Abstract
Neurotransmitter release at the synapse requires membrane fusion. The SNARE complex, composed of the plasma membrane t-SNAREs syntaxin 1A and SNAP-25 and the vesicle v-SNARE synaptobrevin, mediates the fusion of 2 membranes. Synaptic vesicles contain unusually high cholesterol, but the exact role of cholesterol in fusion is not known. In this study, cholesterol was found to stimulate SNARE-mediated lipid mixing of proteoliposomes by a factor of 5 at a physiological concentration. Surprisingly, however, the stimulatory effect was more pronounced when cholesterol was on the v-SNARE side than when it was on the t-SNARE side. Site-directed spin labeling and both continuous wave (CW) and pulsed EPR revealed that cholesterol induces a conformational change of the v-SNARE transmembrane domain (TMD) from an open scissors-like dimer to a parallel dimer. When the TMD was forced to form a parallel dimer by the disulfide bond, the rate was stimulated 2.3-fold even without cholesterol, supporting the relevance of the open-to-closed conformational change to the fusion activity. The open scissors-like conformation may be unfavorable for fusion and cholesterol may relieve this inhibitory factor.
Keywords: EPR, membrane fusion, syntaxin, synaptobrevin
In the neuron, synaptic vesicles fuse with plasma membrane to release neurotransmitters into the synaptic cleft (1–4). Membrane fusion involves extensive bilayer remodeling, which requires free energy or an effective catalyst (5–6). SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) are believed to be the fusion machine that serves as the energy source (7–8) or catalyst that lowers the fusion energy barrier (9). At the onset of fusion, vesicle associated (v-) SNARE synaptobrevin (or VAMP2) combines with target membrane (t-) SNAREs Syntaxin 1A and SNAP-25 to form a helical bundle that bridges 2 membranes, facilitating fusion (10–14).
Cholesterol is a major component of the plasma and the vesicle membranes, composing as much as 30–40 mol % of the total lipids (15–16). Cholesterol is structurally rigid and known to harden the membrane significantly, making it resistant to being bent or deformed. However, there is evidence that cholesterol accelerates membrane fusion that involves a series of bilayer deformations (17–20). The physical origin of cholesterol's positive role can be found partly in its spontaneous negative curvature such that it prefers to be in the negatively curved (concave) surface but dislikes being in the positively curved (convex) surface (21).
Despite progress in understanding the effects of cholesterol on the bilayer curvature, not much is known about its influence on the lateral distribution of SNAREs in the membrane or on structures of their transmembrane domains (TMDs). A series of recent reports addresses the importance of the clustering of SNAREs at the fusion site for successful fusion (19, 22–24). In addition, there is evidence that the SNARE TMDs are essential for formation of the fusion pore (25–27). Considering cholesterol's structural rigidity and its nonideal mixing with phospholipids (28), we speculate that both the lateral distribution and the conformation of the SNARE TMDs are affected significantly by the presence of cholesterol. Indeed, it has been shown that cholesterol is a key component to induce lateral clustering of Syntaxin 1A (19).
In this work, we attempt to correlate the conformational changes of the SNARE TMDs induced by cholesterol to the fusion activity of neuronal SNAREs. The fluorescence lipid-mixing assay was used to investigate the effect of cholesterol on SNARE-mediated membrane fusion. The results show that lipid mixing was promoted significantly when cholesterol is on the v-SNARE-carrying vesicles but not as much when it is on t-SNARE-carrying vesicles. Using site-directed spin labeling (SDSL) and both continuous wave (CW) and pulsed-EPR spectroscopy, we found a significant conformational change of the TMD dimers of v-SNARE synaptobrevin (or VAMP2) that is induced by cholesterol. This change resembles a scissors-type motion of the TMD dimers. The relevance of this structural change to the fusion activity was verified by using the disulfide cross-linking method.
Results
Physiological Cholesterol Stimulates Lipid Mixing.
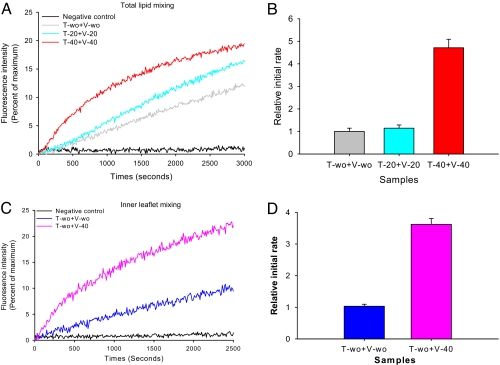
To investigate the effect of cholesterol on SNARE-mediated lipid mixing, we incorporated appropriate amounts of cholesterol into the POPC/DOPS vesicles to achieve 3 different cholesterol concentrations of 0, 20, and 40 mol %. We kept the negatively charged DOPS level at 15 mol % for all 3 samples. The t-SNARE complex was preformed by mixing Syntaxin 1A and SNAP-25 in the 1:1 molar ratio. The t-SNARE complex was then reconstituted into the vesicles (t-SNARE vesicles). The v-SNARE protein VAMP2 was incorporated into a separate population of vesicles that contained 1.5 mol % each of NBD-PS (1,2-dioleoyl-sn-glycero-3-phosphoserine-N-(7-nitro-2–1,3-benzoxadiazol-4-yl) and rhodamine-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) for the fluorescence detection of lipid mixing (v-SNARE vesicles). The lipid-to-protein ratio was kept at 200:1 for all samples (except as noted). To follow the kinetics of lipid mixing, we mixed t-SNARE vesicles and v-SNARE vesicles at 37 °C in the 9:1 ratio. When SNAP-25 was not included, no lipid mixing was detected, as expected (black trace in Fig. 1A). However, significant lipid mixing was observed when SNAP-25 was included in the t-SNARE vesicles (Fig. 1A).
Fig. 1.
Lipid-mixing assays at several different cholesterol concentrations. (A) The changes of NBD fluorescence reflecting lipid mixing. The black trace is the control run with the t-liposomes reconstituted with only Syntaxin 1A (without SNAP-25). The gray trace shows lipid mixing for both v- and t- liposomes without cholesterol. The cyan trace is the lipid-mixing assay with 20 mol % cholesterol on both sides. The red trace represents the lipid-mixing assay with 40 mol % cholesterol on both liposomes. (B) The relative initial rates of lipid mixing. The initial rates were determined by fitting the rise of the fluorescence intensity with the linear line near t = 0. (C) The inner leaflet mixing assay. The gray trace is the negative control without SNAP25. The blue trace is the fluorescence change representing inner leaflet mixing for the liposomes without cholesterol. The pink trace represents inner leaflet mixing for the liposomes with 40 mol % cholesterol. (D) The relative initial rates for inner leaflet mixing. The error bars were obtained from at least n = 4 independent measurements from 4 different preparations.
When physiological 40 mol % of cholesterol was included in both vesicles, we observed a 5-fold increase of the initial rate of lipid mixing with respect to that in the absence of cholesterol (Fig. 1B). Interestingly, there was only minor stimulation of lipid mixing when 20 mol % of cholesterol was incorporated in both membranes. Therefore, the results show that the physiological concentration of cholesterol is necessary to produce significant stimulation of lipid mixing.
SNARE-mediated fusion transits through the hemifusion state in which the outer leaflets are merged but the inner leaflets remain intact (29–31). To further dissect the effect of cholesterol on hemifusion (outer leaflet mixing) and full fusion (inner leaflet mixing), we performed the inner leaflet lipid-mixing assay (Fig. 1C). We used the modified Meer's method (29) that uses dithionite to selectively reduce the fluorescence donor NBD on only the outer leaflet, which allows the selective detection of inner leaflet mixing. The results show that the rate of inner leaflet mixing was enhanced by a factor of 4 at 40 mol % cholesterol (Fig. 1D), indicating that cholesterol accelerates both hemifusion and full fusion similarly.
Cholesterol Is More Effective When It Is on the v-SNARE Vesicle.
A lipidomic study by Jahn and coworkers (16) revealed a surprising result that cholesterol is highly enriched at the synaptic vesicles, constituting ≈40 mol % of the total lipids, which is higher than the cholesterol content of the plasma membrane. Thus, we wonder whether the high cholesterol content in the vesicle has any functional implications on SNARE-mediated lipid mixing.
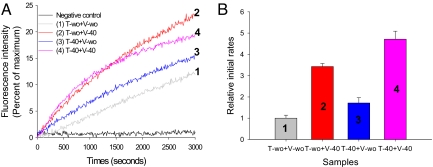
To address this question, we analyzed lipid mixing between the cholesterol-containing v-SNARE vesicles and the cholesterol-absent t-SNARE vesicles and vice versa (Fig. 2A). When 40 mol % of cholesterol was included in the v-SNARE vesicles with no cholesterol on the t-SNARE vesicles, the rate of lipid mixing was ≈70% of that for 40 mol % cholesterol on both vesicles (Fig. 2B). In contrast, when 40 mol % of cholesterol was loaded on the t-SNARE vesicle with no cholesterol on the v-SNARE vesicle, we observed stimulation of lipid mixing by a factor of only 1.7 with respect to the control without cholesterol on both sides. Thus, the result shows that cholesterol on the t-SNARE vesicle is less effective than it is on the v-SNARE side. Such asymmetric effects of cholesterol on SNARE-dependent lipid mixing may not be fully accounted for by the cholesterol's membrane curvature-modifying role. The results may suggest that its impact on the protein structure, particularly on the structure of v-SNAREs, also has important functional implications.
Fig. 2.
Lipid-mixing assays with asymmetric distribution of cholesterol. (A) The changes in the NBD fluorescence intensity reflecting lipid mixing. The black trace is the control run with the t-SNARE liposome reconstituted with only Syntaxin 1A (without SNAP-25). The gray trace is for the case without cholesterol. The red trace is for v-liposome with 40 mol % cholesterol but no cholesterol on t-liposome. The blue trace is vice versa. The pink trace is for 40 mol % cholesterol on both sides. (B) The relative initial rates of lipid mixing. The initial rate without cholesterol on both sides was set as 1. The error bars were obtained from at least n = 4 independent measurements from 4 different preparations.
SDSL EPR Reveals the v-SNARE TMD Forms an Open Scissors-Like Dimer.
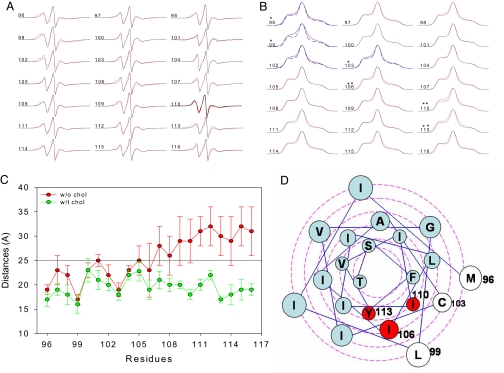
We used SDSL (32–34) and EPR spectroscopy to investigate the conformational changes of the v-SNARE TMD induced by cholesterol. In SDSL, native residues were replaced 1 at a time with cysteines to provide specific labeling sites for the thiol-specific nitroxide spin labels such as methanethiosulfonate spin label (MTSSL) (35, 36). The EPR line shape was sensitive to the motion of the nitroxide as well as to the spin–spin interaction with the neighboring nitroxides. For example, the motional restriction caused by the tertiary interaction with the neighboring helix would broaden the EPR lines. Yet, if the pairs of nitroxides are within 25 Å of one another, the dipolar interaction broadens the EPR lines, from which the interspin distance can be determined (37).
It has been controversial whether the TMDs of v-SNARE VAMP2 exist as monomers or dimers (38–41). To resolve this controversy, we undertook an extensive CW and pulsed EPR study using detergent micelles (as a reference) and liposomes with and without cholesterol. In the case of liposomes, labeled VAMP2 was reconstituted into the vesicles separately with and without 40 mol % cholesterol. EPR spectra were collected at room temperature, and the lipid-to-protein ratio was kept at 200:1 (except as noted). In the absence of cholesterol, the EPR line shapes were moderately broad for all TMD positions (Fig. 3A, black traces), representative of the lipid-exposed nitroxide side chains, and there is little indication of the strong tertiary interaction (29, 42–43). However, when we examined low-temperature absorbance of the EPR spectra, we observed line broadening due to the spin–spin interaction (Fig. 3B, black traces). At low temperatures, the motion of the nitroxide was frozen so that the line broadening was exclusively due to the spin–spin interaction. The interspin distances determined with the Fourier deconvolution method show that the TMDs interact through the N-terminal halves (Fig. 3C) with residues 99 and 103 as the interfacial positions (Fig. 3D) (37, 43). Meanwhile, the distances at the C-terminal halves were >25 Å, which was out of range for the CW EPR method. To address this, pulsed dipolar EPR was used with results also shown in Fig. 3C. Pulsed dipolar spectroscopy represented by DEER and DQC is a proven technique (44–54) to measure distances in the range of 10–80 Å between spin labels introduced in biomolecules and the distributions in distances, P(r). In the case of high flexibility, the distributions are usually broad (45, 54), which was exactly the case of VAMP2 for all residues studied, pointing to the high flexibility of the dimer [supporting information (SI) Fig. S1]. Another aspect of the DEER measurement was to probe the oligomerization order (52, 55). The modulation depth of DEER signals was consistent with just having dimers both with and without cholesterol. Reference studies in detergents and extensive studies in liposomes (Fig. S2) confirmed the dimeric state of the VAMP2 TMD.
Fig. 3.
SDSL and the EPR analysis of the spin-labeled VAMP2 TMD. (A) Room-temperature EPR spectra of spin-labeled VAMP2 mutants shown in the derivative mode. The red spectra are for the mutants reconstituted to liposomes containing 40 mol % cholesterol. The black spectra are for the proteins in the liposomes without cholesterol. (B) Low-temperature EPR spectra of VAMP2 mutants shown in the absorbance mode. For each mutant, the red spectrum is with 40 mol % cholesterol, and the black spectrum is without cholesterol. The blue spectra are the control from yeast Syntaxin-analog Sso1p N227C, which shows no spin–spin interaction (32). The single asterisk represents several dipolar broadened spectra even without cholesterol in the liposome. The double asterisk represents further broadening after the addition of 40 mol % cholesterol. (C) Plot of distances vs. the residue number for the VAMP2 TMD. In the presence of cholesterol, distances (filled circles) were measured by CW-ESR. In the absence of cholesterol, distances (<25 Å) for the residues 96–107 were measured by CW-EPR (open circles). (D) Helical wheel diagram for the VAMP2 TMD. Positions 106, 110, and 113, which showed the shorter distance with 40 mol % cholesterol, are shown in red. Positions 96, 99, and 103, which showed shorter distances even without cholesterol, are shown in white. The lipid mixtures of POPC, DOPS, and cholesterol in molar ratios of 85:15:0 and 45:15:40 were used.
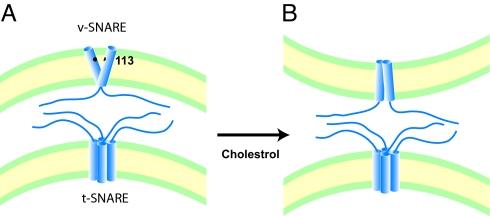
Therefore, the CW and pulsed-EPR results were consistent with the dimer model in which the N-terminal halves were closer, whereas the C-terminal halves were splayed apart, resembling an open scissors (Fig. 5A). However, we note that the helices appeared to be loosely held together and so dynamic that the tertiary interaction could not be picked up in the EPR line shape and the distance distributions were broad (SI Text).
Fig. 5.
Hypothetical models for the VAMP2 TMDs in the membrane in the absence (A) and in the presence (B) of cholesterol. In the absence of cholesterol, the dimer has an open-scissors-like conformation that may force the bilayer to have the negative curvature, which is not favorable for fusion. But in the presence of cholesterol, the dimer changes to the parallel conformation, which may not harbor the unfavorable negative curvature. The Syntaxin TMD was modeled as a tetramer based on our earlier work on yeast SNAREs (24).
Cholesterol Brings the C Termini of the TMDs Closer.
When cholesterol was incorporated in the vesicles, we observed small but clear and unambiguous line broadening at several selected positions including residues 99, 102, 106, 113, and 114 (Fig. 3 A and D, red traces). This was most likely due to the tertiary contacts between the TMDs. Had the line broadening been simply due to the cholesterol-induced decrease of the membrane viscosity, it would have occurred at all positions and not only to a select few positions. In the low-temperature spectra, the line broadening due to the spin–spin interaction became pronounced at several positions (Fig. 3B, red traces). What is particularly interesting was the conspicuous decrease of distances at the C-terminal positions in the presence of cholesterol (Fig. 3C). The distance profile throughout the TMD was quite consistent with the nearly parallel dimer, with positions 96, 99, 103, 106, 110, and 113 at the dimer interface (Fig. 3D). The TMD dimer in the presence of 40 mol % cholesterol appeared to resemble the closed scissors (Fig. 5B), as opposed to the open scissors-like dimer in the absence of cholesterol.
Is the Cholesterol-Induced Change in the TMD Relevant to the Fusion Activity?
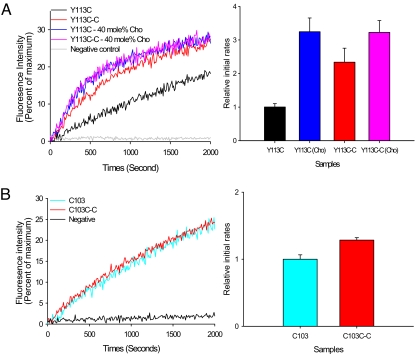
The EPR analysis of spin-labeled VAMP2 showed that cholesterol induces an open-to-closed conformational change in the TMD. To verify whether this conformational change is functionally relevant, we used cysteine–cysteine cross-linking. Cysteine–cysteine cross-linking at a C-terminal position such as Y113C, which is a predicted interfacial position, would bring the C-terminal parts of the dimer into contact, forcing the dimer to take the closed form even in the absence of cholesterol. If the open-to-closed conformational change is functionally important, we should observe the stimulation of lipid mixing with the disulfide-linked dimer in the absence of cholesterol. For the mutant Y113C, 40 mol % cholesterol stimulated the initial rate of lipid mixing by a factor of 3, somewhat less than that for wild-type VAMP2.
When VAMP2 Y113C was cross-linked by Cu(II) phenanthroline (Fig. S3) and subjected to the lipid-mixing assay with the vesicles without cholesterol, we observed the increase of the initial rate by a factor of 2.3 (Fig. 4A), generally supporting our prediction. But the stimulation by cross-linking was less than a factor of 3 and was somewhat short of fully accounting for the effect of cholesterol on lipid mixing. Interestingly, however, when cholesterol (40 mol %) was added in the lipid mixing assay with cross-linked Y113C-C, the further gain was only 25% (Fig. 4A), suggesting that the cholesterol-induced closure of the VAMP TMD dimer accounts for approximately three-fourths of the cholesterol effect on SNARE-dependent lipid mixing. The remaining one-fourth may be attributed to other factors such as the cholesterol's spontaneous curvature. Thus, the analysis suggests that the open-to-closed conformational change in the VAMP2 TMD by cholesterol is the major factor that contributes to the stimulation of lipid mixing.
Fig. 4.
Lipid-mixing assays for cross-linked VAMP2 wild-type C103 or mutant Y113C. (A) The NBD fluorescence change for cross-linked Y113C-C. (Left) The gray trace is a negative control without SNAP-25. The black trace is for non-cross-linked VAMP2 mutant Y113C without cholesterol. The red trace is for the cross-linked VAMP2 mutant Y113C-C without cholesterol. The blue and pink traces are for the non-cross-linked Y113C and cross-linked Y113C-C in the liposomes with 40 mol % cholesterol, respectively. (Right) The relative initial rates. (B) The NBD fluorescence change for cross-linked C103C-C. (Left) The cyan trace is for the lipid-mixing assay for non-cross-linked VAMP2 C103. The red trace is the cross-linked VAMP2 C103C-C. (Right) The relative initial rates.
Yet, when N-terminal position C103 was cross-linked, there was little change in the initial rate when compared with that for the non-cross-linked C103 (Fig. 4B). This was again expected because the cross-linking at the N-terminal positions would not impact the open conformation. Therefore, the results suggest that the open-to-closed conformational change induced by cholesterol has a functional implication on SNARE-dependent fusion.
Discussion
Recently, it has been suggested that regulatory lipids such as sterols, diacylglycerol, and phosphoinositides play critical roles in SNARE-dependent fusion as essential cofactors (56, 57). Cholesterol is also shown to be required for some viral fusion systems such as the fusion by α-virus (58). Although the critical importance of the different lipids in various biological fusion events is recognized, the effect of the regulatory lipids on the structures of the fusion proteins is not well understood.
As an initial attempt to uncover the impact of cholesterol on the structures of SNAREs, we investigated the conformational change of the VAMP2 TMD using SDSL EPR. The EPR analysis showed that the TMDs of v-SNARE VAMP2 formed an open scissors-shaped dimer with the pivot point at the N-terminal region in the absence of cholesterol. The overall shape of the dimeric TMD is likely to resemble an inverse wedge that would fit better to the negatively curved bilayer (Fig. 5A). For fusion to occur, however, the bilayers are required to bend in such a way to create positive curvature toward each other. Therefore, the inverse wedge-shaped TMD dimer would act as an inhibitor for such initial bending. Our results show that physiological cholesterol induces a conformational change from the open to the closed, driving the molecular shape to be more favorable for lipid mixing (Fig. 5B). Cholesterol is shown to have the capacity of thickening the bilayer, perhaps because of the rigid nature of its structure (59). We speculate that the closed conformation in the presence of cholesterol reflects the adjustment of the TMD dimer to the thicker bilayer to maintain the hydrophobic match between the bilayer and the TMD (60).
Cholesterol is generally believed to act as a fusion stimulator (61, 62). Its spontaneous negative curvature has the capacity to stabilize the hemifusion state in which the negatively curved surface is dominant. Moreover, it may drive the expansion of the pore, presumably by destabilizing the early fusion pore that is full of an acutely positively curved surface (17). Interestingly, incorporation of 40 mol % cholesterol on the t-SNARE vesicle has less of an effect on the rate of SNARE-mediated lipid mixing than it does for the v-SNARE vesicles. The results raise the possibility that cholesterol impacts membrane fusion not only through its preference for specific membrane curvature but also through its influence on the TMD conformation in the membrane. Our results suggest that the latter is significantly more important for SNARE-mediated membrane fusion and it is predominantly through its influence on the structure of the v-SNARE TMD.
Although it is thought that the TMD of VAMP2 plays a critical role in membrane fusion, controversy exists concerning its structure in the membrane. Computational and mutagenesis studies predicted that the v-SNARE TMD exists as a dimer with 2 helices oriented with a cross-angle of 38° (38). Furthermore, alanine-scanning mutagenesis indicated that residues 99, 102, 103, 107, 110, and 111 are involved in homodimerization of the TMD. In contrast, the infrared (IR) study by Bowen et al. (41) showed that the VAMP2 TMD exists as monomers in the DMPC bilayer with an unusual 35° tilt with respect to the bilayer normal. The EPR-based structural model in the absence of cholesterol might be similar to the structural model proposed by Bowen et al., except that there is a weak and dynamic helix–helix interaction at the N-terminal region. This weak interaction was not detectable even in the sensitive EPR-line shape analysis. Likewise we suspect that this interaction was not detectable in the IR study. Yet it appears that the EPR-based model in the presence of the cholesterol was quite consistent with the model predicted from the computer modeling and the alanine-scanning experiments. In our model, residues 99, 103, 110, and 113 reside at the dimer interface, which were similar to those positions responsive to alanine mutations.
Perhaps, the monomer–dimer controversy stems from the labile equilibrium between monomers and dimers that may be easily shifted by the changes in conditions such as the lipid composition. Provided the uncertainty of the new DEER method to estimate the stoichiometry of the TMD it is still possible that the monomer–dimer equilibrium exists. Therefore, an alternative scenario might be that cholesterol shifts equilibrium toward the dimer, and the dimerization might be the main event that is responsible for the stimulation of SNARE-dependent lipid mixing by cholesterol.
Although the EPR data are consistent with the dimer model, the periodic distance variation along the sequence (Fig. 3C) is much shallower than normally expected for the well-defined dimer. The results indicate that the dimer may not have a well-defined point of symmetry, and the interaction between 2 monomers may be promiscuous despite some preference given to the helical face consisting of residues 99, 103, 106, and 113 (Fig. 3D). The synaptobrevin TMD is unusual in that it contains as many as 11 β-branched amino acids (Fig. 3D), which may allow neither a well-defined interface between 2 interacting helices nor tight packing, reducing the amplitude of the distance variation significantly along the sequence.
In this work, we used the fluorescence lipid-mixing assays, not the content mixing assay, to investigate the SNARE-dependent membrane fusion. However, the caveats of the lipid-mixing assay are the potential complications arising from liposome instability and leaking (63), lipid flip–flop without fusion (64), and spontaneous lipid dye transfer between apposed membranes (65), warranting further investigations using the content mixing assay which is less vulnerable to these complications.
In summary, we have shown that cholesterol plays a role in modifying the v-SNARE structure in the membrane to be favorable for membrane fusion. Cholesterol induces the conformational change of the v-SNARE TMD dimer, which converts the molecular shape from an open scissors-like to a closed scissors-like shape, thereby modifying the membrane curvature to be favorable for fusion.
Materials and Methods
Plasmid Constructs and Site-Directed Mutagenesis.
Full-length VAMP2 (amino acids 1–116) was inserted into the pGEX-KG vector to make the glutathione S-transferase fusion protein. To introduce unique cysteine in the VAMP2 site-specifically, we changed the native cysteine C103 to alanine. QuikChange site-directed mutagenesis was used to make all cysteine mutants. Recombinant Syntaxin 1A without the Habc domain (SynHT, amino acids 168–288) and recombinant SNAP-25, in which 4 palmitylation sites (cysteines) on the interhelical loop were changed to alanines (amino acids 1–206), were also made as GST fusion proteins. The DNA sequences were confirmed by the Iowa State University sequencing facility.
Protein Expression, Purification, and Spin Labeling.
The experiments were carried out as described (30, 35). The labeling efficiency was calculated by dividing the spin concentration with the protein concentration. For all samples, the labeling efficiency was >85%.
Vesicle Preparation and Membrane Reconstitution.
Large unilamellar vesicles of a 100-nm diameter were prepared as described in refs. 30 and 35. The lipid-to-protein molar ratio was 200:1 (except as noted). The reconstitution efficiency was estimated by determining the protein concentration before and after reconstitution by using EPR. For all samples, the efficiency was ≈85%. For the lipid-mixing assay, the mixture of POPC, DOPS, and cholesterol in a molar ratio of 85:15:0, 65:15:20 or 45:15:40 was used. Additionally, NBD-PS and rhodamine-PE (1.5 mol % each) were added to the v-SNARE vesicles for fluorescence detection of lipid mixing. The t-SNARE proteins SynHT and SNAP-25, in a molar ratio of 1:1, were premixed, and the mixture was left at room temperature for ≈60 min to form the complex before the reconstitution. In all cases the proteins were reconstituted by using the dialysis method described in ref. 66.
Lipid-Mixing Assay.
The v-SNARE liposomes were mixed with the t-SNARE liposomes at a molar ratio of 1:9. The total volume of the reaction solution was 100 μL containing 1 mM lipids. Fluorescence was measured at excitation and emission wavelengths of 465 and 530 nm, respectively. The intensity changes were recorded by the Varian Cary Eclipse model fluorescence spectrophotometer using a quartz cell of 100 μL with 2-mm path length. All of the lipid-mixing experiments were carried out at 37 °C. Approximately 50 min after the reaction, we added 1 μL of 10%-reduced Triton to complete the lipid mixing assay and to obtain the maximum fluorescence intensity (MFI). The inner leaflet-mixing assay was performed following the procedure described in ref. 30.
CW-EPR Data Collection and Analysis.
CW-EPR spectra were collected by using the Bruker ESP 300 spectrometer equipped with a loop-gap resonator (Medical Advances) and a low-noise microwave amplifier (Miteq). Room-temperature (293 K) and low-temperature (130 K) spectra were collected for all reconstituted proteins. To measure interspin distances from CW-EPR line shapes, Fourier deconvolution analysis was used (37).
Distance Measurements with the 4-Pulse Ku-band DEER.
Four-pulse DEER (44–46) experiments were conducted by using a home-built Ku-band (17.35 GHz) pulse FT-ESR/DQC spectrometer as described (52–54). All measurements were conducted at 65 K by using a 15-μL sample volume. All 3 detection mode pulses had 20-ns lengths. The pump π-pulse was typically 12–20 ns. The dipolar signals measured by DEER were analyzed by the L-curve Tikhonov regularization method (67) and refined by MEM (68) to recover distance distributions.
Disulfide Cross-Linking and Lipid-Mixing Assay.
Wild-type VAMP2 has a single native cysteine at position 103. C103 or mutant Y113C was cross-linked with CuSO4 (36 μM) and phenanthroline (72 μM) at room temperature for 40 min. Cross-linked dimers C103C-C and Y113C-C and non-cross-linked C103 and Y113C were separately reconstituted into the fluorescent vesicles, whereas the t-SNARE binary complex was inserted into the nonfluorescent vesicle (POPC/DOPS = 85:15 in moles). The lipid-mixing assay was performed by following the procedure described above except that the 1:1 ratio of v-liposome and t-liposome was used this time. The total volume of the reaction solution was 100 μL containing 0.4 mM lipids.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants R01-51290 (Y.-K.S.), and P41-RR016292 and R01-ER03150 (to J.H.F.). Y.K.-S. was supported by World Class University, Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0813138106/DCSupplemental.
References
- 1.Jahn R, Lang T, Südhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 2.Lin RC, Scheller RH. Mechanism of synaptic vesicle exocytosis. Annu Rev Cell Dev Biol. 2000;16:19–49. doi: 10.1146/annurev.cellbio.16.1.19. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R, Südhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 4.Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- 5.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernomordik LV, Kozlou MM. Mechanism of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 8.Jahn R, Scheller RH. SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;9:31–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 9.Su Z, Ishitsuka Y, Ha T, Shin YK. The SNARE complex from yeast is partially unstructured on the membrane. Structure (London) 2008;16:1138–1146. doi: 10.1016/j.str.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirier MA, et al. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 11.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 12.Lin RC, Scheller RH. Structural organization of the synaptic exocytosis core complex. Neuron. 1997;19:1087–1094. doi: 10.1016/s0896-6273(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 13.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 14.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker SJ, Kinsey WH. Characterization of cortical secretory vesicles from the sea urchin egg. Dev Biol. 1983;96:37–45. doi: 10.1016/0012-1606(83)90309-3. [DOI] [PubMed] [Google Scholar]
- 16.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Biswas S, Yin SR, Blank P, Zimmerberg J. Cholesterol promotes hemifusion and pore widening in membrane fusion induced by influenza hemagglutinin. J Gen Physiol. 2008;131:505–513. doi: 10.1085/jgp.200709932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchward MA, Rogasevskaia T, Höfgen J, Bau J, Coorssen JR. Cholesterol facilitates the native mechanism of Ca2+ triggered membrane fusion. J Cell Sci. 2005;118:4833–4848. doi: 10.1242/jcs.02601. [DOI] [PubMed] [Google Scholar]
- 19.Lang T, et al. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J Cell Biol. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Rand RP. The influence of cholesterol on phospholipids membrane curvature and bending elasticity. Biophys J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua Y, Scheller RH. Three SNARE complexes cooperate to mediate membrane fusion. Proc Natl Acad Sci USA. 2001;98:8065–8070. doi: 10.1073/pnas.131214798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieber JJ, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 24.Lu X, Zhang Y, Shin YK. Supramolecular SNARE assembly precedes hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:700–706. doi: 10.1038/nsmb.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grote E, Baba M, Ohsumi Y, Novick PJ. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J Cell Biol. 2000;151:453–466. doi: 10.1083/jcb.151.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X, Wang CT, Bai J, Chapman ER, Jackson MB. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 28.Shin YK, Freed JH. Dynamic imaging of lateral diffusion by electron spin resonance and study of rotational dynamics in model membranes: Effects of cholesterol. Biophys J. 1989;55:537–550. doi: 10.1016/S0006-3495(89)82847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Zhang F, McNew JA, Shin YK. Hemifusion in S NARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 30.Lu X, Zhang F, McNew JA, Shin YK. Membrane fusion induced by neuronal SNAREs transits through hemifusion. J Biol Chem. 2005;280:30538–30541. doi: 10.1074/jbc.M506862200. [DOI] [PubMed] [Google Scholar]
- 31.Giraudo CG, et al. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J Cell Biol. 2005;170:249–260. doi: 10.1083/jcb.200501093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altenbach C, Marti T, Khorana HG, Hubbell WL. Transmembrane protein structure: Spin labeling of bacteriorhodopsin mutants. Science. 1990;248:1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- 33.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 34.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Mol Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 35.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 36.Kweon DH, Kim CS, Shin YK. Insertion of the membrane-proximal region of the neuronal SNARE coiled coil into the membrane. J Biol Chem. 2003;278:12367–12373. doi: 10.1074/jbc.M211123200. [DOI] [PubMed] [Google Scholar]
- 37.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleming KG, Engelman DM. Computation and mutagensis suggest a right-handed structure for the synaptobrevin transmembrane dimer. Proteins. 2001;45:313–317. doi: 10.1002/prot.1151. [DOI] [PubMed] [Google Scholar]
- 39.Bowen ME, Engelman DM, Brunger AT. Mutational analysis of synaptobrevin transmembrane domain oligomerization. Biochemistry. 2002;41:15861–15866. doi: 10.1021/bi0269411. [DOI] [PubMed] [Google Scholar]
- 40.Roy R, Laage R, Langosch D. Synaptobrevin transmembrane domain dimerization-revisited. Biochemistry. 2004;43:4964–4970. doi: 10.1021/bi0362875. [DOI] [PubMed] [Google Scholar]
- 41.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Shin YK. Transmembrane organization of yeast syntaxin-analogue Sso1p. Biochemistry. 2006;45:4173–4181. doi: 10.1021/bi052178+. [DOI] [PubMed] [Google Scholar]
- 43.Ottemann KM, Thorgeirsson TE, Kolodziej AF, Shin YK, Koshland DE., Jr Direct measurement of small ligand-induced conformational changes in the aspartate chemoreceptor using EPR. Biochemistry. 1998;37:7062–7069. doi: 10.1021/bi980305e. [DOI] [PubMed] [Google Scholar]
- 44.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chem Phys Chem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Borbat PP, Freed JH. Measuring distances by pulsed dipolar ESR spectroscopy: Spin-labeled histidine kinases. Methods Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 46.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q Rev Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 47.Milov AD, Maryasov AG, Tsvetkov YD. Pulsed electron double resonance (PELDOR) and its applications in free-radicals research. Appl Magn Reson. 1998;15:107–143. [Google Scholar]
- 48.Park SY, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 49.Fafarman AT, Borbat PP, Freed JH, Kirshenbaum K. Characterizing the structure and dynamics of folded oligomers: Pulsed ESR studies of peptoid helices. Chem Commun. 2007;4:377–379. doi: 10.1039/b612198e. [DOI] [PubMed] [Google Scholar]
- 50.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-Bound alpha-synuclein forms an extended helix:long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borbat PP, et al. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 2007;5:2211–2219. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Upadhyay AK, Borbat PP, Wang J, Freed JH, Edmondson DE. Determination of the oligomeric states of human and rat monoamine oxidases in the outer mitochondrial membrane and Octyl-β-d-glucopyranoside micelles using pulsed dipolar electron spin resonance spectroscopy. Biochemistry. 2008;44:1554–1566. doi: 10.1021/bi7021377. [DOI] [PubMed] [Google Scholar]
- 53.Berliner LJ, Eaton SS, Eaton GR. Biological Magnetic Resonance. Vol 19. New York: Kluwer Academic/Plenum Publishers; 2000. Distance measurements in biological systems by EPR. [Google Scholar]
- 54.Borbat PP, McHaourab HS, Freed JH. Protein structure determination using long-distance constraints from double-quantum coherence ESR: Study of T4 lysozyme. J Am Chem Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 55.Bode BE, et al. Counting the monomers in nanometer-sized oligomers by pulsed electron-electron double resonance. J Am Chem Soc. 2007;129:6736–6745. doi: 10.1021/ja065787t. [DOI] [PubMed] [Google Scholar]
- 56.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27:2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James DJ, Khodthong C, Kowalchyk JA, Martin TFJ. Phosphatidylinositol 4,5-biphosphate regulates SNARE-dependent membrane fusion. J Cell Biol. 2008;182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umashankar M, et al. Differential cholesterol binding by class II fusion proteins determines membrane fusion properties. J Virol. 2008;82:9245–9253. doi: 10.1128/JVI.00975-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kucerka N, et al. The effect of cholesterol on short- and long-chain monounsaturated lipid bilayers as determined by molecular dynamic simulation and X-ray scattering. Biophys J. 2008;95:2792–2805. doi: 10.1529/biophysj.107.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koehorst RBM, Spruijt RB, Vergeldt FJ, Hemminga MA. Lipid bilayer topology of the transmembrane α-helix of M13 major coat protein and bilayer polarity profile by site-directed fluorescence spectroscopy. Biophys J. 2004;87:1445–1455. doi: 10.1529/biophysj.104.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia F, et al. Inhibition of cholesterol biosynthesis impairs insulin secretion and voltage-gated calcium channel function in pancreatic Beta-cells. Endocrinology. 2008;149:5136–5145. doi: 10.1210/en.2008-0161. [DOI] [PubMed] [Google Scholar]
- 62.Salaün C, James DJ, Chamberlain LH. Lipid rafts and the regulation of exocytosis. Traffic. 2004;5:255–264. doi: 10.1111/j.1600-0854.2004.0162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dennison SM, Bowen ME, Brunger AT, Lentz BR. Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG mediated membrane fusion. Biophys J. 2006;90:1661–1675. doi: 10.1529/biophysj.105.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bai J, Pagano RE. Measurement of spontaneous transfer and transbilayer movement of BODIPY-labeled lipids in lipid vesicles. Biochemistry. 1997;36:8840–8848. doi: 10.1021/bi970145r. [DOI] [PubMed] [Google Scholar]
- 65.Ohki S, Flanagan TD, Hoekstra D. Probe transfer with and without membrane fusion in a fluorescence fusion assay. Biochemistry. 1998;37:7496–7503. doi: 10.1021/bi972016g. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Xu Y, Zhang F, Shin YK. Constitutive versus regulated SNARE assembly: A structural basis. EMBO J. 2004;23:681–689. doi: 10.1038/sj.emboj.7600083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiang Y-W, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172:279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 68.Chiang Y-W, Borbat PP, Freed JH. Maximum entropy: A complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR. J Magn Rson. 2005;177:184–196. doi: 10.1016/j.jmr.2005.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.