Abstract
Previous studies revealed roles for RNA interference (RNAi) in the immediate cellular response to viral infection in plants, nematodes and flies. However, little is known about how RNAi combats viruses during persistent or latent infections. Our analysis of small RNAs cloned from Drosophila cells latently infected with Flock House Virus (FHV) failed to reveal signatures of bulk degradation of the viral genome. Instead, this + strand virus specifically generated Dicer-2-dependent, 21-nucleotide siRNAs that derived in equal proportion from + and − strands. Curiously, luciferase reporters that are fully complementary to abundant viral siRNAs were poorly repressed. Moreover, although the viral siRNAs that were incorporated into an effector complex associated with Argonaute2, bulk FHV siRNAs in latently infected cells were not loaded into any Argonaute protein. Together, these data suggest that direct dicing of viral replication intermediates plays an important role in maintaining the latent viral state. In addition, the denial of bulk viral siRNAs from effector complexes suggests that criteria beyond the structural competency of RNA duplexes influence the assembly of functional silencing complexes.
Keywords: Argonaute2, Dicer-2, RNA interference, virus
Genetic analyses showed that core components of the RNA interference (RNAi) pathway generate viral small interfering RNAs (siRNAs) and restrict virus accumulation in flies (1–5), worms (6–8), and plants (9, 10). Such studies indicate that the cellular defense to viral infection begins when double-stranded RNA (dsRNA) viral genomes or replication intermediates are cleaved by Dicer-class RNase III enzymes into small interfering RNAs (siRNAs). The viral siRNAs are incorporated into Argonaute complexes that subsequently cleave and degrade viral coding RNAs, preventing completion of the viral lifecycle. To counteract the RNAi defense, many viruses have evolved proteins that inhibit various components of the RNAi pathway, thus permitting their successful replication and/or infection (11).
In Drosophila, the RNAi and microRNA (miRNA) pathways are genetically distinct, but have substantial cross-talk (12). The canonical RNAi pathway uses Dicer-2 to process dsRNA into siRNAs, which are mostly loaded into Argonaute2 (AGO2) complex. AGO2-resident RNAs are then methylated at their 3′ ends by the Hen1 methyltransferase (13, 14). The miRNA pathway uses Dcr-1 to process premiRNA hairpins into mature miRNAs, which are mostly loaded into Argonaute1 (AGO1) complex; they remain unmethylated. Recent studies demonstrated that the sorting of diced small RNAs is influenced by the structural features of their duplex precursors. Small RNAs from perfectly double stranded duplexes are favored to enter AGO2, whereas central bulges favor entry into AGO1 (15, 16). However, these rules do not entirely explain the types of RNAs that are resident in AGO1 and AGO2 (17, 18); therefore, there are presumably additional determinants that affect small RNA loading.
Because the studies to date focused on the role of RNAi in combating acute infection, little is known about its role during latent infection. To address this, we exploited cell lines that are persistently infected by Flock House Virus (FHV). RNAi has a significant role in maintaining FHV latency, and the small RNA signatures of this defense provided mechanistic insight. Dicer-2-dependent siRNAs were preferentially generated from particular regions of the bipartite FHV genome. Unexpectedly, the abundant siRNAs derived from latent virus were not effective in silencing reporters bearing complementary sequences. Moreover, although viral siRNAs were preferentially associated with the siRNA effector AGO2 relative to the miRNA effector AGO1, bulk FHV-derived siRNAs were unmethylated and did not substantially associate with either Argonaute. These data suggest that direct dicing of the replication intermediate plays an important role during latent infection, and hint at different activities of the RNAi pathway during acute infection versus persistent infection.
Results
Persistent Infection of Multiple Drosophila S2 Lines by Flock House Virus.
Recent examination of largescale small RNA sequence data indicated the persistent infection of an S2 cell stock (“S2-NP,” from the laboratory of Norbert Perrimon) by Flock House Virus (FHV), a bipartite positive strand + RNA virus (17). FHV RNA1 encodes protein A, the viral RNA dependent RNA polymerase. It also generates RNA3 encoding B2, a dsRNA binding protein that was proposed to mask the viral genome from the RNAi defense (2, 19, 20). The second segment of the FHV genome, RNA2, encodes the capsid proteins.
Although productive FHV infection induces apoptosis and is lethal in adult Drosophila and S2 cells (1, 2, 19), this virus persists in S2-NP seemingly without affecting cell proliferation or survival. We performed Northern blot analysis to confirm the presence of FHV genomic RNAs in these cells, and observed considerable accumulation of RNA2 and RNA3, but little of full length RNA1 (Fig. S1). We collected several other S2 and S2R+ cells from various laboratories and surveyed them for FHV. S2 cells from Ram Dasgupta's laboratory (“S2”) and Phillip Zamore's laboratory (“S2-Z”) were free of FHV (Fig. 1A and Fig. S1), but S2 cells from Gerald Rubin (“S2-GMR”) and S2R+ cells from Dasgupta's laboratory (“S2R+”) both harbored FHV. Therefore, persistent FHV infections are fairly common among Drosophila laboratories.
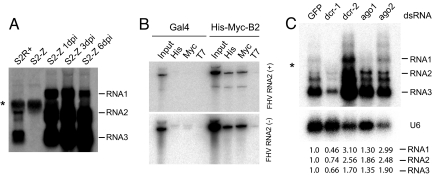
Fig. 1.
Latent FHV infection is maintained by the RNAi pathway. (A) FHV transcripts were detected in S2R+ cells from the DasGupta laboratory, but not in a culture of S2 cells obtained from the Zamore laboratory (S2-Z). Addition of S2R+ conditioned media to S2-Z cells resulted in rapid and high level accumulation of FHV transcripts. (*), background hybridization to ribosomal RNA. (B) Coimmunoprecipitation of both + and − strands of FHV RNA2 with His/Myc-tagged B2. Specificity of the interaction was assessed by background levels of FHV RNA2 recovered in the absence of transfected B2, or after immunoprecipitation with control T7 antibody. (C) Expression of FHV genomic RNAs after depletion of RNAi/miRNA pathway components. (Top) FHV transcripts were up-regulated after depletion of dcr-2 or ago2. (Middle) The blot was stripped and probed for U6 as a loading control; note the slightly unequal loading in the ago2 lane. (*), ribosomal RNA background. Bottom is a quantification of the viral transcripts, normalized first to the U6 level and then expressed as a ratio of the level in cells treated with GFP dsRNA.
Canonical RNAi Factors Restrict Persistent FHV Infection.
In principle, deleterious mutations in persistent FHV strains might account for their reduced potency. For example, latent FHV might simply be mutated for the B2 suppressor, whose loss prevents FHV from mounting a productive infection of WT cells (1, 2, 19). However, assembly of the B2 ORF from the cloned small RNAs in 2 different strains (S2-GMR and S2R+) revealed that it was intact, save for a few nucleotide changes with respect to the reference virus sequence. Furthermore, ectopic expression of functional B2 into latently infected cells did not restore the replication of FHV. Exogenous B2 was able to bind to the viral genome (Fig. 1B), suggesting that the latent strain retained the cis-acting sequences for B2 recognition. Thus, alterations in B2 do not appear to account for the latency of FHV.
We next used a functional test to report directly on the pathogenicity of latent FHV. We prepared conditioned media from latently infected S2R+ cells and added it to cultures of naive S2-Z cells. After as little as 24 h, newly infected cells showed signs of apoptosis, such as membrane blebbing. Northern blot analysis revealed a dramatic accumulation of each of the FHV RNAs in the newly infected cells (Fig. 1A). The abundance of FHV RNA2 and RNA3 was considerably higher in the newly infected cells than in latently infected cells, and importantly strong induction of RNA1 was now seen. Thus, the latent FHV strain is capable of mounting robust infection and replication. The level of RNA1 in the newly infected cells decreased substantially by 6 days postinfection, evidence for a host defense to the viral infection.
Previous studies showed that components of the canonical RNAi pathway mediate defense against various single-stranded RNA and double-stranded RNA viruses (1–4). We tested whether RNAi activity was responsible for maintaining viral latency. We depleted the core components of the RNAi and miRNA pathways (dcr-1, dcr-2, ago1, and ago2) in the latently infected S2R+ cells and assayed virus accumulation by Northern blot analysis. We observed significant accumulation of viral transcripts upon loss of Dicer-2 or AGO2 (Fig. 1C). These data extend a recent quantitative rt-PCR analysis of FHV RNA2 levels after knockdown of small RNA pathway components (17). In particular, the increased abundance of full length RNA1 upon depletion of Dicer-2 or AGO2, as determined by these Northern blot analysis experiments, reported directly upon the derepression of viral replication.
Characteristic Size and Spatial Distribution of Small RNAs Derived from Latent FHV.
We sought further insight into the molecular strategies by which small RNA pathways recognize and degrade these viral genomes. To do so, we analyzed small RNA sequences from latently infected S2-GMR and S2R+ cells using the Illumina platform. To ensure that the quantitative results from these sequencing experiments were reproducible, we analyzed 2 independent small RNA libraries from each celltype yielding ≈30–60,000 FHV reads per library. Because these libraries yielded 3–4 million matches to the reference genome, they contain ≈1–2% FHV reads (Table S1).
Size distribution analysis revealed FHV was predominantly processed into 21 nt species, with similar overall numbers of sense and antisense FHV reads (Fig. 2A). Their strong 21 nt bias suggested that the majority of FHV-derived small RNAs are generated by dicing into siRNAs. Because FHV positive strands outnumber negative strands by at least 50:1 (21), these cloning data suggest that FHV siRNAs were generated from a double-stranded replication intermediate. Notably absent were degradation fragments of the + strand, which might have been expected to be present as a consequence of host defense of the persistent FHV infection. In Drosophila cells, degradation fragments are rapidly phosphorylated resulting in their cloning via 5′ phosphate-dependent protocols (22). Abundant cellular transcripts therefore generate short RNAs of heterogeneous length that span the size window used for cloning (23). In contrast, the vast majority of FHV reads were precisely 21 nt (Fig. 2A).
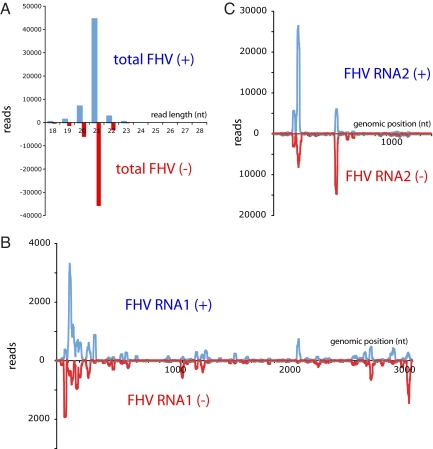
Fig. 2.
Features of FHV-derived small RNAs in latently infected S2R+ cells. (A) Predominantly 21 nt RNAs were generated from both + and − strands of FHV, indicative of siRNA processing. (B) Small RNAs mapped to FHV-RNA1. (C) Small RNAs mapped to FHV-RNA2. Discrete regions of the FHV genome generate abundant siRNAs. These graphs report the combined reads from 2 independently-generated libraries; Fig. S2 shows that similar patterns were seen in the 2 datasets. Moreover, the spatial distribution of siRNAs from the viral genome was similar between independent types of cultured cells bearing latent FHV infection.
Although the entire FHV genome is expected to be double-stranded during + strand replication, the majority of FHV siRNAs mapped to discrete regions of the virus. Most of the viral siRNAs were generated from near the 5′ ends of both FHV genomic RNAs, with certain internal regions also producing a substantial number of siRNAs (Fig. 2 B and C). The spatial patterns of FHV siRNA mappings were highly reproducible between independent samples of S2R+ and of S2-GMR cells, and these different cell lines exhibited similar overall distribution of siRNAs along the FHV genome (Fig. S2). A recent analysis of small RNAs cloned from S2 cells acutely infected with FHV lacking the B2 product revealed a similar spatial pattern of FHV-derived siRNAs (5).
The coincidence of + and − strand siRNAs provided additional evidence of their origin from dicing of the replication intermediate, rather than from intramolecularly base-paired segments. We hypothesize that these siRNA hotspots represent sensitive points in the FHV genome to Dicer-2. Interestingly, we determined that dsRNA binding B2 suppressor protein bound both + and − strands of FHV (Fig. 1C). Based on the viral siRNA cloning data, it seems likely that B2 is bound to the replication intermediate.
Latent FHV-Derived siRNAs Mediate Poor Repression of Complementary Targets.
Based on the known requirement of AGO2 for viral defense (2, 3, 17, 19), a reasonable expectation is that viral siRNAs mediate the silencing of complementary viral transcripts in trans. To test this, we designed luciferase sensors containing 70 nt sense and/or antisense segments of several regions of FHV RNA2 (Fig. 3A). These correspond to 170–240 bp (R1), and 530–600 bp (R3) from which large numbers of siRNAs are derived. We also cloned a companion region from 430 to 500 bp (R2) that did not yield substantial numbers of siRNAs (Fig. 3B). Finally, we generated a matched set of mutant sensors in which the viral sequence was mutated at every 4th position, so as to disrupt the possibility for either siRNA- or miRNA-like regulation by viral siRNAs. These mutant sensors were used to normalize the expression of companion wild-type reporters in FHV-infected (S2R+ and S2-NP) and FHV-free (S2) cells.
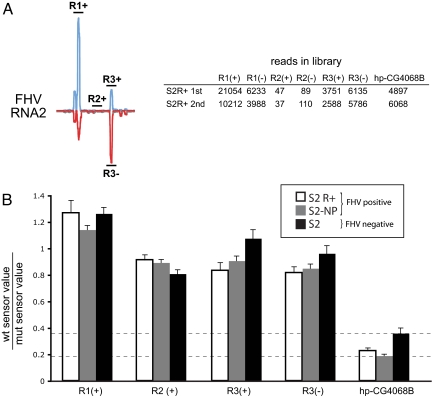
Fig. 3.
FHV-derived siRNAs do not mediate effective target repression. (A) Three ≈70 nt regions of FHV-RNA2 (see Fig. 2C) were selected to generate WT renilla luciferase sensors. Companion mutant sensors contained mutations at every 4th position; sensors were made for both + and − strands of region 3. The table shows the number of reads obtained from the 3 regions of FHV RNA2; the reads matching the endo-siRNA hp-CG4068B are shown for comparison. hp-CG4068B is slightly more abundant than R3+ siRNAs, similar in abundance to R3− siRNAs, and far less abundant than R1+ siRNAs. (B) FHV siRNA sensors were assayed in FHV infected and FHV-free cells. The ratios of normalized WT to mutant sensor activities were plotted, and standard error is depicted. In contrast to the hp-CG4068B sensor, which exhibited 3- to 5-fold repression compared with its mutant sensor (24), FHV siRNAs did not mediate substantial target repression. The dotted line provides a reference for the expected level of repression mediated by siRNAs whose expression is in the range of hp-CG4068B.
Active small RNA-mediated repression would yield ratios of wild-type to mutant sensor activities that are <1, but we would expect their values to be equalized in cells lacking the cognate small RNAs. Surprisingly, the ratios of WT sensors complementary to the abundant siRNAs derived from R1 and R3 were nearly equivalent to their mutant counterparts in latently infected cells (Fig. 3B). If anything, the sensor complementary to the extremely abundant R1 siRNAs was slightly elevated compared with its mutant control sensor. In addition, the normalized sensor values from the non-siRNA producing region 2 were not markedly different from the siRNA-complementary sensors, even though the level of R1 siRNAs was 370-fold that of R2 siRNAs. Finally, no substantial differences were observed with R1 and R3 sensors in infected versus virus-free cells (Fig. 3B).
To confirm the viability of these sensor assays, we performed parallel experiments with WT and mutant sensors for the endo-siRNA hp-CG4068B (24). As expected from previous studies (24), we observed 3- to 5-fold repression of this sensor by endogenous hp-CG4068B. The level of endogenous hp-CG4068B is similar to, or indeed much lower than, the various FHV siRNAs assayed, and these quantitative comparisons were stable over multiple libraries and cell types (Fig. 3 and Table S1). Therefore, these results indicate that the abundant viral siRNAs produced during latent viral infection are largely ineffective at silencing complementary transcripts.
Bulk FHV-Derived siRNAs from Latent Infection Are Not Loaded into Argonaute Proteins.
We sought to explain the disconnect between viral siRNA production and activity by examining their biogenesis and biochemical properties. FHV siRNAs derived from the 170–240 bp (R1) region of RNA2 in latently infected cells were detected on Northern blots using a combination of 4 LNA probes. We used this assay to examine viral siRNA accumulation after the depletion of core miRNA/siRNA factors. The efficacy of knockdowns was assessed by the behavior of a previously tested miRNA (bantam) and endo-siRNA (hp-CG4068B) (17, 18, 24, 25). As expected, mature bantam levels were decreased after knockdown of dcr-1 or ago1, whereas the levels of mature hp-CG4068B were decreased after knockdown of dcr-2 or ago2 (Fig. 4A).
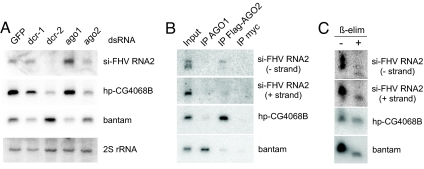
Fig. 4.
Biogenesis and terminal properties of FHV-derived siRNAs. (A) Accumulation of FHV siRNAs after depletion of components of the miRNA/RNAi pathways. Blotting for the bantam miRNA and hp-CG4068B endo-siRNA indicate loss of mature miRNA after dcr-1 or ago1 depletion, and loss of mature siRNA after dcr-2 or ago2 depletion, as expected. In contrast to hp-CG4068B, FHV-derived siRNAs are lost only after depletion of dcr-2, and not with ago2; these were detected with a mixture of sense and antisense FHV probes. (B) Coimmunoprecipitation of small RNAs with AGO1 or Flag-AGO2. Bantam miRNA is predominantly associated with AGO1, whereas hp-CG4068B endo-siRNA is predominantly associated with AGO2. FHV siRNAs that are loaded in effector complex associated primarily with AGO2, but most of the viral siRNAs did not associate with either AGO protein. Sense and antisense FHV siRNAs were detected separately in these experiments; input lane = 25% of input used for IP. These data are quantified in Fig. S3. (C) Terminal nucleotide properties of small RNAs. Endo-siRNAs are resistant to β-elimination, whereas miRNAs are sensitive and migrate more quickly. FHV-RNA2 siRNAs are similarly sensitive to β-elimination.
Using these validated knockdown samples, we observed that mature FHV-siRNAs were highly dependent on dcr-2, even moreso than hp-CG4068B. In contrast, FHV-siRNAs exhibited far less dependence on ago2 than did hp-CG4068B. This correlated with the failure of FHV-derived siRNAs to mediate substantial target repression (Fig. 3), and suggested that the majority of FHV-derived siRNAs might not be loaded into functional effector complexes. There was an apparent increase in FHV-derived siRNAs after ago1 knockdown. It is not clear whether this reflects an indirect effect of increased viral replication in the absence of this miRNA effector, or whether loading into AGO2 is improved if there is less AGO1 present.
To test for the presence of unloaded FHV siRNAs, we examined the association of FHV-derived siRNAs with AGO1 and AGO2. A recent report of largescale sequencing of AGO1- and AGO2-associated RNAs in S2-NP revealed that the incorporation of FHV-derived RNAs into AGO2 was strongly preferred to AGO1 (17). The AGO2-IP dataset GSM280087 harbors 42,364 FHV-derived reads relative to 916,834 reads that mapped to the dm3 assembly of the Drosophila melanogaster genome (≈4.41% si-FHV), whereas the AGO1-IP dataset GSM280088 contains only 3567 FHV-derived reads compared with 2,094,408 reads that mapped to the Drosophila genome (≈0.170% si-FHV). Thus, there are ≈26 times more FHV RNAs in AGO2 than in AGO1.
To directly verify the preferred sorting of viral siRNAs to AGO2, we immunoprecipitated AGO1 or FLAG-tagged AGO2 from S2R+ cells and probed their contents (Fig. 4B). As shown previously, mature bantam miRNA was mostly associated with AGO1, whereas the endo-siRNA hpCG4068B was mostly associated with AGO2 (24, 25). To our surprise, although a minor fraction of FHV-derived siRNA was complexed with AGO2, consistent with previous observations, bulk viral siRNAs from the + or − strands did not associate with either Argonaute (Fig. 4B). Quantitation of relative incorporation of the various siRNAs and miRNAs into AGO1 and AGO2 are provided in Fig. S3. Therefore, although there is indeed preferred sorting of viral siRNAs to AGO2, the majority of them are denied entry into AGO proteins.
Of Drosophila small regulatory RNAs in the 21–24 nt pool, only those that are incorporated into AGO2 silencing complex are subject to 3′ modification by the Hen1 methyltransferase; bulk miRNAs are unmethylated (13). Unmethylated RNAs are sensitive to periodate oxidation and β-elimination at the terminal nucleotide, and consequently migrate as lower molecular weight species (13, 26, 27). As shown previously, most of the mature hp-CG4068B was resistant to β-elimination (18, 24), reflecting its efficient incorporation into AGO2 complex (Fig. 4C). Almost all mature bantam was sensitive to β-elimination, reflecting the efficacy of this treatment and the fact that very little of this miRNA is sorted to AGO2. This assay revealed that bulk viral siRNAs from both the + and − strands were similarly sensitive to β-elimination, confirming their inefficient loading into AGO2 complexes (Fig. 4C). Altogether, these data demonstrate that although the RNAi pathway maintains FHV latency in S2 cells, this is poorly correlated with the activity of bulk AGO2-loaded viral siRNAs as transacting inhibitory species.
Discussion
Role of Dicing During Maintenance of Drosophila Virus Latency.
Here, we have examined how latency of FHV in persistently infected cells is maintained by the RNAi pathway. Viral siRNAs were abundant in small RNA libraries, and shown by Northern blot analysis to be produced in a Dicer-2-dependent fashion. Unexpectedly, our data reveal that bulk viral siRNAs in these cells do not program active effector complexes. These siRNAs fail to repress complementary sensor transcripts, associate with AGO2 rather weakly, and are sensitive to β-elimination. Taken together, our data suggest that in addition to the well-appreciated role for dicing to generate siRNAs that cleave viral transcripts, the direct dicing of double stranded replication intermediates appears to play an appreciable role in maintaining FHV in a latent state.
Although the relative contributions of antiviral dicing and slicing remain to be fully elucidated, it is worth considering whether there might be distinct phases and activities of the RNAi pathway during antiviral response. Upon onset of active infection, siRNAs are both generated and used to target coding RNAs, causing destruction of viral RNAs. However, if the infection can be adequately controlled, the necessity to sort viral siRNAs into RISC might conceivably become less critical, whereas Dicer-2 remains active in turning over the remaining persistent viral RNAs. Dicer-2 was also recently shown to induce the antiviral Vago protein, apparently in an AGO2-independent manner, demonstrating that the role ofDicer-2 extends beyond simply the generation of antiviral RISC (28). Nevertheless, there is clearly a role for AGO2 in suppressing latent FHV (17), as with acute FHV infection (2, 5, 19). It is possible that Dicer-2 requires its effector AGO2 to efficiently clear siRNA duplexes. Alternatively, it might be that a very small amount of AGO2 programmed with viral siRNA suffices to maintain FHV latency.
A caveat of our studies is that a pool of latently infected virus may have undergone genomic rearrangement, which might alter its properties. Previous work described the spontaneous appearance of attenuated, internally deleted FHV isoforms (29, 30), whose destruction by Dicer-2 might account for our ability to detect siRNAs via sequencing and Northern blot analysis. However, this scenario does not account for the robust derepression of full-length viral RNA accumulation upon the depletion of dcr-2. In addition, we demonstrated that latent FHV retains most of its function and can readily infect and kill naïve cells. Future studies on the dynamics and kinetics of de novo infection by previously latent virus may help to establish the parameters of successful establishment of the latent state, and address whether there might be shifting influences of viral dicing and slicing during this transition.
Strategy of Virus Defense and Counterdefense.
Latent infection of FHV, a single stranded RNA virus, was associated with the relatively equivalent accumulation of siRNAs from positive and negative strands. Because − strand transcripts usually account for only 1–5% of corresponding + strands, these siRNA signatures indicate that the double stranded FHV replication intermediate is the substrate for Dicer-2-mediated defense. However, the entirety of the FHV replication intermediate was not equally susceptible to dicing. Instead, the punctuated spatial distribution of FHV-derived siRNAs suggested these as entry locations that are especially sensitive to dicing. Similar findings were recently reported by Ding and colleagues using an acute FHV infection system (5).
The recent report that FHV replication is spatially restricted to spherules within the mitochondrion offers a picture in which the host RNAi machinery may have limited access to replicating virus (21). FHV encodes a double-stranded RNA binding protein (B2) that protects it from the host RNAi response (1, 2, 19, 20). We showed that + and − strands of FHV are associated with B2. These data suggest that B2 protects FHV by binding the Dicer-2-susceptible replication intermediate, as opposed to an intramolecularly double-stranded region of the coding viral genome (see also ref. 5). In light of these spatial constraints, future studies of virus defense will need to address cell biological aspects of virus recognition and evasion. This seems reminiscent of the recent appreciation of the cell biological constraints that compartmentalize miRNA-mediated repression within the cell (31).
Implications of Latent Viral siRNA Biogenesis for Small RNA Sorting.
We showed that FHV-derived siRNAs were preferentially incorporated into AGO2 relative to AGO1, consistent with recent sequencing data from Argonaute immunoprecipitates from latently infected cells (17). However, the bulk of siRNAs were not loaded, and were not 3′ modified as expected for AGO2-loaded small RNAs (13, 14). Similarly, Ding and colleagues recently detected a pool of FHV-derived siRNAs generated from an attenuated form of FHV lacking the B2 suppressor of RNAi were not loaded into either AGO1 or AGO2 effector complexes, and lacked a 3′ modification (5). Moreover, the Ding laboratory showed that viral siRNAs generated during an acute infection also did not depend on AGO2 for their accumulation; in fact they observed over-accumulation of FHV-derived siRNAs after infection of ago2 mutants with attenuated FHV (5). The accumulation of unloaded viral siRNAs during acute and latent infections contrasts strongly with the behavior of diverse classes of endo-siRNAs, for which Northern analysis demonstrates them to require AGO2 for their stable accumulation (17, 18, 23, 24, 32).
Why can the generation and loading of viral siRNAs, unlike that of endo-siRNAs, be uncoupled? In principle, the abundant production of virus during acute infection might saturate the loading machinery, potentially yielding a population of unloaded siRNAs. However, the pool of viral siRNAs produced during latent infection is perhaps <5% the pool of endo-siRNAs that are loaded into AGO2, indicating that the failure to load latent viral siRNAs is not a consequence of saturating the loading machinery. Indeed, the klarsicht locus alone accounts for ≈16% of the contents of AGO2 in S2-NP and S2-GMR cells (17, 23), demonstrating that the highly abundant production of siRNAs from a single locus is possible. Instead, these observations might suggest the existence of licensing measures that select appropriate siRNAs for loading and/or retention in effector complex.
Indeed, mechanisms for siRNA selection were hinted at by recent studies of Drosophila endo-siRNAs. We and others recently characterized hairpin RNAs, which are long inverted repeat transcripts that are processed by Dicer-2 into siRNAs that preferentially load AGO2 (12). Although the small RNA cloning patterns from some hpRNAs revealed signature evidence for the processive action of Dicer-2 to yield phased siRNA duplexes (17, 18, 24), some hpRNAs generated abundant siRNAs from isolated regions embedded in the midst of long double-stranded precursors (24). Such patterns suggest that bulk siRNAs generated by dicing across a long substrate were not loaded into effector complex, and were instead discarded. In at least some systems, siRNA-bearing AGO proteins have been shown to be limiting for siRNA accumulation (33). Consequently, licensing mechanisms for siRNA loading might be important to ensure the optimal occupancy of AGO proteins with the desired siRNAs. In principle, an improved understanding such siRNA selection mechanisms will improve the ability to design effective artificial siRNAs for designed purposes.
Materials and Methods
Library Construction and Analysis.
Independent 50-μg samples of total RNA from S2R+ cells or S2-GMR were fractionated on polyacrylamide gels, and the ≈18–28 nt fraction was cloned according to the protocol of Hannon (17). Each library was subjected to a single lane of sequencing using the Illumina platform. The reads were clipped of the 3′ linkers, and submitted to the National Center for Biotechnology Information Gene Expression Omnibus under the accessions GSM343832 and GSM343833 (S2R+, first and second biological replicates) and GSM361908 (S2-GMR) cells. We also used data from an independent biological replicate of S2-GMR reads that we reported in GSM272652 (32).
Using Flock House Virus genomic sequences obtained from National Center for Biotechnology Information, we extracted ≥18 nt reads that mapped perfectly to the viruses for analyses of the size and spatial distribution of viral small RNAs. To assemble the FHV genome from small RNA reads, we also considered ≥18 nt reads with 1 or 2 mismatches/1 nt deletion/1 nt insertion to the reference FHV sequence, discarding those reads that also mapped perfectly to the dm3 assembly of the D. melanogaster genome.
RNA Molecular Biology.
A mixture of 4 32P-labeled DNA probes were used to detect FHV RNA1, RNA2 and RNA3 genomic RNAs by agarose Northern blot analysis. Two probes were complementary to the shared + strand of RNA1 and RNA3 (5′-ACCTCTGCCCTTTCGGGCTAGAAC-3′ and 5′-TCACTTCCGGTTGTTGGAAGGC-3′), whereas the other two were complementary to the + strand of RNA2 (5′-AGGAGGACACTTGATCGGATCTGG-3′ and 5′-GGGATCGGTGTTGAAGTCAGGTG-3′). A DNA probe complementary to U6 (5′-GCAGGGGCCATGCTAATCTTC-3′) was used to determine relative loading.
β-elimination and detection of small RNAs by acrylamide Northern blot analysis was performed as described (13, 34). We used 4 LNA probes that are complementary to FHV RNA2 + strand (5′-GGGAAAGCGCCGCCATATTCATGCC-3′ and 5′-GTGCGAAGGCACACTTGAGAAACGC-3′) or FHV RNA2 − strand (5′-GCGTTTCTCAAGTGTGCCTTCGCAC-3′ and 5′-GCATGAATATGGCGGCGCTTTCCCG-3′). hp-CG4068B was detected by an LNA probe (5′-GGAGCGAACTTG TTGGAGTCAA-3′), and bantam was detected by a DNA probe (5′-AATCAGCTTTCAAAATGATCTCA-3′). A DNA probe complementary to 2S rRNA (5′-TACAACCCTCAACCATATGTAGTCCAAGCA-3′) was used to determine equal loading.
To analyze B2-associated RNAs, we cotransfected UAS-6xHis-3xMyc-FHV-B2 (35) with Ub-Gal4 into S2R+ cells. After 48 h cells were lysed in 20 mM Tris (pH 7.4), 200 mM NaCl, 2.5 mM MgCl2, 0.05% Nonidet P-40 with Complete Protease inhibitors (Roche). Immunoprecipitation was carried out with mouse anti-6xHis or mouse anti-Myc (Santa Cruz Biotechnology), and T7 antibodies (Novagen) bound to GammaBind G Sepharose (GE Healthcare). RNAs were isolated from beads by phenol choloform extraction followed by ethanol precipitation. RNAs were separated on a 6% Urea acrylamide gel and blotted onto nylon membranes (Genescreen). 32P end-labeled DNA probes were used to detect the 3′end of the FHV RNA2+ strand (5′-ACCTTAGTCTGTTGACTTAAACTGTTTGGG-3′) or the 3′ end of the FHV RNA2 − strand (5′-GTAAACAATTCCAAGTTCCAAAATGGTCAAC-3′).
Immunoprecipitation of AGO complexed RNAs was carried out in cells stably transfected with Flag-HA-tagged AGO2 (17). These cells were lysed in 30 mM Hepes-KOH (pH 7.3), 150 mM K acetate, 2 mM Mg acetate, 0.1% Nonidet P-40, and 5 mM DTT with Complete Roche Protease inhibitors. Flag-HA-AGO2 was immunoprecipitated using Anti-Flag M2-Agarose (Sigma). Rabbit anti-AGO1 (Abcam) and mouse anti-myc (Santa Cruz Biotechnology) were immobilized on GammaBind G Sepharose (GE Healthcare) before immunoprecipitation. RNAs were extracted as described above and analyzed on 15% Urea acrylamide gel and Northern blotting.
Luciferase Assays.
Luciferase assays were performed as described in ref. 34 using dual Renilla/Firefly sensors and the Dual-Glo luciferase kit (Promega). Three different 70 nt regions of the + or − strand of FHV RNA2 were cloned into a modified version of psiCheck2 (Promega) (34): 169–239 bp (region1), 433–503 bp (region 2), and 530–599 bp (region 3). Mutant controls corresponding to each segment were made with changes at every 4th base and similarly cloned into psiCheck2. Values from wild-type sensors were normalized to mutant controls to determine the relative repression. This experiment was performed in 3 cell lines, S2R+, S2-NP, and S2.
Supplementary Material
Acknowledgments.
We thank Gregory Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for Flag-HA-AGO2 plasmid; Norbert Perrimon (Harvard Medical School, Boston, MA), Ram Dasgupta (New York University, New York), Gerald Rubin (Janelia Farm, Ashburn, VA), Philip Zamore (University of Massachusetts Medical School, Worcester, MA), Lucy Cherbas (Indiana University, Bloomington, IN), and Sue Celniker (Lawrence Berkeley National Laboratory, Berkeley, CA) for Drosophila cell cultures and/or RNA samples. Illumina sequencing was performed by the British Columbia Genome Science Centre (Vancouver). Katsutomo Okamura performed preliminary experiments and provided helpful discussion. This work was supported by the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund and National Institutes of Health Grants R01-GM083300 and U01-HG004261.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813412106/DCSupplemental.
References
- 1.Galiana-Arnoux D, et al. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 2.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8:880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 5.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, et al. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 8.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 11.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forstemann K, et al. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czech B, et al. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 20.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 21.Kopek BG, et al. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravin A, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 23.Okamura K, et al. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila. Nat Struct Mol Biol. 2008;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 28.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 29.Ball LA, Li Y. cis-acting requirements for the replication of flock house virus RNA 2. J Virol. 1993;67:3544–3551. doi: 10.1128/jvi.67.6.3544-3551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Ball LA. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J Virol. 1993;67:3854–3860. doi: 10.1128/jvi.67.7.3854-3860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 32.Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 34.Okamura K, et al. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou YT, Tam B, Linay F, Lai EC. Transgenic inhibitors of RNA interference in Drosophila. Fly. 2007;1:311–316. doi: 10.4161/fly.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.