Abstract
Chronic infection with hepatitis C virus (HCV) affects 130 million people worldwide and is a major cause of liver cirrhosis and liver cancer. After translation of the HCV RNA genome into a polyprotein, 2 viral proteases process its non-structural protein (NS) region. While the essential chymotrypsin-like serine protease NS3–4A mediates all cleavages downstream of NS3, the NS2–3 cysteine protease catalyzes a vital cleavage at the NS2/3 site. Protease activity of NS2–3 has been described to require, besides NS2, the N-terminal 181 aa of NS3. The latter domain corresponds to the NS3 serine protease domain and contains a structural Zn2+-binding site with functional importance for both viral proteases. The catalytic triad of the NS2–3 protease resides in NS2; the role of the NS3 part in proteolysis remained largely undefined. Here we report a basal proteolytic activity for NS2 followed by only 2 amino acids of NS3. Basal activity could be dramatically enhanced by the NS3 Zn2+-binding domain (NS3 amino acids 81–213) not only in cis but also in trans which, however, required a more extended N-terminal part of NS3 downstream of NS2 in cis. Thus, this study defines for the first time (i) NS2 as a bona fide protease, (ii) NS3 as its regulatory cofactor, and (iii) functional subdomains in NS3 that cooperate in NS2 protease activation. These findings give new mechanistic insights into function and regulation of the NS2 protease and have important implications for the development of anti-HCV therapeutics.
Keywords: autoprotease, hepacivirus/HCV, protease cofactor, zinc binding domain
Hepatitis C virus (HCV) is an important human pathogen affecting about 3% of the human population. Chronic infections are frequent and a major cause of liver cirrhosis and liver cancer. HCV is a member of the genus Hepacivirus, which is grouped together with the genera Pestivirus and Flavivirus in the Flaviviridae family. Upon infection, the replication cycle of HCV begins with translation of the viral polyprotein from the single-stranded messenger sense RNA genome. The relative order of the proteins is NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COO− (1). The N-terminal part of the polyprotein encompassing the structural proteins (C, E1, E2) and p7 is processed by cellular, ER-resident peptidases. The viral non-structural proteins NS2 to NS5B are released by 2 virus-encoded proteases. The NS2–3 cysteine protease catalyzes the cleavage between NS2 and NS3, which is essential for viral RNA replication (2–4). The chymotrypsin-like serine protease domain residing in the N-terminal 180 aa of NS3 requires for full activity NS4A as cofactor (5, 6). The assembled NS3–4A protease generates the C terminus of NS3 and processes all downstream junctions in the polyprotein.
The catalytic triad of the NS2–3 cysteine protease is composed of histidine 952 (H952), glutamic acid 972 (E972), and cysteine 993 (C993) and resides entirely in NS2 (2, 3). However, it has been published that cleavage at the NS2/3 site (L1026/A1027) essentially requires the N-terminal 181 aa of NS3 in addition to NS2 (7). Although this NS3 part covers the entire NS3 serine protease domain, neither its catalytically active residues nor NS4A are required for processing at the NS2/3 site. Crystallographic data revealed that 3 cysteines (amino acids 1123, 1125, and 1171) and 1 histidine (amino acid 1175), located in NS3 between residues 97 and 149, coordinate 1 Zn2+ ion (8, 9). The presence of Zn2+ has been shown to be of functional importance for the activities of both the NS2–3 and the NS3–4A protease (3, 10). Biochemical and crystallographic data favor a structural rather than a catalytic role for the coordinated Zn2+ ion (7, 11); however, no structure is available for uncleaved NS2–3 to resolve this point of debate. The recently established crystal structure of the cysteine protease domain of NS2 uncovered its dimeric nature and a novel composite architecture of the active sites (12). Each monomer contributes the active site histidine and glutamic acid to 1 catalytic triad, whereas the active site cysteine is provided by the second monomer. Thus a single NS2 monomer on its own has no functional active center, but the homodimer displays 2 active sites. Together with in vitro cleavage studies these findings explained the earlier observation that cleavage may occur also in trans (13).
The crystal structure represents the postcleavage state of the NS2 protease domain in the absence of any NS3-derived sequences; thus, the role of the NS3 part of the NS2–3 protease in NS2–3 cleavage remains unclear.
In the related pestivirus bovine viral diarrhea virus (BVDV), processing at the NS2/3 site is catalyzed in noncytopathogenic strains by a cysteine protease in NS2 (14). Similar to HCV NS3, BVDV NS2 encompasses a putative Zn2+-binding site, which overlaps with the catalytic triad of the enzyme and is critical for NS2 protease activity. Interaction between a cellular chaperone and 2 independent binding sites in BVDV NS2 is a prerequisite for protease activity. This feature allowed an artificial split of the autoprotease into a protease and a substrate domain. Both domains reassembled into a functional protease in the presence of the chaperone (15).
We therefore hypothesized that, in analogy to the work on BVDV, it might be possible to split the NS2–3 protease of HCV and thereby define functional subdomains.
In the present study we observed for the first time that NS2 followed by only the first 2 aa of NS3 displays an intrinsic basal proteolytic activity. Furthermore our experiments identified amino acids 81–213 of HCV NS3, containing the Zn2+-binding site, as a functional NS2 protease-activating region (in the following termed NS3 Zn2+-binding domain). When supplemented in trans, this domain can functionally complement the Zn2+-binding domain of the NS2–3 protease and stimulate the basal activity of the NS2 protease. To be stimulated in trans, NS2 essentially requires the N-terminal 60 aa of NS3 in cis. In line with this finding, a deletion of amino acids 61–80 of the NS3 part of the NS2–3 protease did not interfere with cleavage at the NS2/3 site.
Accordingly, this study identifies for the first time an intrinsic proteolytic activity of NS2 and establishes NS2 as a bona fide protease with NS3 acting as a regulatory cofactor. In addition, we mapped functional subdomains in NS3, which cooperate in NS2 protease activation.
Results
A Proteolytically Active Split NS2–3 Protease.
In the crystal structure of HCV NS3 established by Kim et al. (8), a synthetic NS4A cofactor peptide interacts intimately with the N-terminal β-barrel domain, especially with β-strands A0 and A1 (Fig. S1). We hypothesized that this property could define this part as an autonomous functional subunit of NS3, which is required for the activity of the NS2–3 cysteine protease. For a functional test, the NS2–3 protease was thus split between residues 40 and 41 of its NS3 part. Expression plasmid pFlag-NS2-NS3(1–40)-GST encodes under the control of the T7 promoter a Flag epitope, NS2, and the N-terminal 40 aa of NS3 followed by GST (Fig. 1). According to the structure, the NS3 part in this construct encompasses β-strand A0, helix α0, β-strand A1, and the first 3 aa of the connecting loop to β-strand B1 (Fig. S1). The second plasmid pmyc-NS3(41–213) encodes the remainder of the NS3 fragment, namely amino acids 41–213, preceded by a myc epitope. Construct pFlag-NS2-NS3(1–213)-GST as well as pFlag-NS2-NS3(1–213)-GST/C993A, the latter with an exchange of the NS2 active site cysteine to alanine, served as controls. The HCV polyprotein fragments were transiently expressed by the Vaccinia virus MVA-T7pol system in Bsr cells, a BHK derivative. Metabolically labeled proteins were enriched by radioimmunoprecipitation (RIP), separated by SDS/PAGE, and detected by autoradiography.
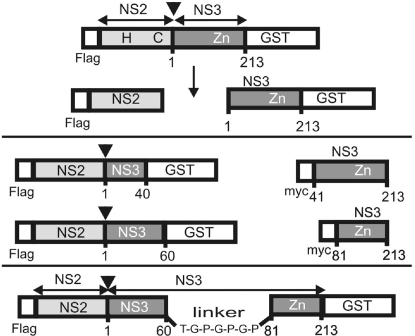
Fig. 1.
Scheme of selected HCV polyprotein fragments expressed in this study (for a complete set, see Figs. S7–S9). Numbers below the bars indicate the first and last aa position of NS3 contained in the construct. (Top) Flag-NS2-NS3 (1–213)-GST, including NS2 and a complete NS3 serine protease domain in cis. H and C indicate residues H952 (NS2 amino acid 143) and C993 (NS2 amino acid 184), 2 of the catalytic residues situated in NS2; Zn shows the location of the NS3 Zn2+-binding region. (Middle Left) Flag-NS2-NS3-GST derivatives with truncated NS3 regions. (Middle Right) Corresponding myc-NS3 fragments used for stimulation of Flag-NS2-NS3-GST processing in trans. (Bottom) Internal replacement of NS3 (amino acids 61–80) in NS2–3 by a flexible linker.
Upon transfection of pFlag-NS2-NS3(1–213)-GST, proteins with molecular weights expected for uncleaved Flag-NS2-NS3(1–213)-GST and Flag-NS2 or NS3(1–213)-GST were pulled down by the Flag or the GST mAb, respectively (Fig. 2Top and Middle, lane 2); this protein pattern indicates efficient but incomplete cleavage at the NS2/NS3 site. No cleavage products were detected after transfection of pFlag-NS2-NS3(1–213)-GST/C993A (Fig. 2 Top and Middle, lane 3), verifying the specificity of the processing event at the NS2/NS3 site. Accordingly, the tagged NS2–3 protease was active and thus suited to study its proteolytic function.
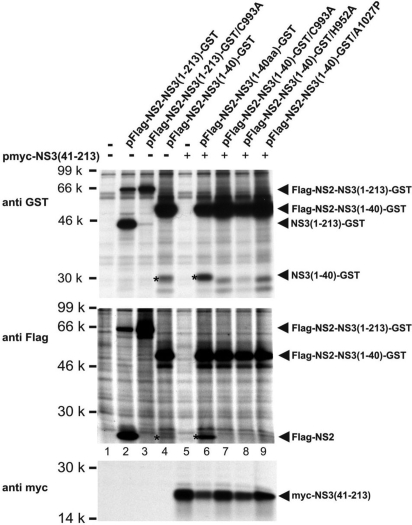
Fig. 2.
In vitro assay of Flag-NS2-NS3-GST processing. (Top, Middle) The plasmids indicated above each lane were transfected alone (lanes 2 to 4) or together with pmyc-NS3(41–213). Proteins were metabolically labeled and radioimmunoprecipitated under denaturing conditions with the antibodies indicated on the left. Proteins were separated by SDS/PAGE and detected by autoradiography. Asterisks indicate the cleavage products Flag-NS2 and NS3(1–40)-GST resulting from processing of Flag-NS2-NS3(1–40)-GST. Products of basal cleavage are shown in lane 4, products of stimulated cleavage in lane 6. Lanes 7–9 show controls with constructs containing mutations in either the catalytic residues of the NS2 protease (lane 7 and 8) or at the NS2/3 cleavage site (lane 9). (Bottom) Expression controls for myc-NS3(41–213).
Upon transfection of pFlag-NS2-NS3(1–40)-GST, unexpectedly, small amounts of Flag-NS2 and NS3(1–40)-GST (Fig. 2 Top and Middle, lane 4) were detected besides the uncleaved polypeptide. The amount of these cleavage products significantly increased when plasmid pmyc-NS3(41–213) was cotransfected (Fig. 2 Top and Middle, lane 6). Variants of pFlag-NS2-NS3(1–40)-GST carrying mutations at the active site residues (C993A or H952A) or the NS2/3 cleavage site (A1027P) (13) served as controls. These mutants did not show any cleavage at the NS2/3 site neither when expressed alone nor on coexpression of myc-NS3(41–213) (Fig. 2 Top and Middle, lanes 7–9).
Thus, processing of NS2-NS3(1–40)-GST occurs at the NS2/3 site, depends on the activity of the NS2–3 cysteine protease, and can be stimulated significantly by a NS3 fragment [myc-NS3(41–213)] in trans.
Stimulation of NS2–3 Cleavage by NS3 in Trans.
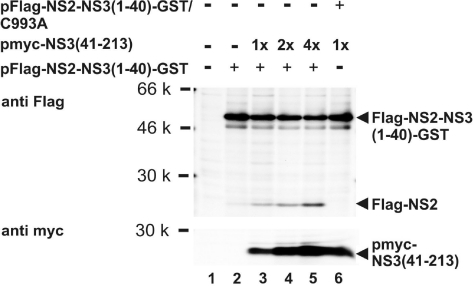
To further investigate stimulation of cleavage at the NS2/3 site by NS3 in trans, increasing quantities of pmyc-NS3(41–213) were cotransfected with a constant amount of pFlag-NS2-NS3(1–40)-GST. As demonstrated by the appearance of increasing amounts of cleavage product Flag-NS2, processing at the NS2/3 site increased in a dose-dependent manner with the molar amount of pmyc-NS3(41–213) cotransfected (Fig. 3Upper, lanes 2–5) [for PhosphorImager (Typhoon 9200; Amersham Biosciences) analysis, see Fig. S2]. No cleavage was observed upon cotransfection of pmyc-NS3(41–213) with the NS2–3 construct encompassing an active site mutant (Fig. 3 Upper, lane 6).
Fig. 3.
Stimulation of Flag-NS2-NS3-GST cleavage by myc-NS3(41–213) in trans. (Upper) Increasing molar amounts of plasmid pmyc-NS3(41–213) are cotransfected with pFlag-NS2-NS3(1–40)-GST. (Lower) The resulting increase in protein expression is shown. The metabolically labeled cleavage product Flag-NS2 is enriched by denaturing RIP and detected by autoradiography (see also Fig. 2).
Next the N-terminal border of the minimal activating NS3 fragment was determined. Upon coexpression of Flag-NS2-NS3 (1–40)-GST and myc-NS3(51–213) no enhancement of cleavage was observed (Fig. S3). C-terminal truncation of the cotransfected NS3 fragment to amino acid 180 resulted in a less stable, but cleavage-activating protein. Further C-terminal truncation to amino acid 160 abolished cleavage enhancement (Fig. S4) in agreement with previous mapping data for the NS2–3 protease (2). Therefore, NS3 fragments terminating at amino acid 213 were used in all subsequent experiments. Thus, in this setup, the N-terminal NS3 fragment could be split into 2 subdomains for stimulation of cleavage in trans but the entire NS3 protease domain was required.
These experiments revealed 2 unexpected results: (i) an NS2–3 protease encompassing only 40 aa of NS3 displayed basal intrinsic proteolytic activity; and (ii) cleavage efficiency of this protease could be strongly enhanced by coexpression of myc-NS3(41–213) in trans.
A Protease Activity of NS2 Followed by 2 Amino Acids of NS3.
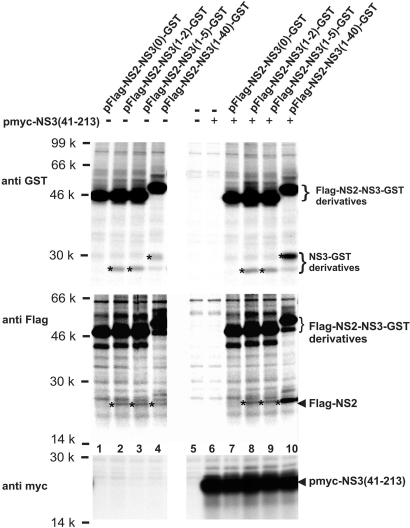
The surprising result described above, that NS2 followed by 40 aa of NS3 shows basal cleavage at the NS2/3 site, prompted us to determine the minimal part of NS3 required for protease activity. To this end Flag-NS2-NS3-GST derivatives varying in the length of their NS3 parts were tested. Flag-NS2 followed by 2, 5, or 40 aa of NS3 and GST still catalyzed cleavage at the NS2/3 site (Fig. 4Top and Middle, lanes 2 to 4); in contrast, fusion of GST directly to the C terminus of NS2 abolished processing (Fig. 4 Top and Middle, lane 1).
Fig. 4.
Minimal part of NS3 required in the Flag-NS2-NS3-GST context for basal cleavage activity and for cleavage stimulation in trans. The C-terminally truncated Flag-NS2-NS3-GST constructs contain 40, 5, 2, or 0 aa of NS3, respectively, followed by the tripeptide TGF and GST. Asterisks indicate the cleavage products as detected by RIP (see also Fig. 2).
Importantly, enhancement of cleavage by coexpression of myc-NS3(41–213) was restricted to Flag-NS2-NS3(1–40)-GST (Fig. 4 Top and Middle, lane 10) and not observed for Flag-NS2-NS3-GST variants with shorter NS3 parts in cis (Fig. 4 Top and Middle, lane 7 to 9).
Thus, an NS3 part as short as 2 aa is sufficient for basal intrinsic NS2–3 protease activity; however, myc-NS3(41–213) expressed in trans could enhance this protease activity only when the NS2–3 protease comprised no less than 40 aa of NS3 in cis.
Minimal NS3 Subdomain in Trans Capable of Activating Cleavage at the NS2/3 Site.
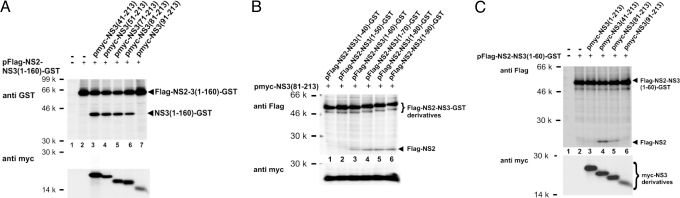
Encouraged by these results, a systematic search for split versions of the NS2–3 protease was started. A NS2–3 protease with 160 aa of NS3 in cis was tested for activation by different NS3 fragments in trans. Upon transfection of pFlag-NS2-NS3(1–160)-GST, only basal cleavage at the NS2/3 site was observed (Fig. 5A, lane 2), which could, however, be strongly stimulated by cotransfection of constructs expressing NS3 fragments N-terminally truncated to amino acid positions 41, 51, 71, or 81 (Fig. 5A, lanes 3–6). Surprisingly, cleavage enhancement of NS2–3(1–160) by these NS3 fragments demonstrates that overlaps in NS3 do not generally interfere with protease activation in split NS2–3 versions. A further truncation of 10 aa (pmyc-NS3(91–213); Fig. 5A, lane 7) abolished cleavage stimulation by the NS3 fragment in trans.
Fig. 5.
Activation of Flag-NS2–3-GST derivatives by the NS3 Zn2+-binding domain in trans. (A) Flag-NS2-NS3(1–160)-GST was coexpressed with different myc-NS3 fragments containing NS3 residues (41–213) to (91–213). (B) Minimal length of the NS3 fragment required in cis for cleavage activation by myc-NS3(81–213) in trans. (C) Minimal NS3 fragment sufficient for cleavage activation of Flag-NS2–3(1–60)-GST in trans (see also Fig. 2).
In the following the NS3 part of the NS2–3 protease required in cis for activation by myc-NS3(81–213) in trans was determined. For this purpose, the shortest NS3 peptide activating in trans identified so far, namely myc-NS3(81–213), was coexpressed with NS2–3 proteases varying in the lengths of their NS3 parts. Upon cotransfection of pFlag-NS2-NS3(1–90)-GST, pFlag-NS2-NS3(1–80)-GST, pFlag-NS2-NS3(1–70)-GST, or pFlag-NS2-NS3(1–60)-GST with pmyc-NS3(81–213), efficient cleavage was observed (Fig. 5B, lanes 3–6); cotransfection of pmyc-NS3(81–213) with pFlag-NS2-NS3(1–50)-GST and pFlag-NS2-NS3(1–40)-GST did not result in enhancement of NS2–3 processing (Fig. 5B, lanes 1 and 2). This was also in agreement with the result that NS2 with 40 aa of NS3 in cis requires a larger NS3 part for activation in trans, namely NS3 (41–213) (see Fig. 4).
Thus, NS2 with 60 aa of NS3 in cis can be stimulated in its proteolytic activity by myc-NS3(81–213)—here termed NS3 Zn2+-binding domain—in trans.
Finally, coexpression of Flag-NS2-NS3(1–60)-GST with a series of myc-NS3 fragments confirmed that the N-terminal border of an NS3 fragment with the capacity to stimulate cleavage at the NS2/3 site is position 81 as encoded by pmyc-NS3(81–213) (Fig. 5C, lane 5) because cotransfection of pmyc-NS3(91–213) had no stimulatory effect on processing (Fig. 5C, lane 6). To exclude that this negative result was not due to the lower amount of myc-NS3(91–213) detected in this assay, the experiment was repeated by using a 6 times higher molar amount of the respective plasmid; despite efficient myc-NS3(91–213) expression, no enhancement of NS2–3 processing was observed (see Fig. S5).
Interestingly, coexpression of myc-NS3(1–213) had almost no stimulatory effect on NS2–3 processing (Fig. 5C, lanes 4 and 3, respectively). The fact that the NS3(1–213) serine protease domain with a complete N terminus had a drastically reduced ability to stimulate processing at the NS2/3 site for NS2–3(1–60) in trans may indicate an intramolecular interaction between the N-terminal NS3 subdomain and the Zn2+-binding domain.
Amino Acids 61–80 of NS3 Are Dispensable for the Activity of the NS2–3 Protease.
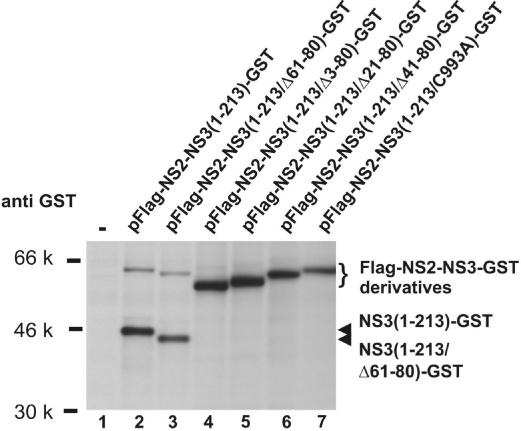
The experiments shown in Fig. 5 B and C demonstrated that amino acids 61–80 of NS3 were not required for stimulation of NS2/3 cleavage by the Zn2+-binding domain in trans. We therefore tested whether this peptide can also be deleted from the NS2–3 protease in cis. In the construct pFlag-NS2-NS3(1–213/Δ61–80)-GST the sequence coding for NS3 amino acid 61–80 was replaced by one encoding a short flexible linker peptide. Expression of this construct led to efficient processing of NS2–3 indicated by the appearance of NS3(1–213/Δ61–80)-GST (Fig. 6, lane 3). In a series of additional constructs, namely pFlag-NS2-NS3(1–213/Δ3–80)-GST, pFlag-NS2-NS3(1–213/Δ21–80)-GST, pFlag-NS2-NS3(1–213/Δ41–80)-GST, this linker coding sequence replaced sequences coding for larger parts of the N-terminal NS3 region. None of these proteins displayed cleavage at the NS2/3 site (Fig. 6, lanes 4–6), as it was the case for the active site mutant pFlag-NS2-NS3(1–213)-GST/C993A (Fig. 6, lane 7).
Fig. 6.
Autocleavage of Flag-NS2–3-GST with internal deletions in NS3. The amino acids of NS3 indicated in brackets were replaced by the flexible linker peptide TGPGPGP. Catalytically inactive Flag-NS2-NS3-GST/(C993A) served as control (see also Fig. 2).
Discussion
The replication of all positive-strand RNA viruses involves translation and proteolytic processing of polyproteins. This process is of vital importance for these viruses because it generates the functional polypeptides required for viral propagation. Moreover, fine-tuning of proteolytic processing by protease cofactors, either virus-encoded or cell-derived, is of major regulatory importance for the replication cycles of this large group of viruses.
The vital cleavage event at the NS2/3 site of HCV requires, according to the literature, besides NS2 at least 181 aa of NS3(1). Because all active site residues are located in NS2, the reason for the requirement of NS3 for cleavage remained elusive. Recently, the postcleavage structure of the HCV NS2 protease domain has been solved at the atomic level (12). Superimposition of the catalytic triad of the active site of the HCV protease with, for example, poliovirus 3C protease or Sindbis virus capsid protease suggested that the structure could represent that of an active enzyme even in the absence of NS3 sequences. This hypothesis is now confirmed by our experimental demonstration of an enzymatic activity of NS2 followed by only 2 aa of NS3, which for the first time defines NS2 itself as a bona fide protease (see Fig. 4). In addition, this important finding classifies NS3 as a cofactor of the NS2 protease. Although a function in catalysis had been ruled out previously for the active site residues of the NS3 serine protease in NS2–3 cleavage (2, 3), the precise role of the Zn2+ ion coordinated by NS3 is still under discussion (7). Our result that the Zn2+-binding domain defined in this study is not essential for basal NS2 protease activity but rather for NS3 cofactor activity may now end this long-lasting debate.
Although NS2–3 can undergo autocleavage at a basal rate with a minimum of 2 aa of NS3 expressed in cis, the NS3 cofactor dramatically enhances its enzymatic activity. The question remains how NS3 promotes NS2–3 cleavage. The unique composite architecture of NS2 suggests a crucial role of dimerization for the activity of the NS2–3 protease (12). A previous study reported an NS2/NS2 interaction in mammalian cells by coimmunoprecipitation also in the absence of NS3 (16). This indicates that NS3 is not essential for basic dimerization (or oligomerization) of NS2. Hence, the NS3 cofactor promotes in a more subtle way the efficient formation of the proteolytically active conformation of the NS2–3 dimer, possibly by promoting the correct positioning of the cleavage site. On the basis of in vitro studies, a role for soluble cytoplasmatic factor(s) in the activation of the NS2–3 protease has been postulated (17) implying that NS3 could recruit necessary chaperones into the folding process. A direct role for NS3 in supporting the NS2–3 dimer to achieve its active conformation without being actively involved in the catalysis of the cleavage reaction implies mechanistic parallels to intramolecular chaperones. This class of protein domains is often found in proteases (e.g., subtilisin or α-lytic protease) where they are called prosequences and precede the protease domain in the polypeptide chain (18). Several prosequences actively assist in folding and thereby protease activation, which may even occur after addition in trans (18). After protease activation, prosequences are cleaved off and dispensable for subsequent enzyme function. In contrast to prosequences, the NS3 protease domain fulfills essential functions in viral replication after processing.
This study defines for the first time functional subdomains within NS3, which cooperate in NS2–3 protease activation. For activation in trans, the N-terminal NS3(1–60) fragment located downstream of NS2 has to be complemented by the NS3 Zn2+-binding domain (amino acids 81–213), indicating that amino acids 61–80 of NS3 are dispensable. This was confirmed by the successful replacement of this NS3 region by a linker sequence in the context of NS2–3 (see Fig. 6). However, larger deletions within the N-terminal NS3 region strongly interfered with NS3-mediated stimulation of proteolysis both in the NS2–3 context (in cis) and in trans. These results imply that the N-terminal NS3 subdomain (amino acids 1–60) has a function beyond a protein/protein interaction platform merely required to ensure proximity between NS2 and the NS3 Zn2+-binding domain because this property should have been functionally substituted by the linker peptide in the deletion mutants.
Within NS3, an interaction between the N-terminal NS3 subdomain and the NS3 Zn2+-binding domain is supported by the fact that NS3 fragments already containing both subdomains were drastically reduced in their capacity to activate the NS2 protease in trans (Fig. 5C). Accordingly, intramolecular interactions within NS3 may inhibit functional interactions of the NS3 Zn2+-binding domain with an N-terminal NS3 subdomain offered in trans. In contrast, a large overlap in NS3 between NS2–3(1–160) and activating NS3 fragments in trans did not interfere with activation by the NS3 Zn2+-binding domain in trans (see Fig. 5A). This might indicate that this incomplete NS3 Zn2+-binding domain, which is also inactive as NS2 protease cofactor (see Fig. 5A), does not adopt a functional conformation.
Our coimmunoprecipitation experiments did not reveal a stable interaction between the NS3 Zn2+-binding domain and the N-terminal NS3 subdomain whether the latter is preceded by NS2 or not. This obviously does not rule out transient or weak interactions not detected by this technique. The absence of a stable interaction between the NS2 protease and the activating NS3 cofactor subdomains is in sharp contrast to the situation in the NS3–4A protease of HCV, where the activating NS4A cofactor peptide is an integral part of the enzyme and contributes 1 β-strand to the N-terminal β-barrel domain of the chymotrypsin-like serine protease fold (8).
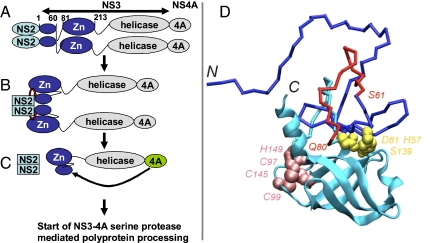
The result that the deletion of amino acids 61–80 from NS3 in the context of NS2–3 did not interfere with the activity of the NS2–3 protease also raises another interesting aspect. Although this peptide is dispensable for the function of NS3(1–213) as cofactor of the NS2 protease, it clearly represents an essential structural element in the already crystallized NS3 serine protease where it is crucial for the correct spacing between the active site histidine and aspartate residues (NS3 amino acids 57 and 81, respectively; see Fig. 7D and Fig. S1). Our findings thus demonstrate that the NS3 part of the NS2–3 protease fulfills 2 distinguishable functions as a (i) protease cofactor or (ii) serine protease. Although both functions involve large parts of the NS3 serine protease domain, those still significantly differ with respect to their sequence requirements and thus most likely also in terms of the required protein conformation. Our finding thus might be a first hint for a major structural change in NS3 necessary for the switch between NS2 protease cofactor and serine protease function. Other indications for different folds might be the lower affinity of uncleaved NS2–3 for NS4A when compared with NS3(19), structural changes observed in NS3 in the presence of the activating peptide of NS4A, (8, 9) and differences observed between the Zn2+ requirements for NS2–3 cleavage and NS3 serine protease activity (20). Structural changes should occur after activation of the NS2–3 protease by the NS3 Zn2+-binding domain resulting in NS4A binding, which is strongly facilitated by NS2–3 cleavage (19) and leads to the final serine protease fold. Mutual stimulation of the NS3 helicase and serine protease domain has been described recently (21). These inter-domain cooperations might compete with the ability of the NS3 protease domain to interact with NS2. In agreement with this assumption, an NS3 fragment starting with amino acid 41 and containing the entire downstream part of NS3 was not capable of activating cleavage of Flag-NS2–3(1–40)-GST in trans (Fig. S6). Accordingly, the data set available up to now suggests an ordered cascade of events as exemplified in Fig. 7. Upon translation, NS2 dimerization and/or folding of the NS3 Zn2+-binding domain may be rate limiting because both are required for induction of NS2–3 cleavage (Fig. 7 A and B); this processing event is necessary to allow efficient NS4A binding to NS3, which in turn initiates processing at all NS3/4A dependent cleavage sites and thereby the formation of active replication complexes (Fig. 7C).
Fig. 7.
Mechanistic model of the sequential contribution of NS3 subdomains to processing of the NS2–3-4A region. (A) NS2 (light blue) is translated from the viral genomic RNA; an NS2 dimer may already be formed by 2 polyprotein chains translated simultaneously from 1 viral RNA. Each NS2 monomer is followed by a growing NS3 serine protease domain (dark blue). At this stage, the 2 activating domains in NS3, namely the N-terminal subdomain (amino acids 1–60) and the Zn2+-binding domain (amino acids 81–213), connected via a dispensable linker sequence (amino acids 61–80), fold into the “NS2 cofactor conformation” of the NS3 protease domain. (B) NS3(1–60) in concert with the Zn2+-binding domain activates NS2 and thereby induces NS2-NS3 cleavage (red arrow). (C) After cleavage, a conformational change is likely to occur in NS3 enabling efficient association of NS4A, which in turn completes the NS3 chymotrypsin-like serine protease fold. (D) Tentative structural model of the N-terminal NS3 subdomain (amino acids 1–180). The well folded C-terminal β-barrel subdomain is represented by ribbon diagrams and colored cyan. Less stable or unfolded structures (NS3 amino acids 1–95, including the N-terminal β-barrel subdomain in the absence of the stabilizing NS4A peptide) are represented by sticks colored in blue, except segment 61–80 colored in red. Side-chain atoms of the catalytic triad (His-57, Asp-81, and Ser-139) are highlighted as yellow spheres. Atoms of Cys-97, Cys-99, Cys-145, and His-149 involved in Zn2+ binding are represented as pink spheres. For modeling procedure see SI Materials and Methods.
Both HCV proteases, NS2–3 and NS3–4A, catalyze vital processing events. Therefore these enzymes represent important targets for the still insufficient therapy of chronic HCV infections, with an estimated 130 million cases worldwide. However, viral autoproteases such as HCV NS2–3 often display very fast cleavage kinetics, which complicates the identification of inhibitory lead structures. Besides defining novel targets in NS3, the findings of this study also may help to circumvent the kinetic problem because they allow study of proteolysis in “slow motion.” In the absence of the activating NS3 Zn2+-binding domain, only a few of the NS2–3 protease molecules undergo cleavage; this allows incubation of the uncleaved precursors with inhibitor candidates before triggering cleavage via the NS3 Zn2+-binding domain in trans.
Thus the definition of functional subdomains within the NS2–3 protease of HCV contributes to the mechanistic understanding of this crucial step in viral replication and opens new avenues for the development of therapeutics against this important pathogen.
Materials and Methods
Cells.
Bsr cells, a BHK derivative, were grown in Dulbecco's modified Eagle's medium with 10% FCS. Cells were maintained at 37 °C and 5% CO2. Vaccinia virus modified virus Ankara-T7pol (22) was generously provided by G. Sutter (Paul-Ehrlich-Institut, Langen, Germany).
Transient Expression and Metabolic Labeling of Proteins.
The applied procedures have been described (15). The T7-vaccinia virus system was used in Bsr cells for protein expression. For transfection of plasmid DNA (3 μg), Superfect (Qiagen) was applied. For labeling of 1 × 106 cells, 140 μCi (1 Ci = 37 GBq) of [35S]methionine/-cysteine (35S-ProMix; Amersham Biosciences) were used.
Antibodies and Antisera.
The anti-Flag tag, anti-GST (GST), and anti-myc tag monoclonal antibodies (mAb) were purchased from Sigma-Aldrich. Secondary species-specific antibodies were purchased from Dianova.
RIP and SDS/PAGE.
Cells were lysed in radioimmunoprecipitation (RIP) assay (RIPA) buffer [50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1% (vol/vol) Nonidet P-40, 1% (wt/vol) deoxycholate, 2% (wt/vol) SDS, and 0.5 mM PefablocSC (Merck)]; afterwards, boiling samples were diluted 1:10 in RIPA buffer without SDS. Protein A-Sepharose (Sigma-Aldrich) and RIPA buffer were used for RIP. Proteins were separated in polyacrylamide-tricine gels with 8% or 10% polyacrylamide.
Expression Plasmids.
All expression plasmids are based on pCITE (Novagen), which encompasses the internal ribosomal entry site (IRES) of encephalomyocarditis virus downstream of the T7 RNA polymerase promoter. The cDNA sequence encoding NS2–3 of HCV BK (23) is the one used by Pallaoro et al. (24). The encoded parts of the HCV BK polyprotein are indicated by the names of the constructs (see Fig. 1 and SI Materials and Methods and Figs. S7–S9).
pFlag Constructs.
The aa sequence encoded by all pFlag constructs upstream of the HCV NS2 sequence is MDYKDDDDKLEPG. The constructs encode the 3 aa TGF followed by GST (GST) downstream of the HCV polypeptides.
In the deletion mutants within the NS3 gene (e.g., pFlag-NS2-NS3(1–213/Δ61–80)-GST), a sequence coding for the peptide TGPGPGP replaces the deleted NS3 sequence indicated in the plasmid name.
pmyc Constructs.
The aa sequence MPEQKLISEEDLAM is encoded upstream of the HCV NS3 polyprotein fragment indicated by the plasmid name.
Supplementary Material
Acknowledgments.
We thank S.-E. Behrens (Martin-Luther-Universität, Halle, Germany) for donating the cDNA of HCV BK, G. Sutter for providing vaccinia virus modified virus Ankara-T7pol, K.-K. Concelmann (Ludwig-Maximilians-Universität, Munich) for the Bsr cells, I. Lorenz for critical reading of the manuscript, and H.-J. Thiel for continuous generous support. This study was financed by Sonderforschungsbereich 535 and Grant TA 218/1-1 of the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810950106/DCSupplemental.
References
- 1.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins (LWW); 2007. pp. 1101–1152. [Google Scholar]
- 2.Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice C. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hijikata M, et al. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Failla C, Tomei L, de Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welbourn S, Pause A. The hepatitis C virus NS2/3 protease. Curr Issues Mol Biol. 2007;9:63–69. [PubMed] [Google Scholar]
- 8.Kim JL, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 9.Love RA, et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 10.Thibeault D, Maurice R, Pilote L, Lamarre D, Pause A. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J Biol Chem. 2001;276:46678–46684. doi: 10.1074/jbc.M108266200. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Yao N, Le HV, Weber PC. Mechanism of autoproteolysis at the NS2-NS3 junction of the hepatitis C virus polyprotein. Trends Biochem Sci. 1998;23:92–94. doi: 10.1016/s0968-0004(98)01180-3. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. Structure of the catalytic domain of the hepatitis C virus NS2–3 protease. Nature. 2006;442:831–835. doi: 10.1038/nature04975. [DOI] [PubMed] [Google Scholar]
- 13.Reed KE, Grakoui A, Rice CM. Hepatitis C virus-encoded NS2–3 protease: Cleavage-site mutagenesis and requirements for bimolecular cleavage. J Virol. 1995;69:4127–4136. doi: 10.1128/jvi.69.7.4127-4136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lackner T, et al. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J Virol. 2004;78:10765–10775. doi: 10.1128/JVI.78.19.10765-10775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackner T, Thiel HJ, Tautz N. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc Natl Acad Sci USA. 2006;103:1510–1515. doi: 10.1073/pnas.0508247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieroni L, et al. In vitro study of the NS2–3 protease of hepatitis C virus. J Virol. 1997;71:6373–6380. doi: 10.1128/jvi.71.9.6373-6380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinde U, Inouye M. Intramolecular chaperones: Polypeptide extensions that modulate protein folding. Semin Cell Dev Biol. 2000;11:35–44. doi: 10.1006/scdb.1999.0349. [DOI] [PubMed] [Google Scholar]
- 19.Welbourn S, et al. Hepatitis C virus NS2/3 processing is required for NS3 stability and viral RNA replication. J Biol Chem. 2005;280:29604–29611. doi: 10.1074/jbc.M505019200. [DOI] [PubMed] [Google Scholar]
- 20.Tedbury PR, Harris M. Characterisation of the role of zinc in the hepatitis C virus NS2/3 auto-cleavage and NS3 protease activities. J Mol Biol. 2007;366:1652–1660. doi: 10.1016/j.jmb.2006.12.062. [DOI] [PubMed] [Google Scholar]
- 21.Beran RK, Pyle AM. Hepatitis C viral NS3–4A protease activity is enhanced by the NS3 helicase. J Biol Chem. 2008;283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 23.Takamizawa A, et al. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallaoro M, et al. Characterization of the hepatitis C virus NS2/3 processing reaction by using a purified precursor protein. J Virol. 2001;75:9939–9946. doi: 10.1128/JVI.75.20.9939-9946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.