Abstract
The essential and non-trivial role of the denatured state of proteins in their folding reaction is being increasingly scrutinized in recent years. Single molecule FRET (smFRET) experiments show that the denatured state undergoes a continuous collapse (or coil-to-globule) transition as the concentration of a chemical denaturant is decreased, suggesting that conformational entropy of the denatured state is an important part of the free energy of folding. Such observations question the validity of the classical Tanford transfer model, which suggests that the folding free energy can be understood solely based on the difference in amino acid solvation between the folded state and a fixed unfolded state. An alternative to the transfer model is obtained here from a polymer theoretical analysis of a series of published smFRET data. The analysis shows that the free energy of denatured-state collapse has a linear dependence on denaturant concentration, an outcome of the interplay between enthalpic and entropic contributions. Surprisingly, the slope of the free energy of collapse agrees very well with the respective slope of the free energy of folding. This conformity of values obtained from two very different measurements shows that it is the collapse transition in the denatured state which mediates the effect of denaturants on folding. The energetics of folding are thus governed by the competition of solvation and conformational entropy in the denatured state.
Ever since the seminal work of Anfinsen 1, it is known that protein molecules can spontaneously fold into their native form. Thus, under native conditions, the folded state (N) is thermodynamically favored over the denatured (or unfolded) state (U), i.e. the free energy of folding ( ) is negative. For typical proteins is about 5–15 kcal/mol 2. This marginal stability seems to be important for enzymatic catalysis 3, as well as other biological processes such as protein degradation 4. It also affects the probability for misfolding and aggregation 5, which are the cause of amyloidal diseases such as Alzheimer and Creutzfeldt-Jakob disease. It is therefore of great interest to understand the properties of proteins affecting their stability under various biologically relevant conditions, and to get a measure, even if indirect, of .
Chemical denaturants such as urea or guanidinium chloride (GdmCl) have been traditionally used to study protein stability and denaturation. They disrupt the native structure of proteins, although the microscopic mechanism by which this is achieved remains a matter of some controversy 6–12. By gradually increasing the concentration of a denaturant while observing a spectroscopic signature such as tryptophan fluorescence or circular dichroism, one can follow the complete process of protein denaturation. For small proteins (typically less than 100 amino acids) the ensuing "denaturation curve" is fitted well with a model having only two states 13. The free energy of folding between two denaturant concentrations D and D′ can be extracted from this analysis, and is often found to depend linearly on the molar concentration of denaturant 14,15:
| [1] |
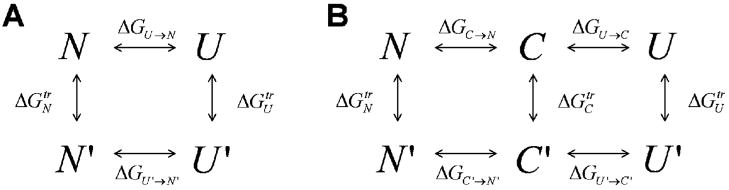
where m is the slope of this linear relation, and primes denote quantities at D′. Extrapolating Equation 1 to D = 0, one obtains a measure for the protein stability . The m-value is related to the steepness or cooperativity of the folding transition. Despite its obvious importance in many aspects of protein chemistry, the physical interpretation of this m-value is not entirely clear. The well-known transfer model (TM), suggested originally by Tanford 14,16 almost 50 years ago, relates the change in ΔGU→N to the transfer free energy of the native and unfolded states ( and , respectively), using the thermodynamic cycle in scheme 1A. Tanford’s TM model, which assumes that the transfer free energy involves only changes in solvation of amino acid residues, can be written approximately as:
| [2] |
Scheme 1.
Thermodynamic cycles for unfolding. A. The well-known thermodynamic cycle relating two states, native (N) and unfolded (U), at two denaturant concentrations D (top) and D′ (bottom). We use the notation U and U′ for the unfolded state at the two concentrations, to stress that in fact a different ensemble of conformations is sampled in each. Thus, the transfer energy is composed of the enthalpy of transfer and the change in the conformational entropy of the unfolded state ensemble. B. For the calculation of the free energy of collapse, we introduce a new reference state, which is designated the maximally compact denatured state (C). Notice that in general this state does not equal the unfolded state at zero D.
Equation 2 relates the transfer free energy of folding to the average transfer free energy of the amino acid residues of the protein from water to 1M denaturant solution ( ), and the average change in (fractional) solvent accessible surface area (SASA) between the folded state and the unfolded state ( ). n is the number of amino acids in the protein. This equation provided a route for obtaining a theoretical estimate for the m-value of a protein, which has been exploited by several authors over the years. The calculation of SASA for the folded state is straight forward from available crystal structures. On the other hand, it is not trivial to decide what SASA should be used to represent the ensemble of conformations we refer to as the unfolded state. Myers et al. assumed an extended β-conformation for the unfolded state 17, while Auton et al. used the mean SASA of two conformations for the unfolded state – an extended conformation and a more “compact” conformation calculated from excised peptide fragments of folded structures 18. Since denaturation experiments show that the m-value is a constant, must be a constant too and therefore the unfolded state SASA cannot depend on D. in other words, changes in the conformational entropy of the chain with denaturant concentration are ignored. One premise of this paper, based on the analysis of single-molecule fluorescence resonance energy transfer (smFRET) experiments, is to show that this assertion is incorrect.
It has been realized for some time that the folding transition may be preceded by a fast collapse into a compact structure 19–21. A comprehensive discussion of the theoretical foundation and experimental evidence for denatured-state collapse can be found in a recent Perspective 22. Unlike the folding reaction, the collapse of denatured proteins has been barely studied by equilibrium methods, perhaps due to the inability to separate the denatured-state population from the folded state population under solution conditions where the two states co-exist. smFRET experiments are beginning to change this situation. In smFRET experiments individual protein molecules labeled with two fluorophores (typically close to their termini) are studied one by one as they diffuse in and out of a focused laser beam. It was shown that for most proteins studied with this method, the average FRET efficiency (Ē) of the denatured state (inversely related to the overall dimensions of the denatured protein) decreases monotonically as D increases, signifying that the denaturant causes the unfolded state to expand 23–32. Clearly, then, both the SASA and change with D. How can the m-value be constant?
In this paper we propose a solution to this conundrum. We analyze experimental smFRET data of several different proteins using a theory originally developed to explain the coil-to-globule (CG) transition in homopolymers. It is found that in essentially all of the smFRET experiments analyzed here the free energy of collapse is linear over the whole range of D, due to a subtle balance of enthalpy and conformational entropy terms; the latter is completely neglected in the TM. The denaturant dependence of the free energy of collapse is the same as the denaturant dependence of the free energy of folding, which implies that denaturants affects ΔGU→N by modulating the energetics of the denatured state. Using a high D approximation to our model we propose an alternative model for the m-value.
Analysis and Results
Extracting thermodynamics from single-molecule FRET experiments
The CG transition in polymers has been extensively studied experimentally and theoretically for over 50 years 33. Most of the theories of CG transition are either applicable only in one of the two phases (i.e. the expanded state or the collapsed states), or have too many parameters to be useful for analyzing experimental data. The mean-field Sanchez theory 34, on the other hand, quantitatively describes the chain expansion throughout the CG transition and has just one free parameter. We modify this theory here to analyze the CG transition in denatured proteins. The theory models the probability distribution function of the radius of gyration S as:
| [3] |
Here is the Flory-Fisk empirical distribution for the radius of gyration of an ideal chain of n monomers (i.e. amino acid residues) 35, and g(φ,ε), the excess free energy per monomer with respect to the ideal chain, is given by:
| [4] |
where φ is the volume fraction occupied by the chain, and ε is the difference in mean-field solvation energy between a residue within the maximally-collapsed chain and in free solution. Inter-residue interactions can be of varied origins (e.g. hydrobophic, electrostatic and even backbone-mediated 36), and in principle ε includes all of these in an average way The first term in Equation 4 describes how attraction between residues (ε >0) favors compaction of the chain, while the second term is the entropy arising from excluded volume interactions, which favors expansion. The volume fraction is related to the radius of gyration through φ = SN3/S3, where SN is the radius of the gyration of the fully-compact native state (with φN = 1 by definition). One necessary parameter is the radius of gyration of the ideal chain, . This parameter is fixed by requiring that , which was derived using Landau’s theory of phase transitions 34.
Recent experiments 37–39 suggest that the end-to-end distance dynamics for a small denatured protein are on the microsecond (or even nanosecond) time scale, while smFRET data are averaged over millisecond fluorescence bursts. Under such circumstances the shape of the denatured-state FRET distribution (due mainly to photon shot-noise) does not contain useful information. Ē of the denatured-state distribution can, however, be ascribed to the following average over the end-to-end distance distribution, P(Ree):
| [5] |
Here R0 is the Förster radius for energy transfer and is the FRET efficiency at an end-to-end distance Ree. In the past either the P(Ree) of a Gaussian chain or of a worm-like chain were used to fit smFRET experiments 24,26,30. However, these probability distribution functions do not explicitly include information about the CG transition and its energetics. A model for P(Ree) based on equation 3 corrects this deficiency. Formally, the end-to-end distance distribution P(Ree) may be related to the distribution P(S) through the relation P(Ree) = ∫ P(Ree|S)P(S)dS, for which the conditional probability P(Ree|S) needs to be specified. Here we adopt a simple model for this conditional probability, in the spirit of the mean-field approximation inherent in Sanchez’s theory, namely that P(Ree|S) is given by the probability to find the two chain ends within a sphere with radius g · S. A single value of the constant g is used for all proteins (g ≡ 2.3), and it is selected to reproduce correctly the Gaussian distribution expected when P(S) = P0 (S) (see Supporting Text). In practice, the denatured state structure is expected to be somewhat aspherical, as found experimentally 40 and theoretically 41,42. However, the quality of current experiments does not merit such detailed modeling of the denatured-state structure. Further, the effect of anisotropy on P(Ree|S) is not expected to significantly affect the results of our analysis because of the FRET kernel in equation 5.
We are now in the position where we can evaluate ε from experimental data through Equations 3–5. We best fit ε for each protein and for each denaturant concentration to match the measured Ē of the denatured state. One final point that requires attention is the calculation in the case that labeling in was not done close to the termini. In this case we replace Ree in the FRET efficiency kernel of Equation 5 by , where l is the actual number of residues separating the donor and acceptor.
Once ε is obtained for a protein at each denaturant concentration we can readily calculate different properties of the denatured state of the protein, such as its root mean-squared radius of gyration, 〈S2〉1/2, which is convenient to present in terms of the expansion factor α2 = 〈S2〉/〈S2〉0. The free energy can be either calculated by using G(ε) = −kBT〈ln P(S)〉, where 〈 〉 stands for an average over the distribution at ε, or by using G(ε) = n · g(φ̄, ε), in which φ̄ is the mean volume fraction. The two calculations give essentially identical results.
All proteins exhibit a continuous expansion
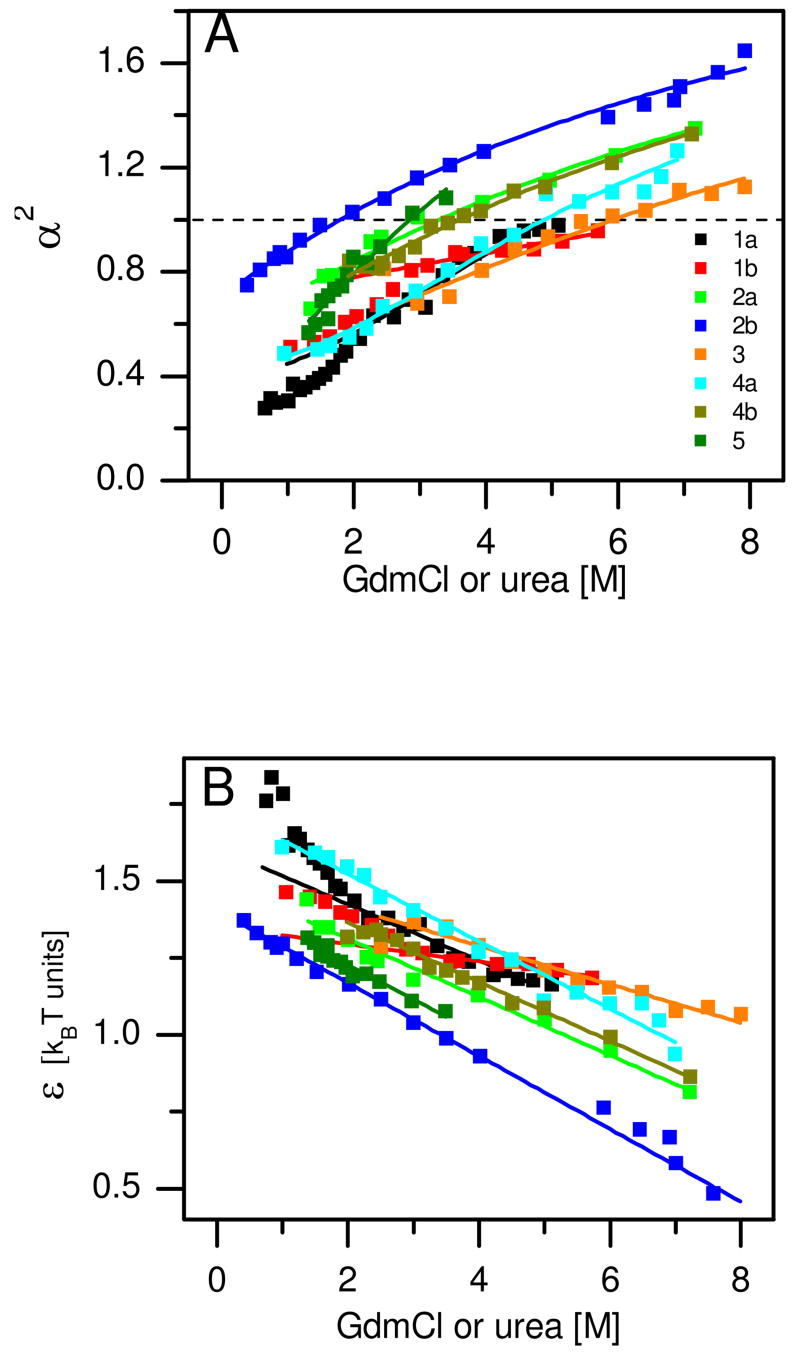
We analyzed twelve data sets measured on five different proteins (see Table 1 for details on the proteins) 25–27,29,30,32 using the method introduced above. Two of these proteins were measured in two different labs, namely protein L 26,29 and CspTm 29,30. Schuler and coworkers obtained FRET efficiency distributions for five different variants of CspTm, each labeled on a different pair of sites 30; we analyzed all five variants (Table 1). Most data sets were measured using GdmCl as the denaturant; two of the data sets were measured with urea (Table 1). Figure 1A shows the expansion factor α2, which describes the change in size as a function of solution conditions, calculated for all proteins (for clarity only one of the CspTm variants from reference 30 is shown). There is a continuous expansion of the denatured state of these proteins over the whole range of D. Furthermore, most proteins span both sides of the α2 = 1 line (the CG transition occurs formally at this point for very long polymers), while some remain below the transition point even at the highest D used, although they will likely cross this line at a slightly higher concentration. The D values matching the CG transition point (DCG, see Table 1 and Supporting Text) are thus broadly distributed, but are always above that of the folding transition (D50, Table 1).
Table 1.
Analysis of denaturant-induced CG transition from smFRET experiments
| Number | Protein | n2 | Labeling positions 3 | Denaturant | D504[M] | DCG5[M] | SN6[Å] | SU,0M7[Å] | SU,D8[Å] | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1a–1b | Barstar | 90 | 12, 89 | GdmCl Urea |
1.24 2.33 |
6.4 11.7 |
13.3 | 18.2 24.5 |
31.6 30.3 |
32 |
| 2a | CspTm | 66 | 1, 66 | GdmCl | 2.0 | 5.3 | 12.7 | 20.4 | 30.2 | 29 |
| 2b–2f | CspTm | 67 | 2, 67 10, 67 21, 67 22, 67 34, 67 |
GdmCl | 2.0 1.7 2.3 1.9 2.4 |
3.4 5.1 3.8 4.4 3.4 |
12.7 | 22.5 18.2 20.8 19.8 23.0 |
32.4 30.7 32.1 31.5 32.3 |
30 |
| 3 | Im9 | 86 | 23, 81 | Urea | 3.83 | 8.3 | 15.5 | 22.8 | 36.9 | 27 |
| 4a | Protein L | 64 | 1, 64 | GdmCl | 1.64 | 6.8 | 13 | 17.1 | 28.9 | 26 |
| 4b | Protein L | 65 | 1, 65 | GdmCl | 2.6 | 5.8 | 13 | 19.8 | 30.3 | 29 |
| 5 | RNaseH | 155 | 3, 135 | GdmCl | 1.37 | 3.7 | 17.1 | 24.8 | 50.4 | 25 |
Protein abbreviations: Barstar - Ribonuclease inhibitor protein from Bacillus Amyloliquefaciens (PDB: 1BTA), CspTm – Cold-shock protein from Thermotoga Maritima (1G6P), Im9 - immunity protein for colicin E9 (1IMP), Protein L – IgG binding domain B1 of protein L from Peptostreptococcus magnus (1HZ6), RNaseH - Ribonuclease Hi From Escherichia Coli (1RCH).
Number of residues.
As reported in original references.
Denaturation midpoint, as reported in original references, or obtained from a fit of reported denaturation data to a two-state model.
CG transition midpoint, defined as the point at which α=1+19/22·φ0 (see Supporting Text).
Radius of gyration of native state, calculated using HydroPro from published structures obtained from the Protein Data Bank.
Radius of gyration of unfolded state at 0 M denaturant, calculated by extrapolation.
Radius of gyration of unfolded state at 6 M GdmCl (8M urea), calculated by extrapolation.
Figure 1.
The CG transition in denatured proteins. Results of Sanchez theory analysis of smFRET experiments (see Table 1 for a full list of all proteins analyzed). A. The expansion factor α2 ≡ 〈S2〉/〈S2〉0 as a function of denaturant concentration. The dashed line marks the CG transition (α2 = 1). Results calculated from the experimental data using the analysis outlined in the text are shown as squares. Expansion factor values were recalculated based on the linear fits of panel B, and are shown as solid lines. The numbers in the legend match the numbers in Table 1. B. the mean-field interaction energy ε as a function of denaturant concentration. Linear fits are shown as solid lines. In all proteins (apart from barstar) the fits encompass the whole range of denaturant concentrations. Color code as in A.
The mean-field interaction energy ε obtained from the analysis is shown in Figure 1B. It is positive (i.e. attractive), and decreases linearly in all proteins apart from barstar, for which it is linear only at high D. We therefore fitted ε to a linear model, taking only the high concentration points in the case of barstar (fits are shown in Figure 1B). We used the fitted lines to re-calculate the expansion factors, which are presented as solid lines in Figure 1A. The agreement of the solid lines with the experimental data shows that indeed the expansion is captured by the linear analysis of ε. We can use this analysis to extrapolate the experimental results and calculate the radii of gyration under native conditions (SU,0M) and highly denaturing conditions (SU,D). These values are given in Table 1. The extrapolated values indicate that under native conditions the size of the unfolded state of the proteins used for this calculation may be 30–50% larger than the size of their native state, correlating well with previous estimates 43. Further, in all proteins studied there exists a remarkable expansion of the denatured state (50–100%) as solution conditions are changed from native to highly denaturing.
The free energy of collapse is linear with denaturant concentration
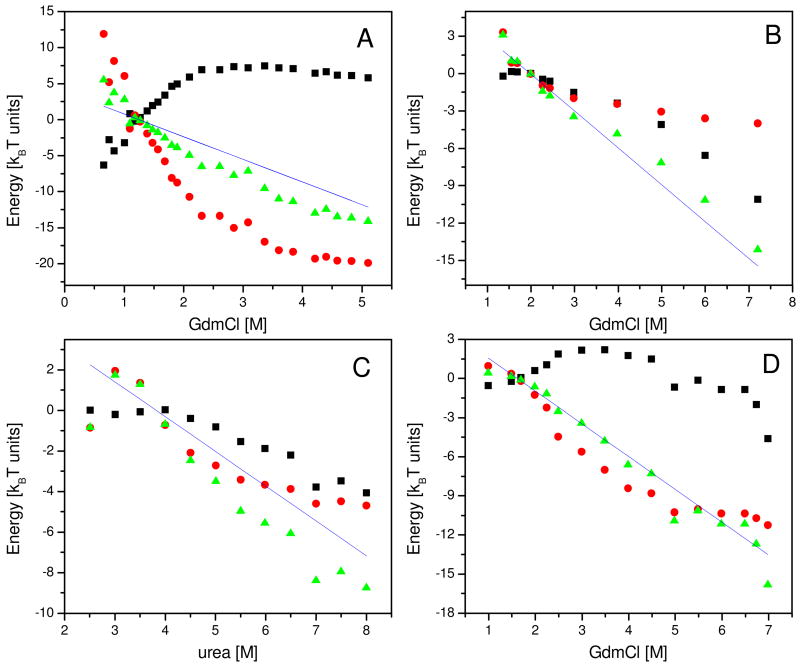
To calculate the free energy of collapse we define a new thermodynamic reference state which is the maximally collapsed denatured state (C). By definition, this state has volume fraction 1. This state may differ from U even under native conditions, as we have shown in the previous section. We calculate the free energy of collapse, ΔGU →C as well as the enthalpy and entropy of collapse, ΔEU →C and ΔSU →C respectively, for each protein and D. Figure 2 shows the results for four proteins, while the rest are shown in Supporting Figure 1. Surprisingly, despite some differences between different proteins, in all cases neither ΔEU →C nor ΔSU →C show clear linear dependence on denaturant, but the sum of the two terms ΔGU →C is clearly linear over a very broad range of denaturant concentrations, and over about 15kBT (~25 kcal/mol) in energy. In most of the proteins, this linearity originates from compensating enthalpic and entropic contributions. The reason for the variation of the D-dependence of ΔEU →C or ΔSU →C between proteins is beyond the scope of this paper.
Figure 2.
Thermodynamic functions of collapse calculated from smFRET results for four different proteins. Values of the free energy of collapse, − ΔGU →C, as a function of denaturant concentration, are shown as green triangles. The enthalpic contributions −ΔEU →C and the entropic contributions kBT · ΔSU →C are shown as black squares and red circles, respectively. The interpolated value of each thermodynamic function at D50 is subtracted from all points. Solid lines show the denaturant dependence of the free energy of folding, ΔGU →N, as published in the literature. A. Results for protein L denatured in GdmCl (labeled 4a in Table 1). B. Im9 denatured in urea (3 in Table 1). C. CspTm denatured in GdmCl (2a in Table 1). D. Barstar denatured in GdmCl (1a in Table 1).
Denaturants affect protein stability by modulating denatured-state collapse
Comparing the slope of the free energy of collapse with the function −m · D for each protein (solid lines in Figure 2 and Supporting Figure 1), an excellent agreement is found essentially in all cases. Thus the change in the free energy of collapse with denaturant is described by the same m-value as the change in the free energy of folding. This surprising finding, which is the main result of this paper, is particularly intriguing in view of the fact that two very different experiments lead to the two curves. ΔGU →C is obtained from smFRET experiments, which measure the change in the size of the denatured state. m · D, on the other hand, is obtained from standard denaturation experiments. Furthermore, these two measurements characterize two different phenomena, the second-order (continuous) coil-globule transition vs. the first-order folding transition, respectively.
Using the thermodynamic cycle depicted in Scheme 1B we can write
| [6] |
The remarkable result of our analysis of the experimental data implies that ΔGU →N − ΔGU′ →N′ = ΔGU →C − ΔU′ →C′. Equation 6 is therefore valid only if ΔGC′ →N′ = ΔGC→N, and observation of Scheme 1B shows that this immediately leads to . Thus the maximally collapsed state (C), which was defined a-priori only by demanding that its volume fraction be set to 1, is found to have the same constant SASA and overall distribution of exposed amino acids as the native state (N). The important conclusion from this analysis is that all the effect of denaturants can be accounted for by that part of the cycle containing the U and C states. Thus the denaturant effect on folding is mediated through the collapse transition, i.e. through changes in the dimensions of the denatured state.
Expression for the m-value at high D
When both D and D ′ are high, typically (but not necessarily) above ~6 M, almost all proteins reach their maximum expansion, and φ becomes nearly constant (see Supporting Figure 2). Thus, the entropic terms in ΔGU′→C′ and ΔGU′→C′ become essentially identical, and only the enthalpic terms differ. Using φ̄U, which is the average volume fraction at high D, and employing the substitution ε = ε0 − m̄D, which is based on the finding in Figure 1B that ε is linear with D, we get from Equations 4 and 6 that
| [7] |
Comparing Equations 1 and 7 we get the relation
| [8] |
Equation 8 suggests a physical interpretation for the m-value, which is alternative to the TM (Equation 2). It relates the linear dependence of ΔGU →N with D to the change (slope) of the mean-field interaction energy (m̄) and the volume fraction of the denatured state in the highly expanded state (φ̄U). We can validate this relation with the experimental data. As shown in Supporting Figure 2, φ̄U is between 0.1 and 0.2. Taking an average value of 0.15 for this variable, the mean 1/2m̄(1−φ̄U) for the set of proteins used in this study 44 is (2.8±0.1)×10−2 kcal/mol/M. For the same set of proteins we have m/n = (2.3 ± 0.3) × 10−2 kcal/mol/M. It is important to note again that these very similar values on the right and left sides of Equation 8 come from two totally different types of measurements, namely the denaturation curve in the case of the m-value and the denatured-state size variation in the case of the m̄-value.
Discussion and Conclusion
In this work we used smFRET data of denatured-state expansion from a series of proteins to calculate the free energy of denatured-state collapse. It was found that ΔGU →C is linear over the entire range of D used in these experiments, with essentially the same slope as ΔGU →N. The similarity of the slopes suggests that changes in the free energy of folding with denaturant concentration are due to changes in the free energy of the denatured state. These changes, which are related to the CG transition of the denatured protein, have an enthalpic part, due to modulation of the solvation of amino-acid residues, but they also have a significant entropic part, due to an overall change in the size of the denatured protein. Interestingly, the analysis validates the linearity of ΔGU →N (through the linearity of ΔGU →C) over a much larger range of denaturant concentration then experimentally available in standard denaturation experiments, in which it can be measured only a few kcal/mol below and above D50.
The expansion of the denatured state of proteins with increasing D poses a question on the validity of Tanford’s TM. We have shown that the TM is, in fact, not valid for low or moderate D, the range over which the conformational entropy changes continuously. Rather, we expect the TM to be strictly correct only for transfer of a protein between two denaturant concentrations at which the chain is already fully expanded and the change in conformational entropy is negligible. As was shown recently, however, by applying the TM separately to each individual member in the ensemble of denatured-state conformations, one can overcome the conformational entropy problem and reproduce many experimental results, including a protein’s denaturation curve and the measured m-value 45.
The theory used in this paper to describe the CG transition assumes a non-specific collapse, so that a mean-field energy term can account for the interaction between monomers in the chain. It is possible that in some proteins the collapse transition is driven by specific interactions between protein residues. This is beyond the simple picture of the Sanchez theory, but may still be incorporated into it by parameterizing ε in terms of a more detailed model of the protein. Alonso and Dill employed this route 46 and wrote an expression that allowed them to predict the value of ε from the ratio of hydrophobic to polar amino acids in a protein and their distribution within its folded structure. It will be interesting in the future to apply a similar calculation to the results presented in this work.
Our analysis pertains of course only to the thermodynamics of collapse and folding, and does not have immediate implications for the kinetics of folding. Kinetic experiments suggest that the free energy barriers for folding (ΔGU →TS) and unfolding (ΔGN →TS) are also linear with D. This can be explained by two alternative models. First, if the transition state (TS) has a fixed size, and therefore a constant SASA (which is larger than that of the native state, but smaller than that of the denatured state at all conditions), ΔGU →TS and ΔGN →TS will both be linear. On the other hand, if the transition state undergoes a continuous expansion with D similar to that of the denatured state, a sufficient condition for linearity of the folding barriers is that ΔGU →TS is linearly-dependent on ΔGU →C. Molecular dynamics simulations suggest the TS is, in fact, rather compact 47,48. However, there is no evidence to support or dispel denaturant-induced changes in TS size.
There have been suggestions, based on time-resolved SAXS experiments, that in some proteins collapse may not temporally precede folding 49,50. However, it is still possible that in equilibrium experiments, yet to be conducted, the denatured state of these particular proteins will show a collapse transition. Indeed, the collapse of protein L has been demonstrated independently by two labs in equilibrium experiments 26,29, while not seen in a time-resolved SAXS study 51. For a fuller discussion of this issue see reference 22.
The analysis of smFRET data on denatured-state collapse of proteins in denaturant solutions has led here to a new paradigm for protein folding, in which the thermodynamics of the collapse transition is correlated with the thermodynamics of folding. As D is decreased, it is the change in conformation of the denatured state (from coil to globule) that modulates the energetic relation of this state to the folded state, driving the protein into a conformation more conducive to folding. The conformational entropy of the denatured protein becomes a key player in this process. Indeed, our analysis attributes a large fraction of the denaturant-related free energy of folding to conformational entropy changes. The current work emphasizes the essential role of the denatured state in protein folding. Further, it makes an important connection between the polymer physics of the denatured protein chain and the thermodynamics of folding. Our ability to reveal this connection and further verify the proposed physical interpretation for m-value in terms of denatured-state collapse (Equation 8), depended on the availability of new data on the CG transition from smFRET experiments. This is yet another example where single-molecule experiments and their analysis shed new (and unexpected) light on a familiar problem.
Supplementary Material
Details of computational methods, a table of m and m̄ values, a figure showing the thermodynamics of collapse for additional proteins and a figure showing the denaturant dependence of φ for all proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This research was made possible by the historic generosity of the Perlman Family and by financial support of the NIH (grant no. 1R01GM080515). G.Z. was supported by the Charles Clore fellowship for outstanding scholars in natural sciences.
Abbreviations
- CG
coil-to-globule
- smFRET
single-molecule FRET
References
- 1.Anfinsen CB, Haber E. J Biol Chem. 1961;236:1361–1363. [PubMed] [Google Scholar]
- 2.Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W. H. Freeman and Company; 1999. [Google Scholar]
- 3.Roca M, Liu H, Messer B, Warshel A. Biochemistry. 2007;46:15076–15088. doi: 10.1021/bi701732a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Alai MM, Gallo M, Salame M, Wetzler DE, McBride AA, Paci M, Cicero DO, de Prat-Gay G. Structure. 2006;14:309–319. doi: 10.1016/j.str.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Chiti F, Taddei N, Bucciantini M, White P, Ramponi G, Dobson CM. EMBO J. 2000;19:1441–1449. doi: 10.1093/emboj/19.7.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitney PL, Tanford C. J Biol Chem. 1962;237:PC1735–PC1737. [PubMed] [Google Scholar]
- 7.Schellman JA. Biopolymers. 1987;26:549–559. doi: 10.1002/bip.360260408. [DOI] [PubMed] [Google Scholar]
- 8.Mountain RD, Thirumalai D. J Am Chem Soc. 2003;125:1950–1957. doi: 10.1021/ja020496f. [DOI] [PubMed] [Google Scholar]
- 9.Mason PE, Neilson GW, Enderby JE, Saboungi ML, Dempsey CE, MacKerell AD, Brady JW. J Am Chem Soc. 2004;126:11462–11470. doi: 10.1021/ja040034x. [DOI] [PubMed] [Google Scholar]
- 10.Stumpe MC, Grubmuller H. J Phys Chem B. 2007;111:6220–6228. doi: 10.1021/jp066474n. [DOI] [PubMed] [Google Scholar]
- 11.O’Brien EP, Dima RI, Brooks B, Thirumalai D. J Am Chem Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- 12.England JL, Pande VS, Haran G. J Am Chem Soc. 2008;130:11854–11855. doi: 10.1021/ja803972g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson SE. Folding and Design. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 14.Tanford C. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 15.Tanford C. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 16.Tanford C. J Am Chem Soc. 1964;86:2050–2059. [Google Scholar]
- 17.Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton M, Holthauzen LMF, Bolen DW. Proc Natl Acad Sci U S A. 2007;104:15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SH, Shastry MC, Roder H. Nat Struct Mol Biol. 1999;6:943–7. doi: 10.1038/13311. [DOI] [PubMed] [Google Scholar]
- 20.Hagen SJ, Eaton WA. J Mol Biol. 2000;301:1019–27. doi: 10.1006/jmbi.2000.3969. [DOI] [PubMed] [Google Scholar]
- 21.Kimura T, Uzawa T, Ishimori K, Morishima I, Takahashi S, Konno T, Akiyama S, Fujisawa T. Proc Natl Acad Sci U S A. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziv G, Thirumalai D, Haran G. Phys Chem Chem Phys. 2009;11:83–91. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler B, Lipman EA, Eaton WA. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 24.Laurence TA, Kong X, Jager M, Weiss S. Proc Natl Acad Sci U S A. 2005;102:17348–53. doi: 10.1073/pnas.0508584102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmenkina EV, Heyes CD, Nienhaus GU. J Mol Biol. 2006;357:313–24. doi: 10.1016/j.jmb.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 26.Sherman E, Haran G. Proc Natl Acad Sci U S A. 2006;103:11539–43. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tezuka-Kawakami T, Gell C, Brockwell DJ, Radford SE, Smith DA. Biophys J. 2006;91:L42–4. doi: 10.1529/biophysj.106.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. Proc Natl Acad Sci U S A. 2007;104:2649–54. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Proc Natl Acad Sci U S A. 2007;104:1528–33. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann A, Kane A, Nettels D, Hertzog DE, Baumgartel P, Lengefeld J, Reichardt G, Horsley DA, Seckler R, Bakajin O, Schuler B. Proc Natl Acad Sci U S A. 2007;104:105–10. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang F, Sato S, Sharpe TD, Ying L, Fersht AR. Proc Natl Acad Sci U S A. 2007;104:123–7. doi: 10.1073/pnas.0609717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann H, Golbik RP, Ott M, Hubner CG, Ulbrich-Hofmann R. J Mol Biol. 2008;376:597–605. doi: 10.1016/j.jmb.2007.11.083. [DOI] [PubMed] [Google Scholar]
- 33.Flory PJ. Principles of polymer chemistry. Cornell University Press; New York: 1953. [Google Scholar]
- 34.Sanchez IC. Macromolecules. 1979;12:980–988. [Google Scholar]
- 35.Flory PJ, Fisk S. J Chem Phys. 1966;44:2243. [Google Scholar]
- 36.Tran HT, Mao A, Pappu RV. J Am Chem Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 37.Buscaglia M, Schuler B, Lapidus LJ, Eaton WA, Hofrichter J. J Mol Biol. 2003;332:9–12. doi: 10.1016/s0022-2836(03)00891-x. [DOI] [PubMed] [Google Scholar]
- 38.Chattopadhyay K, Elson EL, Frieden C. Proc Natl Acad Sci U S A. 2005;102:2385–9. doi: 10.1073/pnas.0500127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nettels D, Gopich IV, Hoffmann A, Schuler B. Proc Natl Acad Sci U S A. 2007;104:2655–60. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calmettes P, Roux B, Durand D, Desmadril M, Smith JC. J Mol Biol. 1993;231:840–848. doi: 10.1006/jmbi.1993.1330. [DOI] [PubMed] [Google Scholar]
- 41.Dima RI, Thirumalai D. J Phys Chem B. 2004;108:6564–6570. [Google Scholar]
- 42.Tran HT, Pappu RV. Biophys J. 2006;91:1868–1886. doi: 10.1529/biophysj.106.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mok YK, Kay CM, Kay LE, Forman-Kay J. J Mol Biol. 1999;289:619–638. doi: 10.1006/jmbi.1999.2769. [DOI] [PubMed] [Google Scholar]
- 44.Since only two datasets were measured using urea as denaturant, we use only the proteins denatured with GdmCl here
- 45.O’Brien EP, Jr, Brooks B, Ziv G, Haran G, Thirumalai D. Proc Natl Acad Sci U S A. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso DO, Dill KA. Biochemistry. 1991;30:5974–85. doi: 10.1021/bi00238a023. [DOI] [PubMed] [Google Scholar]
- 47.Guo ZY, Brooks CL. Biopolymers. 1997;42:745–757. doi: 10.1002/(sici)1097-0282(199712)42:7<745::aid-bip1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 48.Veitshans T, Klimov D, Thirumalai D. Fold Des. 1997;2:1–22. doi: 10.1016/S1359-0278(97)00002-3. [DOI] [PubMed] [Google Scholar]
- 49.Jacob J, Dothager RS, Thiyagarajan P, Sosnick TR. J Mol Biol. 2007;367:609–15. doi: 10.1016/j.jmb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Jacob J, Krantz B, Dothager RS, Thiyagarajan P, Sosnick TR. J Mol Biol. 2004;338:369–82. doi: 10.1016/j.jmb.2004.02.065. [DOI] [PubMed] [Google Scholar]
- 51.Plaxco KW, Millett IS, Segel DJ, Doniach S, Baker D. Nat Struct Mol Biol. 1999;6:554–6. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of computational methods, a table of m and m̄ values, a figure showing the thermodynamics of collapse for additional proteins and a figure showing the denaturant dependence of φ for all proteins. This material is available free of charge via the Internet at http://pubs.acs.org.