Abstract
Loss-of-function mutations of SQUINT (SQN)—which encodes the Arabidopsis orthologue of cyclophilin 40 (CyP40)—cause the precocious expression of adult vegetative traits, an increase in carpel number, and produce abnormal spacing of flowers in the inflorescence. Here we show that the vegetative phenotype of sqn is attributable to the elevated expression of miR156-regulated members of the SPL family of transcription factors and provide evidence that this defect is a consequence of a reduction in the activity of ARGONAUTE1 (AGO1). Support for this latter conclusion was provided by the phenotypic similarity between hypomorphic alleles of AGO1 and null alleles of SQN and by the genetic interaction between sqn and these alleles. Our results suggest that AGO1, or an AGO1-interacting protein, is a major client of CyP40 and that miR156 and its targets play a central role in the regulation of vegetative phase change in Arabidopsis.
Keywords: CyP40, HSP90, miR156, phase change, immunophilin
Shoot growth in plants can be divided into juvenile, adult, and reproductive phases according to the character of the lateral organs (leaves and buds) produced during each phase. The juvenile-to-adult transition is known as vegetative phase change and is accompanied by changes in the shape and differentiation of leaves and by an increase in reproductive competence. Screens for mutations that accelerate vegetative phase change in Arabidopsis have produced a large number of genes, many of which encode proteins involved in the biogenesis or function of small RNAs that play key regulatory roles in this process. Although SQUINT (SQN) was one of the first vegetative phase change genes to be identified (1), the basis for its effect on this process remains unknown.
SQN is an orthologue of the immunophilin cyclophilin 40 (Cyp40), a member of a large, evolutionarily conserved class of proteins that possess a peptidyl prolyl cis/trans isomerase (PPIase) domain. These proteins are commonly known as immunophilins because they were originally identified by virtue of their ability to bind the immunosuppressants cyclosporin A or FK506 (reviewed in ref. 2). The 2 major families of immunophilins—cyclophilins and FK506-binding proteins (FKBPs)—have structurally distinct PPIase domains that are unrelated in amino acid sequence. Both families include low-molecular-weight proteins that consist solely of a PPIase domain, as well as larger proteins—such as CyP40—in which this domain is present in association with other functional domains. Arabidopsis possesses more than 50 immunophilins, most of which have no known function (3).
The most intensively studied immunophilins in Arabidopsis are the multidomain FKBP proteins PASTICCINO1 (PAS1) and TWISTED DWARF (TWD)/ULTRACURVATA2 (UCU2). Like CyP40, these FKBP proteins possess an N-terminal PPIase domain and a C-terminal tetratricopeptide repeat (TPR) domain (4). Loss-of-function mutations in PAS1 produce small, distorted seedlings with defects in cytokinin and auxin signaling (5), whereas mutations in TWD/UCU2 produce plants with small, helically rotated roots and leaves, which may be due to reduced auxin transport (6, 7).
In mammals, CyP40 is a major component of the mature estrogen receptor complex and a minor component of the progesterone and glucocorticoid receptor complexes (8, 9). Studies in yeast and mammals have shown that the protein has both PPIase and protein chaperone activity (10–12) and that it binds to HSP90 via its TPR domain (13–15). In Saccharomyces cerevisiae, CyP40-like proteins are encoded by Cpr6 and Cpr7 (16). Cells lacking Cpr6 do not have an obvious phenotype, but cells lacking Cpr7 have a reduced growth rate (17), which has been attributed to a defect in the expression of the thiamine biosynthetic enzyme Thi4p (18).
Previous studies in both Arabidopsis and maize have shown that constitutive expression of the microRNA (miRNA) miR156 prolongs the expression of the juvenile phase of vegetative development and increases the rate of leaf initiation (19–23). In Arabidopsis, miR156 regulates 10 members of the SPL family of transcription factors (24). An analysis of the effect of sqn on gene expression revealed that several SPL transcripts were elevated in sqn and encouraged us to study the function of this gene in more detail. Our results suggest that SQN promotes the activity of miR156 by promoting the activity of ARGONAUTE1, the protein primarily responsible for miRNA-directed posttranscriptional silencing in Arabidopsis (25, 26).
Results
Null Alleles of SQN.
Seven loss-of-function mutations of SQN have been identified in genetic screens conducted in our laboratory and by other investigators (1, 27) [supporting information (SI) Fig. S1]. These mutations produce major defects in the SQN protein and have identical phenotypes, characterized by the precocious production of adult leaves, a delay in leaf initiation, aberrant phyllotaxis in the inflorescence, and an increase in carpel number (1). The severe nature of these mutations and their phenotypic similarity strongly suggest that they represent null, or nearly null, alleles. The experiments described in this article were conducted with the nonsense mutation sqn-1, hereafter referred to as sqn.
SQN Is Required for miR156-Directed Gene Silencing.
To identify genes that contribute to the phenotype of sqn, we profiled transcripts in 10-day-old mutant and wild-type seedlings using the Affymetrix 8K microarray. Among the 45 transcripts whose level was elevated more than 1.5-fold in sqn were AGO1, SPL2, and SPL3. Recognizing that these mRNAs are targets of miRNAs, we examined the effect of sqn on other miRNA-regulated transcripts. Quantitative RT-PCR performed with primers that flank the miRNA target site confirmed the microarray results for these 3 genes and revealed that CUC2, ARF17, SPL4, SPL5, SPL6, SPL9, and SPL11 are also elevated 1.5-fold or more in sqn; there was no major change in the abundance of the other miRNA-targeted transcripts we examined (Fig. 1A).
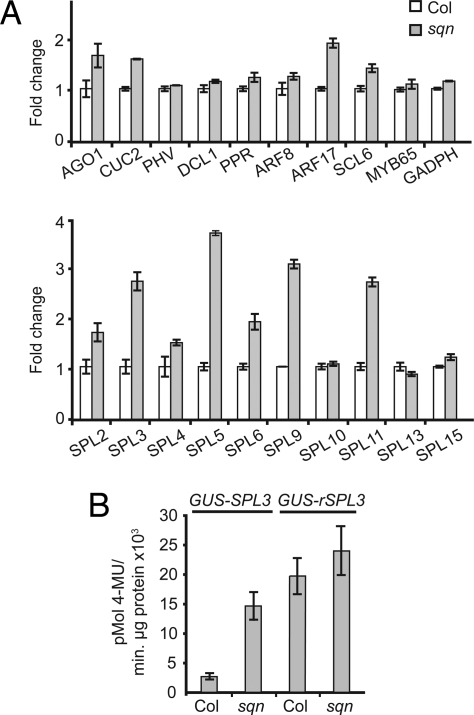
Fig. 1.
sqn interferes with miRNA-mediated silencing. (A) Quantitative RT-PCR analysis of miRNA-targeted transcripts in 12- (top graph) and 18-day-old (bottom graph) sqn seedlings, relative to wild-type Col. GADPH was used as a nontarget control. Values were normalized to actin-2, then to the value of wild-type Col plants, which was fixed to 1. Average of 3 replicates (±SEM). (B) GUS activity in extracts of 30 primary transformants of wild-type Col and sqn plants transformed with miR156-sensitive (35S::rSPL3) or miR156-resistant (35S::rSPL3) reporter genes (±SEM).
To determine whether SQN is involved in the transcriptional or posttranscriptional regulation of these genes, we took advantage of miR156-sensitive and miR156-resistant reporter constructs that have been used previously to study the regulation of SPL3 by miR156 (19). These constructs express a β-glucoronidase-SPL3 fusion transcript (GUS-SPL3) under the regulation of the Cauliflower Mosaic Virus 35S promoter. One construct (35S::GUS-SPL3) encodes a wild-type SPL3 transcript, which is repressed by miR156, whereas the other (35S::GUS-rSPL3) expresses an SPL transcript with a mutated target site, rendering it insensitive to miR156. These reporter genes were introduced into wild-type and sqn plants by Agrobacterium-mediated transformation, and GUS activity was measured in 25–30 primary transformants of each genotype 2 weeks after germination (Fig. 1B). 35S::GUS-rSPL3 was more highly expressed than 35S::GUS-SPL3 in both wild-type and sqn plants, demonstrating that SPL3 is repressed by miR156 in both genotypes. However, 35S::GUS-rSPL3 was expressed at nearly the same level in both genotypes, whereas 35S::GUS-SPL3 was expressed nearly 5 times higher in sqn than in wild-type plants. This result suggests that sqn interferes with either the production or the activity of miR156.
The Precocious Phenotype of sqn Is a Consequence of the Elevated Expression of miR156-Regulated SPL Genes.
miR156-regulated members of the SPL family of transcription factors are among the most highly upregulated genes in sqn (Fig. 1A). To determine whether the phenotype of sqn is attributable to the misexpression of these genes, we crossed a 35S::miR156a transgene into sqn, because previous work has shown that 35S::miR156 strongly represses the expression of all SPL genes (19, 21, 22, 24). Hemizygosity for 35S::miR156a nearly completely suppressed the effect of sqn on leaf shape, abaxial trichome production, and the rate of leaf production (Fig. 2A, B, D, E, and G), demonstrating that the vegetative phenotype of sqn is largely attributable to genes regulated by miR156.
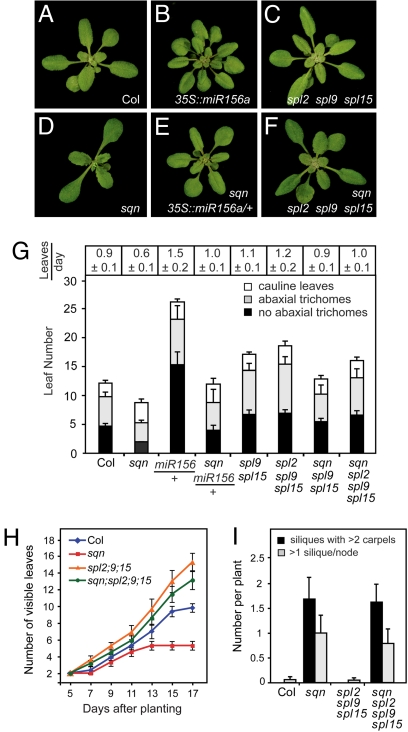
Fig. 2.
The vegetative phenotype of sqn is attributable to the overexpression of genes normally repressed by miR156. (A–F) 21-day-old rosettes. (A) Wild-type Col, (B) 35S::miR156, (C) spl2–1 spl9–4 spl15–1, (D) sqn, (E) sqn 35S::miR156a/+, (F) sqn, spl2–1 spl9–4 spl15–1. (G) Rate of leaf initiation and number of juvenile (abaxial trichomes), adult (no abaxial trichomes), and cauline leaves in various genotypes (±SEM). (H) The rate of leaf initiation in wild-type Col, sqn, spl2–1 spl9–4 spl15–1, and sqn spl2–1 spl9–4, spl15–1 (±SEM). (I) spl2–1 spl9–4 spl15–1 does not rescue the effect of sqn on carpel number or silique spacing.
Loss-of-function mutations in various SPL genes were crossed to sqn to identify the targets of miR156 that contribute to the sqn phenotype. spl2–1, spl9–4, and spl15–1 have either no (spl2–1) or a very small (spl9–4 and spl15–1) effect on shoot development as single mutations but produce a significant delay in abaxial trichome production and an increase in the rate of leaf production in multiple mutant combinations (22, 23) (Fig. 2 C, G, and H). Plants homozygous for sqn and spl9–4 spl15–1, or sqn and spl2–1 spl9–4 spl15–1, had essentially the same leaf morphology and timing of abaxial trichome production as spl9–4 spl15–1 and spl2–1 spl9–4 spl15–1, respectively. However, the rate of leaf production in these lines was intermediate between that of sqn and the spl parent (Fig. 2 F, G, and H). Thus, SPL9, SPL15, and SPL2 are required for the effect of sqn on leaf shape and abaxial trichrome production and contribute to—but are not entirely responsible for—its effect on leaf initiation. In contrast to their effect on vegetative morphology, spl2–2 spl9–4 spl15–1 had no effect on the carpel number and phyllotaxis defects of sqn (Fig. 2I), indicating that these reproductive phenotypes are controlled by other genes. The carpel phenotype of sqn may be attributable to a reduction in the activity of the floral homeotic gene AGAMOUS (AG) (27). This would occur if sqn interfered with the activity of miR172, leading to an increase in the expression of the miR172 target APETALA2, which represses AG (28).
SQN Is Functionally Related to AGO1.
Null alleles of AGO1 produce growth-arrested, severely radialized seedlings, whereas weak alleles produce small, semisterile plants with highly serrated leaves (29, 30). Two recessive missense mutations of AGO1 (ago1–45 and ago1–46) were identified in screens for Ethylmethane sulfonate (EMS)-induced mutants that resemble sqn (Fig. 3 A–F). These newly identified alleles have weaker phenotypes than previously identified hypomorphic mutations of AGO1, such as ago1–25 and ago1–27 (30) (Fig. 3 B, G, and H), and are the weakest alleles of AGO1 described to date. The phenotype of ago1–45 is nearly identical to sqn; ago1–46, ago1–27, and ago1–25 have stronger phenotypes than sqn but resemble sqn in many ways. Like sqn, all of these ago1 alleles produced elliptical, unusually large first leaves and delay leaf initiation. They also reduced the rate of leaf expansion, reduced the number of rosette leaves lacking abaxial trichomes, increased the number of cauline leaves, and produced irregularly spaced siliques (Fig. 3 B, E–H). They differed from sqn only in having no effect on carpel number (data not shown). The phenotypic similarity between these weak alleles of AGO1 and null alleles of SQN strongly suggests that the phenotype of sqn is largely attributable to a reduction in the activity of AGO1.
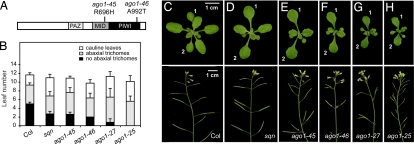
Fig. 3.
Weak alleles of AGO1 resemble sqn. (A) ago1–45 and ago1–46 are missense mutations in the MID and PIWI domains of AGO1. (B) sqn and ago1 mutations reduce the number of juvenile leaves (leaves with no abaxial trichomes), have little or no effect on the number of adult leaves (leaves with abaxial trichomes), and increase the number of cauline leaves (±SEM). (C–H) Eighteen-day-old rosettes and immature inflorescences of (C) wild-type Col, (D) sqn, (E) ago1–45, (F) ago1–46, (G) ago1–27, and (H) ago1–25. Leaves 1 and 2 are indicated. Rosettes are at the same magnification, as are the inflorescences.
To test this hypothesis we examined the genetic interaction between sqn, ago1, and mutations in several genes (DCL1, HEN1, and HYL1) involved in the biogenesis of miRNAs (Fig. 4). Plants homozygous for sqn and dcl1–7 (Fig. 4 F and N), hen1–6 (Fig. 4 G and O), or hyl1–2 (Fig. 4 H and P) had a much more severe phenotype than these single mutants, but the strongest interaction was observed between sqn and hypomorphic mutations of AGO1. Plants heterozygous for sqn and homozygous for ago1–45, ago1–46, or ago1–27 had fewer, smaller, and more elongated leaves than the ago1 single mutants (Fig. 4Q; data not shown), whereas plants homozygous for sqn and these hypomorphic ago1 alleles had the severe phenotype typical of ago1 null mutations: these double mutants had radialized cotyledons, short hypocotyls, very-slow-growing shoots, and were rarely more than 5 mm long (Fig. 4 B–E, J–M). Plants doubly mutant for sqn and the strong allele ago1–47 (SALK_089073) were indistinguishable from ago1–47 (Fig. 4 E and M). This latter result is particularly significant because it suggests that SQN operates with AGO1; if these proteins operated independently, sqn would be expected to interact synergistically with this putative null ago1 allele.
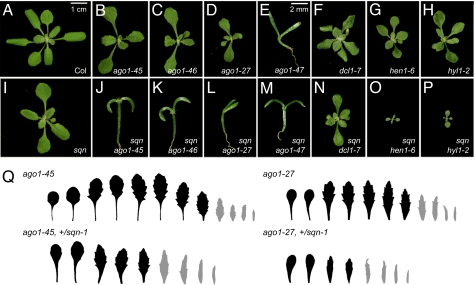
Fig. 4.
sqn interacts synergistically with genes in the miRNA pathway. (A–P) Two-week-old wild-type Col (A), and plants singly and doubly mutant for sqn (I) and ago1–45 (B and J), ago1–46 (C and K), ago1–27 (D and L), ago1–47 (E and M), dcl1–7 (F and N), hen1–6 (G and O), and hyl1–2 (H and P). sqn ago1 double mutants have the same severe phenotype as the ago1 null allele, ago1–47. Double mutants between sqn and dcl1–7, hen1–6, and hyl1–2 were smaller, grew more slowly, displayed earlier onset of abaxial trichomes, and had more severe floral defects and lower fertility than the single mutants. A–D, F–I, and N–P are at the magnification indicated in A. E and J–M are at the magnification indicated in E. (Q) Successive rosette (black) and cauline (gray) leaves of an ago1–45 and ago1–27 plant, and plants homozygous for ago1–45 or ago1–27 and heterozygous for sqn. sqn dominantly enhances the phenotype of these ago1 mutations.
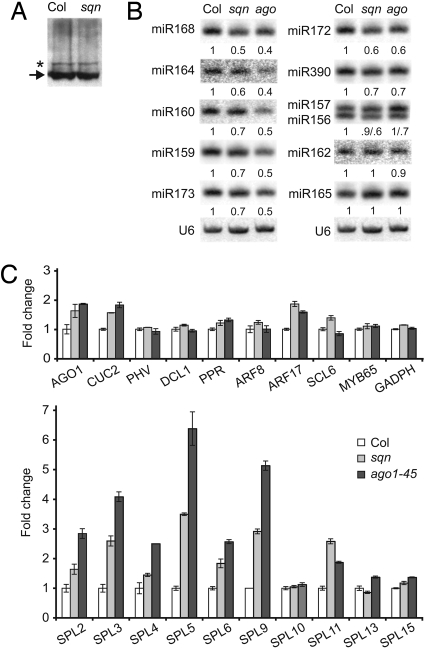
Western blots of protein extracts from sqn and wild-type plants probed with an antibody to AGO1 revealed no difference in the amount of AGO1 protein in these genotypes (Fig. 5A). To explore the possibility that SQN promotes the activity of AGO1, we compared the molecular phenotypes of sqn and ago1–45, the ago1 allele whose morphology most closely resembles sqn. In floral buds, ago1–45 produced a slight decrease in the accumulation of miR168, miR164, miR160, miR159, miR173, miR172, and miR390 but had little or no effect on miR156/157, miR162, and miR165 (Fig. 5B). sqn had a similar phenotype, although it produced a smaller decrease in miRNA levels than ago1–45. sqn and ago1–45 had similar effects on the accumulation of miRNA-regulated transcripts as well (Fig. 5C). Like sqn, agol-45 produced a large increase in the abundance of SPL2, SPL3, SPL4, SPL5, SPL6, SPL9, and SPL11, a smaller increase in the abundance of AGO1, CUC2, and ARF17, and had little or no effect on the level of PHV, DCL1, PPR, ARF8, SCL6, MYB65, SPL10, SPL13, and SPL15.
Fig. 5.
sqn and ago1–45 have similar effects on gene expression. (A) Western blot of protein isolated from Col and sqn seedlings probed with an antibody to AGO1; the background band indicated by the asterisk served as a loading control. (B) Northern blot of low-molecular-weight RNA isolated from floral buds of wild-type, sqn, and ago1–45 plants and hybridized sequentially with oligonucleotides complementary to the functional strand of the indicated miRNA. U6 was used as a loading control. (C) The abundance of miRNA-regulated transcripts in ago1–45 and sqn, relative to wild type. The values for sqn are the same as those in Fig. 2 and are presented here again for comparison. Quantitative RT-PCR was performed on RNA isolated from 12- (top graph) and 18-day-old (bottom graph) seedlings using primers specific for each gene. GADPH was used as a nontarget control. Values were normalized to actin-2, then to the value of wild-type Col plants, which was fixed to 1. Average of 3 replicates (±SEM).
In addition to mediating miRNA-directed transcript cleavage, AGO1 is required for the sense-silencing of some transgenes, the best-studied example being the 35S::GUS insertion in the L1 line (31). To determine whether SQN is required for this process, we compared the effects of sqn, ago1–45, and ago1–27 on the expression of LI (Fig. S2A). As has been previously reported (30, 31), ago1–27 completely blocked L1 silencing (n = 25). ago1–45 had a variable phenotype: 63% of plants expressed GUS at the same level as ago1–27, whereas the remaining plants were completely silenced (n = 32). L1 was silenced at the same frequency in sqn (97%; n = 30) as in wild-type plants (92%; n = 25). Thus, SQN is not required for L1 silencing. sqn also had no effect on the abundance of transcripts silenced by transacting siRNAs (Fig. S2B), indicating that it is not required for the production or function of this class of endogenous siRNAs.
Does SQN Interact with HSP90 or AGO1?
CyP40 co-immunoprecipitates with HSP90 in both mammals (13, 14) and S. cerevisiae (32) and is thought to act in association with this protein (8). We initially explored the possibility that SQN interacts with Arabidopsis HSP90 using a yeast 2-hybrid assay, because this method has been used to study the interaction between human CyP40 and HSP90 (33). No interaction was observed between the full-length SQN and HSP90 proteins under either low- or high-stringency selection (data not shown); the C-terminal TPR fragments of HSP90 and SQN produced false-positive interactions with control proteins and therefore could not be assayed by this method. We were also unable to test the interaction between SQN and AGO1 because constructs expressing AGO1 could not be propagated in yeast. As an alternative approach, we searched for proteins that co-immunoprecipitate with SQN-GFP in plants transformed with a 35S::SQN-GFP construct that rescues the sqn phenotype. SQN-GFP was immunoprecipitated from seedling extracts with an antibody to GFP and subjected to nano-liquid chromatography coupled to tandem mass spectrometry (nLC-MS/MS) analysis. Although SQN was readily detectable in 2 independent experiments, with 39 independent peptide hits totaling approximately 60% coverage of the protein, no other proteins were enriched in the immunoprecipitate compared with the same precipitate from nontransgenic plant extracts. Western blots of immunoprecipitated SQN-GFP probed with antibodies against HSP90 or AGO1 also failed to reveal these proteins (Fig. S3; data not shown). Thus, if SQN binds to HSP90 or AGO1, this interaction is either transient or quite weak.
Plants with reduced levels of HSP90 have a highly pleiotropic phenotype that resembles the phenotype of strong ago1 alleles (34). To determine whether HSP90 plays a role in miRNA-related processes, we characterized the phenotype and genetic interactions of hsp90.2–3, a point mutation in the ATP binding domain in 1 of the 4 genes encoding cytoplasmic HSP90 (34, 35). Northern analysis revealed that miR168 and miR172 were reduced, whereas miR160 was elevated in hsp90.2–3 (Fig. S4A), indicating that HSP90 plays a role in miRNA-related processes but that its function is distinct from that of SQN. The morphology of hsp90.2–3 resembled sqn in some respects but not in others. hsp90.2–3 plants had slightly elongated juvenile leaves, an increased numbers of juvenile, adult, and cauline leaves, and produced flowers with extra sepals (Table 1 and Fig. S4 B, F, and L). A significant number of seeds doubly mutant for hsp90.2–3 and sqn failed to germinate, or produced seedlings with aborted roots, short hypocotyls, distorted or multiple cotyledons, and delayed development of the shoot apex (Table 1 and Fig. S5). The remaining sqn hsp90.2–3 plants had dark green, narrow, slow-growing leaves, an increased number of cauline leaves and associated lateral inflorescences, open flowers with an increased number of sepals, petals, and carpels, and irregularly spaced, sterile siliques (Table 1 and Fig. S4 G, M, and S). Plants doubly mutant for hsp90.2–3 and ago1–45 had nearly this same vegetative phenotype (Table 1, Figs. S4 B, H, N, and T, and S5) but differed from hsp90.2–3 sqn-1 in producing relatively normal flowers. This synergistic interaction suggests that HSP90 regulates some of the same processes as AGO1 and may act directly on this protein. Unfortunately, because the paralogues of HSP90.2 are tightly linked, it is difficult to generate the HSP90 null genotype necessary to rigorously test this hypothesis.
Table 1.
hsp90.2-3 interacts synergistically with sqn-1 and ago1-45
| Genotype | Ungerminated seeds (%)* | Abnormal seedlings (%)* | Flowers with 5 sepals (%)† | Siliques with 3 carpels (%)† | Leaves without abaxial trichomes (± SEM)‡ | Leaves with abaxial trichomes (± SEM)‡ | Cauline leaves (± SEM)‡ |
|---|---|---|---|---|---|---|---|
| Col | 0 | 0 | 0 | 0 | 4.9 ± 0.1 | 5.8 ± 0.1 | 2.9 ± 0.1 |
| sqn-1 | 2 | 0 | 0 | 20 | 2.1 ± 0.1 | 5.7 ± 0.1 | 4.3 ± 0.1 |
| ago1-45 | 0 | 0 | 0 | 0 | 2.2 ± 0.1 | 7.0 ± 0.1 | 3.8 ± 0.1 |
| hsp90.2-3 | 0 | 0 | 13 | 0 | 6.3 ± 0.2 | 9.8 ± 0.2 | 4.4 ± 0.1 |
| sqn-1, hsp90.2-3 | 13 | 13 | 80 | 100 | 2.9 ± 0.1 | 15.5 ± 0.2 | 6.0 ± 0.2 |
| ago1-45, hsp90.2-3 | 12 | 6 | 4 | 0 | 3.2 ± 0.2 | 9.7 ± 0.1 | 6.2 ± 0.2 |
*n = 50 for single mutants and 500 for double mutants.
†n = 25 flowers or siliques.
‡n = 22 plants.
Discussion
The biochemical properties of CyP40 have been intensively studied for many years, but the biological function of this protein remains unknown (9). Our results indicate that in Arabidopsis CyP40 promotes miRNA-mediated gene repression, probably by promoting the activity of AGO1. In addition, we showed that the precocious vegetative phenotype of sqn is largely attributable to a defect in the activity of a single miRNA, miR156.
Evidence that SQN is required for the activity of miR156 is provided by the observation that sqn had little or no effect on the level of miR156 but caused a significant increase in the accumulation of transcripts regulated by this miRNA and by the observation that an miR156-sensitive reporter gene was expressed at a significantly higher level in sqn than in wild-type plants. The discovery that hypomorphic alleles of AGO1 have a morphologic and molecular phenotype strikingly similar (and in at least 1 case, nearly identical) to that of sqn, strongly suggests that the effect of sqn on miR156 activity can be attributed to a defect in the activity of AGO1. Further support for this hypothesis is provided by the observation that sqn dominantly enhanced the phenotype of hypomorphic alleles of AGO1, which suggests that the activity of AGO1 is exquisitely sensitive to the dose of SQN. Consistent with this hypothesis, we found that plants doubly mutant for sqn and weak or strong loss-of-function mutations of AGO1 have a phenotype that closely resembles the phenotype of null AGO1 mutations. The simplest interpretation of these results is that SQN and AGO1 have related functions.
How are SQN and AGO1 functionally related? sqn does not affect the amount of the AGO1 protein, indicating that it is not required for the stability of AGO1. An alternative possibility is that SQN promotes the activity of AGO1. Studies of mammalian CyP40 (9) suggest that SQN may assist in the folding of AGO1 or a protein that interacts with AGO1, or may hold these proteins in an active conformation. However, SQN cannot be absolutely required for AGO1 activity because null alleles of AGO1 have a much stronger phenotype than null alleles of SQN. This observation raises the possibility that SQN is more important for some of the functions of AGO1 than for others. AGO1 promotes miRNA-directed transcript cleavage (25, 26) and translational repression (36) and is also required for at least some types of siRNA-mediated posttranscriptional gene silencing. If SQN is only required for some of these functions, mutations in SQN would be expected to have a more limited phenotype than mutations in AGO1. This latter hypothesis is supported by the observation that sqn had no effect on L1 silencing. However, we suspect this result is attributable to the relatively small effect of sqn on the overall activity of AGO1 rather than to the functional specificity of SQN because of the stochastic effect of ago1–45 on L1 silencing. This stochastic phenotype contrasts with the uniform and fully penetrant morphological phenotype of ago1–45 and suggests that AGO1 is more important for miRNA-mediated silencing than for sense-silencing.
Much remains to be learned about the mechanism by which SQN promotes the activity of AGO1. In both yeast (32) and mammals (10, 11, 13–15), CyP40 has both PPIase and protein chaperone activity and is thought to act in association with HSP90. It is reasonable to assume that SQN has at least some of the activities that have been assigned to CyP40 because it is 65% identical and 80% similar to human CyP40 within the PPIase domain, and the 5 aa within the TPR domain that are critical for the interaction of CyP40 and HSP90 (33) are conserved in SQN. Furthermore, the synergistic interaction between sqn and hsp90.2–3 suggests that these proteins may be functionally related. But whether SQN actually acts together with or independently of HSP90, as well as the specific mechanism by which it promotes the activity of AGO1, will need to be answered by future studies.
Materials and Methods
Genetic Stocks and Growth Conditions.
All of the genetic stocks used in this study were in a Columbia (Col) background. ago1–45 and ago1–46 were identified in a screen of M2 families from EMS-mutagenized plants. Seeds of ago1–25, ago1–27, and the L1 line were provided by H. Vaucheret (Institut National de la Recherche Agronomique, Versailles, France); seeds of dcl1–7 were obtained from Animesh Ray (Keck Graduate Institute, Claremont, CA); hen1–2 (SALK_090960), hyl1–2 (SALK_064863), spl2–1 (SALK_022235), spl9–4 (SAIL_150_B05), spl15–1 (SALK_074426), and ago1–47 (SALK_089073) were obtained from the Arabidopsis Biological Resource Center. Information about growth conditions, phenotypic analysis, and the identification of double mutants is proved in SI Materials and Methods. GUS assays were performed as previously described (37).
Microarray Analysis, RNA Blots, and Quantitative Real-Time PCR.
Total RNA was isolated using TRIzol (Invitrogen) from 10–18-day-old seedlings or from floral buds grown under long-day (16 h light, 8 h dark) conditions. For microarray analysis, RNA for 10-day-old seedlings was further purified with RNAeasy (Qiagen) columns and subsequently processed by the University of Pennsylvania microarray facility. Quantitative real-time PCR was performed on DNase-treated seedling or floral bud RNA, using primers (Table S1) that flank the miRNA target site. More detailed information is provided in SI Materials and Methods.
Co-immunoprecipitation and Western Blots.
An SQN-GFP fusion construct was generated using an SQN cDNA and introduced into sqn plants by Agrobacterium transformation. Protein extracts from plants homozygous for this transgene were used for co-immunoprecipitation, mass spectrometry, and Western blots, as described in SI Materials and Methods. To assay the effect of sqn on AGO1 proteins levels, total protein extracts from mutant and wild-type floral buds were run on an SDS-PAGE gel, transferred overnight at 30V at 4 °C, and probed with antibody to AGO1 provided by Y. Qi (National Institute of Biological Sciences, Beijing, China).
Supplementary Material
Acknowledgments.
We thank Pam Green and members of the Poethig laboratory for helpful comments on the manuscript; and Herve Vaucheret for providing seeds of some of the lines used in this study. This work was supported by National Institutes of Health Grant RO1 GM051893.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812729106/DCSupplemental.
References
- 1.Berardini TZ, Bollman K, Sun H, Poethig RS. Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science. 2001;291:2405–2407. doi: 10.1126/science.1057144. [DOI] [PubMed] [Google Scholar]
- 2.Galat A. Peptidylproline cis-trans-isomerases: Immunophilins. Eur J Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- 3.He Z, Li L, Luan S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004;134:1248–1267. doi: 10.1104/pp.103.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrar Y, Bellini C, Faure JD. FKBPs: At the crossroads of folding and transduction. Trends Plants Sci. 2001;6:426–431. doi: 10.1016/s1360-1385(01)02044-1. [DOI] [PubMed] [Google Scholar]
- 5.Vittorioso P, Cowling R, Faure JD, Caboche M, Bellini C. Mutation in the Arabidopsis PASTICCINO1 gene, which encodes a new FK506-binding protein-like protein, has a dramatic effect on plant development. Mol Cell Biol. 1998;18:3034–3043. doi: 10.1128/mcb.18.5.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Perez JM, Ponce MR, Micol JL. The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 2004;134:101–117. doi: 10.1104/pp.103.032524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchard R, et al. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem. 2006;281:30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- 8.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 9.Ratajczak T, Ward BK, Minchin RF. Immunophilin chaperones in steroid receptor signalling. Curr Top Med Chem. 2003;3:1348–1357. doi: 10.2174/1568026033451934. [DOI] [PubMed] [Google Scholar]
- 10.Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: Chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 11.Pirkl F, Buchner J. Functional analysis of the Hsp90-associated human peptidyl prolyl cis/trans isomerases FKBP51, FKBP52 and Cyp40. J Mol Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 12.Mok D, et al. The chaperone function of cyclophilin 40 maps to a cleft between the prolyl isomerase and tetratricopeptide repeat domains. FEBS Lett. 2006;580:2761–2768. doi: 10.1016/j.febslet.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann K, Handschumacher RE. Cyclophilin-40: Evidence for a dimeric complex with hsp90. Biochem J. 1995;307:5–8. doi: 10.1042/bj3070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratajczak T, Carrello A. Cyclophilin 40 (CyP-40), mapping of its hsp90 binding domain and evidence that FKBP52 competes with CyP-40 for hsp90 binding. J Biol Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 15.Owens-Grillo JK, et al. The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- 16.Duina AA, Marsh JA, Gaber RF. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast. 1996;12:943–952. doi: 10.1002/(sici)1097-0061(199608)12:10<943::aid-yea997>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Duina AA, Kalton HM, Gaber RF. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J Biol Chem. 1998;273:18974–18978. doi: 10.1074/jbc.273.30.18974. [DOI] [PubMed] [Google Scholar]
- 18.Faou P, Tropschug M. A novel binding protein for a member of CyP40-type cyclophilins: N. crassa CyPBP37, a growth and thiamine regulated protein homolog to yeast Thi4p. J Mol Biol. 2003;333:831–844. doi: 10.1016/j.jmb.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 21.Gandikota M, et al. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwab R, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Prunet N, et al. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell. 2008;20:901–919. doi: 10.1105/tpc.107.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohmert K, et al. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morel JB, et al. Fertile hypomorphic ARGONAUTE mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elmayan T, et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10:1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duina AA, Chang HC, Marsh JA, Lindquist S, Gaber RF. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 33.Ward BK, et al. A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90. J Biol Chem. 2002;277:40799–40809. doi: 10.1074/jbc.M207097200. [DOI] [PubMed] [Google Scholar]
- 34.Sangster TA, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE. 2007;2:e648. doi: 10.1371/journal.pone.0000648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubert DA, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 37.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.