation, and survival of mouse thymocytes. J Immunol. 2002;169:4873–4881. doi: 10.4049/jimmunol.169.9.4873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.