Abstract
Atomic level characterization of proteins and other macromolecules in the living cell is challenging. Recent advances in NMR instrumentation and methods, however, have enabled in-cell studies with prospects for multidimensional spectral characterization of individual macromolecular components. We present NMR data on the in-cell behavior of the MetJ repressor from Escherichia coli, a protein that regulates the expression of genes involved in methionine biosynthesis. NMR studies of whole cells along with corresponding studies in cell lysates and in vitro preparations of the pure protein give clear evidence for extensive nonspecific interactions with genomic DNA. These interactions can provide an efficient mechanism for searching out target sequences by reducing the dependence on 3-dimensional diffusion through the crowded cellular environment. DNA provides the track for MetJ to negotiate the obstacles inherent in cells and facilitates locating and binding specific repression sites, allowing for timely control of methionine biosynthesis.
Keywords: DNA–protein interactions, gene regulation, met repressor, methionine regulon, nonspecific DNA
The environment in which proteins and nucleic acids function within a cell is well known to be crowded and complex (1). Understanding the influence of the numerous large and small molecule components on the biophysical and functional properties of these macromolecules in cellular environments, however, is extremely difficult (2, 3). For example, the detailed mechanism of gene regulation is of great interest, but the study of these interactions within living cells has always been a challenge.
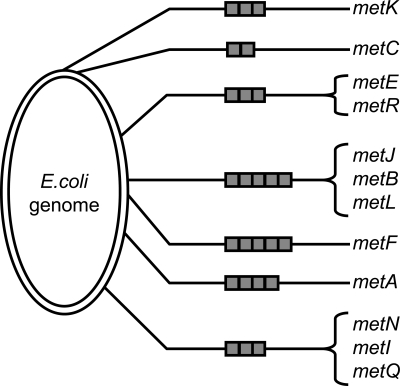
The transcriptional repressor MetJ controls the expression in Escherichia coli of the Met regulon, which is composed of at least 12 genes scattered at 7 sites around the genome (Fig. 1). These genes code for proteins that are involved in methionine biosynthesis and transport (4, 5). The promoters of these genes contain from 2 to 5 tandem repeats of the MetJ binding site, which are called metboxes and have the consensus sequence AGACGTCT (6). When MetJ is activated by S-adenosylmethionine (SAM) it binds tightly to the metboxes and shuts down transcription (7, 8). The different genes repressed by MetJ not only have a variable number of metboxes but also variable sequences. MetJ is thus capable of specific binding to several variations of the consensus sequence in vivo and fine-tuning its repression of each gene (4, 9).
Fig. 1.
Schematic of the E. coli chromosome (4,639,675 bp) showing the locations of the genes regulated by MetJ (not to scale). The gray boxes indicate the number of 8-bp metboxes at each locus, ranging from 2 to 5.
NMR spectroscopy has recently been shown to provide a valuable strategy for studying proteins in living cells (10–13). This in-cell spectroscopy gives atomic level fingerprints and even 3-dimensional spectral data that are used to assign resonances and characterize features of individual components in the cellular milieu (12). The method has been used to show ligand binding (14), protein–protein interactions (15) and in at least 1 case, a gain of structure presumably as a result of molecular crowding inside the cell (13). Here, we report NMR results on both in-cell and cell lysate studies, which show that MetJ associates nonspecifically with genomic DNA in cells, thereby providing an efficient search mechanism for identifying and binding metbox repressor sequences.
Results
Whole Cells and Lysates.
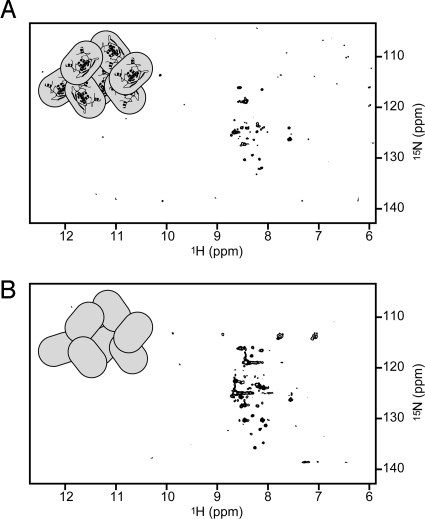
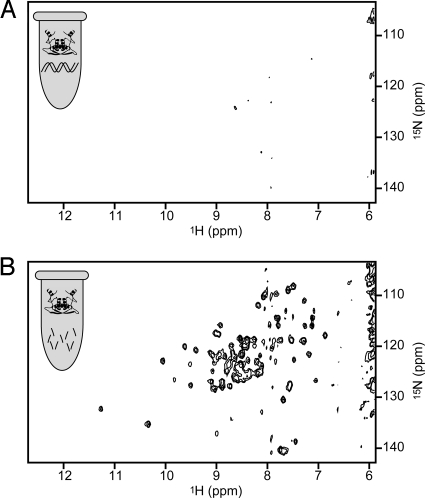
E. coli cells overexpressing MetJ in the presence of 15N-labeled ammonium chloride as the sole nitrogen source were harvested and concentrated for NMR studies as described in the methods section. Proton-nitrogen correlation spectra (15N-HSQC) of intact, living cells showed a rather minimal pattern (Fig. 2A) similar to that for controls in which cells lacking the MetJ plasmid were grown in 15N-labeled media (Fig. 2B). The NMR spectrum observed in preparations of freshly lysed cells containing overexpressed 15N-labeled MetJ is similar to that for whole cells. These spectra are clearly different from the well-dispersed HSQC spectrum of purified MetJ (shown in Fig. 3A). In fact, spectra very similar to those in Fig. 2 have been seen by other researchers and shown to indicate detection of only the labeled background molecules (16).
Fig. 2.
HSQC spectra of intact, living cells does not show the overexpressed MetJ spectrum. Cells overexpressing MetJ (A) show the same pattern as cells without the MetJ plasmid (B) indicating that the spectrum from MetJ is not being detected. The amide region for proton resonances is on the x axis, and amide nitrogen resonances are on the y axis. Icons represent cells with or without the Cα-backbone structure of a MetJ dimer (PDB entry 1CMB).
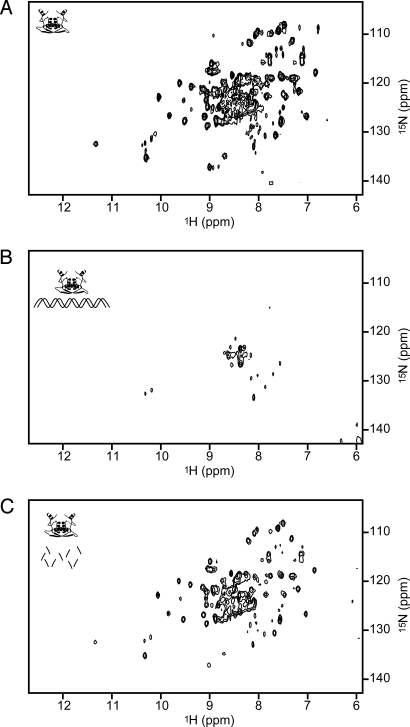
Fig. 3.
The MetJ spectrum can be restored in preparations exhibiting nonspecific DNA binding by DNase treatment. (A and B) The spectrum of purified MetJ dimer (A) is lost after the addition of nonspecific DNA (B). (C) The spectrum is restored with the addition of DNase. Icons represent a MetJ dimer in solution: alone, interacting with nonspecific double-stranded DNA, or with that DNA digested into fragments.
The absence of identifiable protein NMR spectral features within cells or cell lysates can result either from insufficient expression or from association with heavy cell components causing restricted motion and/or intermediate exchange. SDS/PAGE gels of our NMR samples, however, show that MetJ is overexpressed as expected at levels sufficient for NMR detection (≈300 μM after concentration of the cells). Furthermore, we were able to partially purify MetJ from the lysates, using ammonium sulfate fractionation and cellulose phosphate chromatography, a method highly selective for MetJ. An NMR sample produced this way from a 50-ml culture showed the standard MetJ spectra (data are identical to those shown in Fig. 3A). This and gel analysis of the induction demonstrate that there is sufficient MetJ in the lysate to produce strong NMR signals.
MetJ is a DNA binding protein, which suggests that it may be associating nonspecifically with genomic (and plasmid) DNA. This interaction must be nonspecific with overexpressed MetJ, because the number of MetJ molecules in the cell is in several thousandfold excess over the number of natural metbox binding sites. Our in-cell NMR data therefore suggest substantial nonspecific MetJ binding to genomic DNA.
Purified MetJ with Nonspecific and Specific DNA.
To investigate MetJ binding with nonspecific DNA, NMR spectra were acquired on mixtures of purified 15N-labeled MetJ and sonicated salmon sperm DNA to mimic large genomic DNA. This nonspecific DNA is composed of 300- to 2,000-bp fragments. Starting with a solution containing 7.2 mg of MetJ (spectrum in Fig. 3A), nonspecific DNA was titrated in until the characteristic pattern for MetJ disappeared with 120 μg of DNA (Fig. 3B). As the DNA was added, the peaks from the free MetJ progressively weakened but did not shift position, indicating that MetJ interacts with the DNA, and that this nonspecific interaction attenuates detectable MetJ resonances. The loss of the entire MetJ spectrum suggests that MetJ forms a relatively compact structure upon nonspecific DNA binding. This contrasts with its structural homolog, ParG, where a flexible N-terminal domain remains unaffected in the NMR spectrum upon DNA binding (17). DNase and MgSO4 were then added to a final concentration of 25 μg/mL and 5 mM, respectively, to digest the salmon sperm DNA, which restored the MetJ spectrum (Fig. 3C).
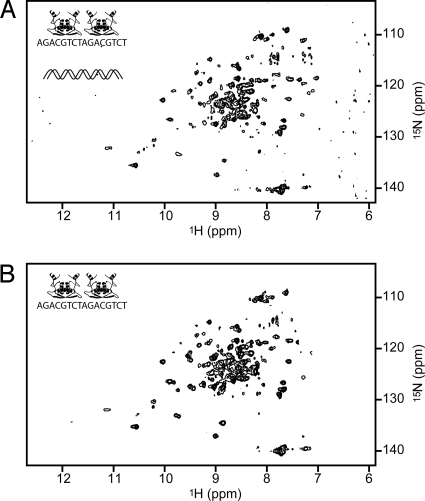
MetJ can also be rescued from nonspecific DNA binding by the addition of “metbox” DNA, a small oligo containing 2 repeats of MetJ's preferred binding sequence. To a starting solution of 15N-labeled MetJ (as in Fig. 3A), salmon sperm DNA was added until the signal disappeared (as in Fig. 3B). Metbox DNA was then added at twice the stoichiometric concentration of MetJ, which restored the MetJ spectrum (Fig. 4A). The pattern that is observed matches that of a purified in vitro sample of MetJ with the same oligo (Fig. 4B), showing that MetJ is forming a complex with metbox DNA that can successfully compete with and displace the nonspecific DNA binding.
Fig. 4.
MetJ can be rescued from nonspecific DNA binding (seen in Fig. 3B) by metbox DNA. Addition of a 20-bp DNA fragment containing 2 adjacent 8-bp metbox sequences restores the HSQC fingerprint of MetJ bound to its recognition site (A) as shown by comparison with the purified complex (B). Icons represent 2 MetJ dimers interacting specifically with metbox DNA, in the presence or absence of nonspecific DNA.
Pure MetJ in Unlabeled Lysates.
Our experiments using salmon sperm DNA showed that MetJ can interact nonspecifically with DNA and that either DNase treatment or competition with specific DNA can restore the MetJ spectrum. To further demonstrate the nonspecific association with genomic DNA, we used purified 15N-labeled MetJ with unlabeled cell lysates. This method has the advantage that only MetJ is 15N-labeled, so there is no background labeling of other cell components, and we can quantitatively control the concentration of MetJ.
To a starting solution of purified 15N-labeled MetJ (as in Fig. 3A), an equal volume of an unlabeled cell lysate was added resulting in nonspecific DNA binding and disappearance of spectral features (Fig. 5A), thus recapitulating the results observed in our first experiments. For this experiment, the lysate from 4 mL of culture (≈1010 E. coli cells) completely eliminated the signal from 7 mg of MetJ. Tenfold excess DNase was then added to a final concentration of 250 μg/mL, which restored the MetJ signal (Fig. 5B).
Fig. 5.
The MetJ spectrum can be restored in preparations exhibiting interactions within cell lysates by DNase treatment. (A) The spectrum of purified MetJ dimer (seen in Fig. 3A) is lost after the addition of an unlabeled E. coli cell lysate. (B) The spectrum is restored by the addition of DNase. Icons represent the MetJ dimer in cell lysates, with nonspecific DNA either intact or digested.
This experiment revealed that nonspecific MetJ-DNA interactions in cell lysates could be attenuated by excess DNase, suggesting similar results might be observed in cells containing endogenous overexpressed MetJ. We investigated this possibility by adding excess DNase to freshly lysed cells overexpressing 15N-labeled MetJ, but were still not able to detect the MetJ signals by NMR. We note, however, that in this experiment the ratio of DNA in the 50-ml culture to overexpressed MetJ (estimated to be 5 mg from gel analysis) is >10 times larger than in the above controlled samples in which lysate from only 4 mL of cells was added to 7 mg of purified, labeled MetJ. This larger excess of DNA would tend to favor even weak binding to the residual DNA fragments.
Discussion
The combination of in-cell NMR and experiments with cell lysates indicates that the MetJ repressor protein in E. coli has substantial nonspecific associations with genomic DNA in the cellular environment. Other evidence for nonspecific MetJ/DNA association comes from surface plasmon resonance kinetic studies with varying sizes of DNA fragments, which show that MetJ can find a target site faster when it is embedded within longer pieces of DNA (18). This work, along with our present study, suggests a mechanism for the enhancement of MetJ's ability to find its specific DNA target genes. There is abundant genomic and plasmid DNA within cells where nonspecific MetJ interactions can occur. The protein concentration in vivo is estimated to be 600 MetJ dimers per cell (7) but even with typical overexpression levels of 600,000 MetJ dimers per cell, our results indicate the 4.6 million bp of genomic DNA per cell are sufficient to ensure that all of the MetJ is associating nonspecifically with DNA. It is noted that overexpression constitutively represses the Met regulon, requiring the addition of exogenous methionine to allow cell growth (7, 19). This overexpression does not lead to any other growth defects, however, indicating that MetJ does not nonspecifically repress other transcription units. This is in contrast to the overexpression behavior of TrpR, the regulator of tryptophan biosynthesis, which will promiscuously repress genes involved in the biosynthesis of Ile, Leu, Val, Thr, Ser, Phe, and Tyr (20).
Although it is always important to consider the consequences of overexpression on any in vivo biological model, in this case the nonselective association of a large excess of MetJ with genomic DNA clearly indicates that at the much lower native concentration, the protein will also be associating with the DNA available in the cell. To further reinforce this conclusion, analytical ultracentrifugation studies in our lab also show nonspecific binding to DNA in cases where the DNA is far in excess of the MetJ concentration.
These results also provide insight into the detailed mechanism used by MetJ to locate metbox sequences, and likely other DNA binding proteins to find their recognition sequences, when functioning within the cell. In general, proteins that work by binding specific DNA sequences, such as transcription factors and restriction enzymes, must sort through millions of bases of DNA to find a small number of target sites. For some DNA binding proteins, target binding is faster than the 3D diffusion rate [e.g., lac repressor (21), EcoR1 endonuclease (22), and integration host factor (23)], suggesting that these proteins use some method of “facilitated diffusion” to efficiently search for a target sequence. Maintaining a close interaction with DNA, rather than depending on free diffusion in solution, provides such a facilitating strategy (24–26).
Various studies have been performed to test this model. Studies with RNA polymerase show sliding along DNA, with an increased lifetime for the nonspecific complex when bound to a mica surface versus free in solution (27). The authors speculate that this surface might be a good model for the viscous interior of the cell. In the case of the restriction enzyme EcoRV, however, linear diffusion occurred only over short distances (<30 bp) before dissociation, and 3D transfer between DNA strands was more important (28). There have also been several reports of single-molecule experiments on DNA binding proteins, using recently developed techniques for imaging, fluorescent-labeling, and sample immobilization (29). One such study used fluorescent lac repressor in living cells and showed good evidence for nonspecific interactions with DNA, linear diffusion along DNA, and transfer from one DNA segment to another (30). For the DNA-repair enzyme hOgg1, single-molecule studies have shown good evidence for extensive linear sliding along DNA (31). In vitro NMR techniques, such as paramagnetic relaxation and exchange analysis, have also been used to observe nonspecific binding and transfer from one piece of DNA to another (32–34). Our present results show that the MetJ repressor binds nonspecifically to DNA in the living cell, and thus can use this mechanism to speed specific binding and efficiently regulate methionine biosynthesis.
The in-cell and other NMR results presented here complement previous work to provide compelling evidence that transcription factors can use efficient diffusion in reduced dimensionality to facilitate the kinetics associated with their function. In the case of MetJ, it is capable of repressing or derepressing target genes within 30 min of a change of methionine concentration (35, 36). Our NMR spectral data clearly indicate that MetJ's nonspecific interactions with genomic DNA are extensive, providing a mechanism for this efficiency.
Materials and Methods
Sample Preparation.
Whole-cell samples were prepared using a protocol developed in our laboratory (12). Briefly, a 50-ml culture of BL21(DE3) cells (Novagen) containing a plasmid with the MetJ gene was grown in minimal media supplemented with [15N]NH4Cl (Cambridge Isotopes). After 4 h of induction with IPTG the cells were harvested by centrifugation at 1000 × g then resuspended in a minimal volume of the spent media (≈20% cells by volume). D2O was added to 10%. NMR spectra were acquired within 30 min of sample preparation.
To have cells available as early as possible in the day for NMR studies, a modified induction protocol was also used whereby a 50-mL culture was grown overnight in media containing only 0.05% glucose. This limiting concentration of glucose arrested cell growth at an OD600 between 0.5 and 0.6. In the morning, glucose was added to a final concentration of 0.3% and 1 mM IPTG was added to induce protein expression. Cells could then be harvested in the early afternoon after 3–5 h. Induction yields were comparable to those using standard methods.
For the lysate samples, the induced cells were pelleted and resuspended in 4 mL/g BugBuster (Novagen) supplemented with 50 μg/mL lysozyme, 25 μg/mL DNase, 5 mM MgSO4, 1 mM DTT, 1 mM protease-inhibitor PMSF and 10% D2O. Lysis was allowed to proceed for 15–30 min, then the cells were spun down and the soluble portion used for the NMR experiments. Unlabeled cell lysates were made from noninduced, overnight cultures grown in rich media (LB broth) and lysed the same way.
Quantitative plating of cells (using a method similar to that described in ref. 37) showed colony production and cell viability did not change in our recombinant cells under NMR sample conditions during a time period well in excess of that needed to collect NMR spectra. We also examined the supernatants of whole cell samples by both NMR and SDS/PAGE for evidence of protein leakage within the time scale of our experiments but found none. For NMR, the supernatant was analyzed with and without DNase treatment, but in both cases the spectra were comparable to the background signals shown in Fig. 2, with no evidence of characteristic MetJ resonances.
Purified MetJ was obtained by published procedures (38). Metbox DNA was purchased from IDT as a 20-bp oligo containing 2 copies of MetJ's preferred binding site flanked by GG and CC bases: 5′ GG-AGACGTCT-AGACGTCT-CC 3′. Sonicated salmon sperm DNA was from Stratagene; DNase and lysozyme were from Sigma.
NMR Conditions.
All spectra were acquired at 600 MHz on a Varian Inova spectrometer equipped with an H,C,N triple-resonance cryogenically cooled probe using gradient-enhanced TROSY HSQC experiments. Spectra were acquired for 10–45 min at 25 °C for samples containing cells and 40 °C for purified MetJ samples. NMR spectra were processed with NMRPipe (39) and analyzed with Sparky (40). All spectra in a given series (e.g., Fig. 2 A and B) are plotted at the same nominal contour level.
Acknowledgments.
We thank Drs. R. C. Greene, P. Zhou, R. A. Venters for their helpful discussions during the course of this work. This work was partially supported by the National Institutes of Health Grant RR-022854. The Duke University NMR Center spectrometers were partially supported by the National Institutes of Health, National Science Foundation, the Howard Hughes Medical Institute, and North Carolina Biotechnology Center.
Footnotes
The authors declare no conflict of interest.
References
- 1.Goodsell DS. Inside a living cell. Trends Biochem Sci. 1991;16:203–206. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. Chemical space and biology. Nature. 2004;432:824–828. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 3.Ellis RJ, Minton AP. Cell biology: Join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 4.Old IG, Phillips SE, Stockley PG, Saint Girons I. Regulation of methionine biosynthesis in the Enterobacteriaceae. Prog Biophys Mol Biol. 1991;56:145–185. doi: 10.1016/0079-6107(91)90012-h. [DOI] [PubMed] [Google Scholar]
- 5.Gal J, Szvetnik A, Schnell R, Kalman M. The metD d-methionine transporter locus of Escherichia coli is an ABC transporter gene cluster. J Bacteriol. 2002;184:4930–4932. doi: 10.1128/JB.184.17.4930-4932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belfaiza J, et al. Evolution in biosynthetic pathways: Two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saint-Girons I, et al. Interactions of the Escherichia coli methionine repressor with the metF operator and with its corepressor, S-adenosylmethionine. J Biol Chem. 1986;261:10936–10940. [PubMed] [Google Scholar]
- 8.Shoeman R, et al. Regulation of methionine synthesis in Escherichia coli: Effect of metJ gene product and S-adenosylmethionine on the expression of the metF gene. Proc Natl Acad Sci USA. 1985;82:3601–3605. doi: 10.1073/pnas.82.11.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marincs F, Manfield IW, Stead JA, McDowall KJ, Stockley PG. Transcript analysis reveals an extended regulon and the importance of protein–protein co-operativity for the Escherichia coli methionine repressor. Biochem J. 2006;396:227–234. doi: 10.1042/BJ20060021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlton LM, Pielak GJ. Peeking into living eukaryotic cells with high-resolution NMR. Proc Natl Acad Sci USA. 2006;103:11817–11818. doi: 10.1073/pnas.0605297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serber Z, Corsini L, Durst F, Dotsch V. In-cell NMR spectroscopy. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- 12.Reardon PN, Spicer LD. Multidimensional NMR spectroscopy for protein characterization and assignment inside cells. J Am Chem Soc. 2005;127:10848–10849. doi: 10.1021/ja053145k. [DOI] [PubMed] [Google Scholar]
- 13.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard JA, MacLachlan LK, King GW, Jones JJ, Fosberry AP. Nuclear magnetic resonance spectroscopy reveals the functional state of the signalling protein CheY in vivo in Escherichia coli. Mol Microbiol. 2003;49:1191–1200. doi: 10.1046/j.1365-2958.2003.03628.x. [DOI] [PubMed] [Google Scholar]
- 15.Burz DS, Dutta K, Cowburn D, Shekhtman A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR) Nat Methods. 2006;3:91–93. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serber Z, Ledwidge R, Miller SM, Dotsch V. Evaluation of parameters critical to observing proteins inside living Escherichia coli by in-cell NMR spectroscopy. J Am Chem Soc. 2001;123:8895–8901. doi: 10.1021/ja0112846. [DOI] [PubMed] [Google Scholar]
- 17.Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon–helix–helix structure. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 18.Lawrenson ID, Stockley PG. Kinetic analysis of operator binding by the E. coli methionine repressor highlights the role(s) of electrostatic interactions. FEBS Lett. 2004;564:136–142. doi: 10.1016/S0014-5793(04)00336-9. [DOI] [PubMed] [Google Scholar]
- 19.Smith AA, Greene RC. Cloning of the methionine regulatory gene, metJ, of Escherichia coli K12 and identification of its product. J Biol Chem. 1984;259:14279–14281. [PubMed] [Google Scholar]
- 20.Bogosian G, Somerville R. Trp repressor protein is capable of intruding into other amino acid biosynthetic systems. Mol Gen Genet. 1983;191:51–58. doi: 10.1007/BF00330889. [DOI] [PubMed] [Google Scholar]
- 21.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 22.Terry BJ, Jack WE, Modrich P. Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J Biol Chem. 1985;260:13130–13137. [PubMed] [Google Scholar]
- 23.Dhavan GM, Crothers DM, Chance MR, Brenowitz M. Concerted binding and bending of DNA by Escherichia coli Integration Host Factor. J Mol Biol. 2002;315:1027–1037. doi: 10.1006/jmbi.2001.5303. [DOI] [PubMed] [Google Scholar]
- 24.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 25.von Hippel PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 26.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthold M, et al. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gowers DM, Wilson GG, Halford SE. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA. Proc Natl Acad Sci USA. 2005;102:15883–15888. doi: 10.1073/pnas.0505378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 30.Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doucleff M, Clore GM. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc Natl Acad Sci USA. 2008;105:13871–13876. doi: 10.1073/pnas.0805050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 34.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc Natl Acad Sci USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emmett MR, Johnson JR. Control of metF gene expression in maxicell preparations of Escherichia coli K-12: Reversible action of the metJ protein and effect of vitamin B12. J Bacteriol. 1986;168:1491–1494. doi: 10.1128/jb.168.3.1491-1494.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collier CD, Johnson JR. The Escherichia coli K-12 metJ193 allele contains a point mutation which alters the hydrophobic pocket responsible for in vitro binding of S-adenosylmethionine: Effects on cell growth and induction of met regulon expression. J Bacteriol. 1990;172:3918–3924. doi: 10.1128/jb.172.7.3918-3924.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckman JS, Siedow JN. Bactericidal agents generated by the peroxidase-catalyzed oxidation of para-hydroquinones. J Biol Chem. 1985;260:14604–14609. [PubMed] [Google Scholar]
- 38.Augustus AM, Reardon PN, Heller WT, Spicer LD. Structural basis for the differential regulation of DNA by the methionine repressor MetJ. J Biol Chem. 2006;281:34269–34276. doi: 10.1074/jbc.M605763200. [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Goddard TD, Kneller DG. SPARKY 3. San Francisco: Univ of California; 2004. [Google Scholar]