Abstract
We have investigated the mechanism of resistance of a HIV type 1 (HIV-1) R5 primary isolate, D1/85.16, to the small molecule CCR5 inhibitor, vicriviroc (VVC). Unlike other viruses resistant to this class of compound, D1/85.16 lacks sequence changes in the V3 region of the gp120 surface glycoprotein. Inspection of env sequences from D1/85.16 compared with those derived from the parental, inhibitor-sensitive virus, CC1/85, revealed a cluster of 3 conservative changes in the fusion peptide (FP) of the gp41 transmembrane glycoprotein that tracked with the resistance phenotype. Studies with engineered Env-chimeric and point-substituted viruses confirmed that these 3 FP residues were substantially responsible for VVC resistance without altering coreceptor usage, as assessed in both peripheral blood mononuclear cells and the TZM-bl cell line. VVC resistance is manifested differently in the 2 cell types, and there are assay-dependent complexities to the dose-response curves for the engineered resistant viruses. To explain them, we created a model for resistance and generated theoretical VVC inhibition curves that closely mimic the experimental data for the resistant viruses. The basis for the model is the existence of distinct forms of CCR5, with varying affinities for small molecule CCR5 inhibitors that are presumed to be present in different proportions on different cell types, and are used selectively by resistant HIV-1 variants when ligated with an inhibitor. Together, the experimental results and theoretical model may help understand how HIV-1 uses CCR5 to enter target cells under various conditions.
Keywords: vicriviroc, virus entry, coreceptor conformation, theoretical model
The small molecule CCR5 inhibitors maraviroc (MVC) and vicriviroc (VVC) are now used to treat infection with HIV type 1 (HIV-1) (1). These and similar compounds target CCR5, a G protein-coupled receptor (GPCR) used by HIV-1 early in the course of the infection (2). HIV-1 entry is mediated by the envelope glycoprotein (Env) complex, a heterotrimer of 3 gp120 surface subunits noncovalently linked to 3 gp41 transmembrane subunits. The gp120 subunits contact first CD4, then CCR5; conformational rearrangements within the trimer drive insertion of the gp41 fusion peptide (FP) region into the host cell membrane, leading to fusion and the initiation of infection (1). MVC or VVC binding within a cavity in the CCR5 transmembrane helices stabilizes the extracellular domains in a conformation that is not efficiently recognized by wild-type gp120; they inhibit entry via a noncompetitive or allosteric mechanism (1).
The clinical use of small molecule CCR5 inhibitors could, theoretically, drive the emergence of more pathogenic HIV-1 X4 variants that use CXCR4 for entry (1). Although expansion of preexisting X4 or dual-tropic (R5X4) variants during therapy has been reported (3), the principal escape mechanism is the evolution of mutations that permit HIV-1 to continue to use CCR5 despite the presence of the inhibitor; fully resistant viruses can use both the inhibitor-CCR5 complex and free CCR5 for entry (4, 5). The genetic determinants of resistance in vitro have been mapped, almost invariably, to the gp120 V3 region (4, 6–9). This localization is consistent with the variable nature of V3 and its function as a component of the CCR5-binding site (1, 10). VVC-resistant viruses generated in vitro contained various sequence changes in gp120 including, but not limited to, V3 (7), and the emergence of VVC-resistance in a patient has also been attributed to V3 changes (11). The V3-route to resistance was followed when a primary isolate, CC1/85, was cultured with AD101, a preclinical precursor of VVC (6, 12). The fully resistant variant, CC101.19, was stable and fit; it did not rapidly revert to sensitivity when cultured in peripheral blood mononuclear cells (PBMC) without AD101 (13). CC101.19 has 4 point substitutions in V3 that are necessary and sufficient for resistance to AD101, VVC, and similar compounds (6, 12). However, when CC1/85 was cultured with VVC, we identified a fully resistant clone with no V3 sequence changes (14).
Here, we have studied the VVC-resistant D1/85.16 virus, to better understand the genetics of resistance and Env structure-function relationships that influence HIV-1 entry and its inhibition. We show that the critical sequence changes that confer resistance, without causing coreceptor switching to CXCR4, are 3 physically proximal substitutions in the gp41 FP region. Thus, there are both V3-dependent and V3-independent genetic pathways to the same phenotype of CCR5 inhibitor-resistance that can be followed by the HIV-1 quasispecies of the same isolate. An additional implication is that the gp120 V3 region and the gp41 FP may become structurally or functionally associated at some stage during virus-cell fusion. Analysis of the behavior of resistant and sensitive clones in PBMC and the TZM-bl cell line led us to develop a model to explain cell type-dependent complexities in resistance to CCR5 inhibitors. The model predicts the existence of at least 2 forms of CCR5 with different affinities for small molecule inhibitors that are present in different relative abundances on different cell types.
Results
VVC Resistance of D1/85.16 Maps to gp41.
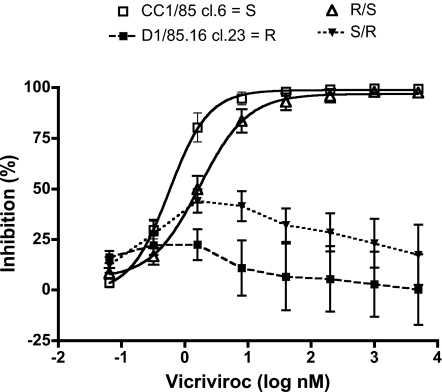
The D1/85.16 isolate arose under VVC selection pressure (14), and uses the inhibitor-CCR5 complex as the basis for its resistance to VVC and related compounds (5). Unlike CC101.19, D1/85.16 lacked a consistent pattern of sequence changes in the gp120 V3 region, and a VVC-resistant clone, D1/85.16 cl.23, had a V3 sequence identical to VVC-sensitive CC1/85 clones (6, 14). To test whether resistance tracked with the gp120 or gp41, we swapped these subunits from D1/85.16 cl.23 (= R) with the corresponding components of the parental clone CC1/85 cl.6 (= S). Therefore, the R/S chimeric virus contains gp120 from R and gp41 from S, and conversely for the reverse chimera, S/R [for virus nomenclature, Table S1]. Both chimeric viruses were replication-competent in PBMC with titers of 104 to 105 TCID50/mL on day 7 postinfection (p24, 3–5 ng/mL). VVC resistance in these cells tracked with gp41. Thus, the R/S chimera behaved identically to the sensitive isolate and clone S, the S/R chimera to the resistant isolate and clone R (Fig. 1). The complex structure to the VVC-inhibition curve for S/R is analyzed further below. Similar results for both chimeric viruses were also obtained when AD101 was used.
Fig. 1.
VVC resistance maps to gp41. Clonal chimeric viruses R/S and S/R (Table S1) bearing gp120 and gp41 subunits derived either from the parental, VVC-sensitive clone S, or the VVC-resistant clone R were tested for VVC sensitivity in a multicycle PBMC-based replication assay measuring p24 production 7 days postinfection. The data shown are mean values derived from 7 to 15 independent experiments ± SEM.
Sequence Changes in the FP Region of D1/85.16.
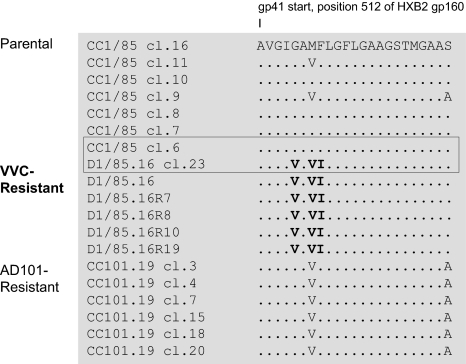
We sequenced the env genes from isolate D1/85.16 and its clone R, as well as from 4 reversion culture isolates (D1/85.16R7, D1/85.16R8, D1/85.16R10, and D1/85.16R19; Fig. S1). These sequences were compared with clones derived from the CC1/85 parental virus and the AD101-resistant CC101.19 isolate (6). The only consistent pattern of changes involved the FP region of gp41. Thus, all 6 D1/85.16 isolates or clones contained 3 changes that were rare or absent from the parental CC1/85 clones: G516V, M518V, and F519I (Fig. 2). Of these, 516V and 519I were absent from all of the CC1/85 or CC101.19 sequences, whereas 518V was present in 2/7 CC1/85 clones and in all 6 CC101.19 clones. This pattern suggests 518V is a naturally occurring polymorphism that becomes enriched under AD101 or VVC selection pressure. All 3 changes involve the conservative substitution of hydrophobic amino acids, consistent with their location in the FP, and each introduces a C-β branched amino acid (Val or Ile). No virus containing all 3 altered amino acids is present in the Los Alamos HIV Sequence Database (www.hiv.lanl.gov/, as per 10.28.2008), implying that their occurrence in D1/85.16 did not arise by chance. All of the CC101.19 clones also contained an Ala substitution at position 534, but 6/7 parental CC1/85 clones and all of the D1/85.16-derived sequences contained Ser, suggesting this position is irrelevant to VVC resistance.
Fig. 2.
Alignment of N-terminal gp41 sequences from VVC-sensitive and VVC-resistant viruses. The first 23 amino acid residues from the gp41 N terminus are shown for 7 clones from the CC1/85 parental virus, 6 VVC-resistant isolates or clones based on D1/85.16, and 6 clones from the AD101-resistant CC101.19 isolate. The sequences are recorded relative to that of CC1/85 cl.16 (top line), with dashes indicating amino acid sequence identity. Amino acid numbering is based on HXB2 Env, with the first residue of gp41 at position 512. The 3 conservative substitutions of hydrophobic residues, G516V, M518V, and F519I, in the FP region of resistant viruses are highlighted in bold. Among the 7 parental clones, S (GenBank accession no. AY357338) is the most closely related to R; these 2 clones, which were used for subsequent genetic studies, are boxed. The env sequences of the D1/85.16-derived viruses have been deposited in GenBank (accession nos. FJ713453–FJ713458).
Site-Directed Mutagenesis of Specific Residues in the FP Region.
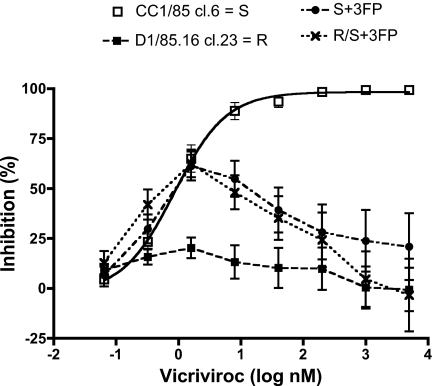
To test whether the G516V, M518V, and F519I changes confer VVC resistance, site-directed mutagenesis was performed on the sensitive parental clone S. This clone was chosen because it was the most closely related to the resistant clone R (14). The same 3 substitutions were also introduced into the sensitive R/S chimera to investigate their actions in a different gp120 context. The engineered clones S + 3FP and R/S + 3FP were replication-competent in PBMC; their titers were several-fold lower than the other test viruses' ranging from 103 to 104 TCID50/mL on day 7 postinfection (p24, ≈3 ng/mL). When the reverse chimera S/R and the S + 3FP and R/S + 3FP engineered clones were tested for VVC sensitivity by using PBMC, they were all resistant to high VVC concentrations (Fig. 3). As previously, the S and R/S viruses were VVC-sensitive, whereas R was fully resistant. The S/R reverse chimera and the S + 3FP and R/S + 3FP mutants were also cross-resistant to AD101. Thus, the 3 FP changes are sufficient to confer VVC (and AD101) resistance on a sensitive virus, and without causing CXCR4 usage (Fig. S2). However, the same 3 FP changes do not have this effect when introduced into a different, VVC-sensitive virus, JR-FL, implying that they act only in a defined Env context (Fig. S3). Studies on the roles played by the individual changes and their combinations in conferring resistance on CC1/85 are now in progress.
Fig. 3.
The 3 gp41 FP amino acid substitutions confer resistance to VVC. Site-directed mutagenesis was performed to introduce the 3 changes (G516V, M518V, and F519I) into the VVC-sensitive parental clone S and the R/S chimera (Table S1). The engineered mutant viruses, S + 3FP and R/S + 3FP, were then tested for VVC sensitivity in a multicycle PBMC-based replication assay measuring p24 production 7 days postinfection. For comparison, S and the VVC-resistant clone R were also tested. In the same experiments, the R/S and S/R chimeras behaved comparably with S and R, respectively, but the curves are not shown, for clarity. The depicted results are the average of 6–10 independent experiments, with the error bars indicating the SEM.
However, there were intriguing aspects to the VVC dose-response curves for the resistant viruses. Thus, the replication of S/R, S + 3FP and R/S + 3FP was partially inhibited (40–60%) by low VVC concentrations (≈0.3–2 nM), but enhanced by higher concentrations (up to 5 μM; see Figs. 1 and 3). This pattern of partial, biphasic inhibition at low VVC concentrations was also observed, albeit less so, with R (Figs. 1 and 3). The replication-enhancing effect of higher VVC concentrations appeared to be more pronounced for the R/S + 3FP virus than for S + 3FP (Fig. 3). Therefore, the gp120 component derived from R, which is present in the former but not the latter virus, may contribute to the resistance phenotype, particularly at high inhibitor concentrations. In contrast, the enhancing effects of higher VVC concentrations were similar for the reverse chimera S/R and the S + 3FP mutant (Figs. 1 and 3); therefore, this phenotypic effect of the gp41 subunit from R may be attributable to the 3 specific FP changes. A possible biological explanation for the biphasic inhibition curve is modeled below.
Cell Type Dependence of VVC-Resistance.
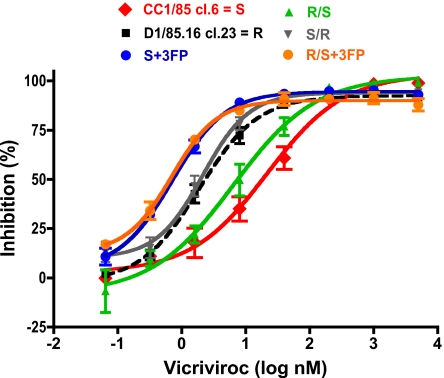
Resistance to small-molecule CCR5 inhibitors varies with the cell type (4, 5, 7). When viruses that are fully resistant to a compound such as VVC and MVC in PBMC are tested in cell lines engineered to express CCR5 (e.g., U87.CD4.CCR5 or TZM-bl), they tend to be inhibited but only up to a maximum value [the maximum percent inhibition (MPI)]. The extent of inhibition is then unaffected by a further increase in inhibitor concentration (“the plateau effect”). The MPI reflects the relative ability of the virus to use the inhibitor-bound and inhibitor-free forms of CCR5 for entry; the higher the MPI value, the less efficiently a resistant virus recognizes the inhibitor-CCR5 complex (4, 5). Therefore, we tested the engineered, clonal infectious viruses for their sensitivity to VVC in TZM-bl cells, to further characterize their resistance profiles (Fig. 4).
Fig. 4.
VVC resistance is manifested differently in a cell line-based assay. The same infectious, clonal viruses studied in PBMC were tested in TZM-bl cells with a luciferase reporter gene endpoint. The VVC inhibition curves (dashed black line for the resistant virus R) asymptote to a plateau that is high, but consistently lower (upper limit of 95% C.I. <100%) for the resistant than the sensitive viruses. The data shown are the average of 3 independent experiments. Resistance-related parameters derived from these experiments are summarized in Table 1.
Each virus was replication-competent in TZM-bl cells, with relative luminescence units (RLU) varying within a 10-fold range (Table 1). The parental clone S and the sensitive chimera R/S behaved similarly; each was fully inhibited by VVC (MPI >99% in both cases), with half maximum (IC50) values of 20 and 7.8 nM, respectively (Fig. 4, Table 1). The R and S/R viruses had MPI values of 93 and 94% with reduced IC50 values of 2.4 and 2.0 nM; although these MPI values are high, the upper limits of their 95% C.I. were consistently <100% (Table 1). The MPI values for the engineered resistant viruses S + 3FP and R/S + 3FP were also consistently <100% (95 and 90%, respectively), but their IC50 values were reduced even further, to 0.70 and 0.66 nM (Fig. 4, Table 1). Similar results for the S, R, and S + 3FP viruses were also obtained by using AD101.
Table 1.
Indicators of VVC-resistance in TZM-bl cells
| Virus | MPI, % (95% C.I.) | IC50, nM | RLU |
|---|---|---|---|
| CC1/85 cl.6 = S | 100 (94–110)* | 20 | 1.4 × 105 |
| D1/85.16 cl.23 = R | 93 (89–96) | 2.4 | 1.6 × 105 |
| R/S | 100 (93–110)* | 7.8 | 6.7 × 104 |
| S/R | 94 (90–98) | 2.0 | 4.1 × 104 |
| S + 3FP | 95 (93–97) | 0.70 | 4.2 × 104 |
| R/S + 3FP | 90 (87–93) | 0.66 | 1.9 × 104 |
Data are mean values derived from 3 independent experiments. *, P < 0.01 for R vs. S and for R vs. R/S MPI values; P > 0.05 between the MPI for R and each other clone.
Therefore, the engineered mutant viruses, which are strongly resistant in PBMC (Fig. 3), are ≈30-fold more sensitive (as judged by IC50 values) than the parental clone to low VVC concentrations when tested in TZM-bl cells (Fig. 4, Table 1). However, these viruses do display the noncompetitive resistance indicator that is characteristic of assays based on engineered cell lines, i.e., MPI significantly <100% (4, 5, 7). Experiments using U87.CD4.CCR5 cells and pseudotyped viruses bearing Env proteins from clones S and R yielded similar findings, including the replication of a small viral fraction in the presence of high VVC concentrations and a reduced IC50 for R compared with S (Fig. S4). Also, the principal finding from both the cell line- and PBMC-based assays is that gp41 has the paramount role in resistance, principally due to the 3 specific FP changes. The gp120 context can have an additional influence, particularly at high inhibitor concentrations. Thus, the MPI values are almost identical for the S/R and S + 3FP viruses, and lower for R/S + 3FP (Table 1). The complexities of CCR5 usage that may underlie these observations are discussed below.
Modeling the Cell Type-Dependency of Resistance.
The dose-response curves for the resistant clones depicted in Figs. 1, 3, and 4 are clearly both complex and cell type-dependent. To try to understand the underlying biology, we developed a model (Fig. S5). We postulate that 2 different subpopulations of CCR5 receptors coexist on the cell surface in proportions that vary between cell types, and that the infectivity of a viral variant is a linear function of the amount of a CCR5 form(s) that can support entry of that variant. One CCR5 subpopulation (designated CCR5-A) is assumed to bind VVC, and related inhibitors, with a low affinity, whereas the second form (CCR5-B) binds them with a higher affinity. Wild-type HIV-1 can have a preference for one of the free forms of CCR5, but cannot use either inhibitor-bound form for entry. A resistant variant can use inhibitor-CCR5-A complexes while losing the capacity to enter via the free form of CCR5-A, but retaining or increasing its ability to use the free form of CCR5-B. This version of the model is the simplest; in reality, there can be >2 subpopulations of CCR5. The mathematical formulation of the model is presented in SI Materials and Methods, and the different scenarios are depicted schematically in Fig. S5. The principles of the modeling results are shown in Fig. S6.
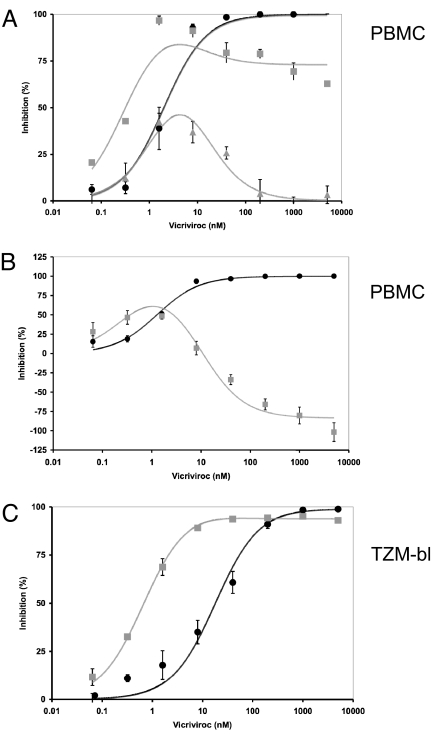
We used nonlinear regression to fit the general function of the model (15) to experimental PBMC and TZM-bl data (Fig. 5). In this analysis, we made no assumptions about the relative proportions of the 2 CCR5 forms, or their absolute affinities for VVC, when fitting the function to the PBMC data. However, to avoid creating excessively wide C.I. when modeling the TZM-bl cell data, IC50 values were estimated for S and S + 3FP from the sigmoid dose-response plots derived experimentally by using these cells. The resulting IC50 ratio of 37 was then used as a constant affinity ratio in a 2-parameter function (SI Materials and Methods). The resulting functions were then fitted to the experimental data (Fig. 5).
Fig. 5.
Fitting of model function to empirical data from PBMC and TZM-bl cell assays. The relative inhibition of virus infectivity is expressed in percentage on the y axis as a function of the VVC concentration in nM on the x axis. The black circles represent data for wild-type virus (S); data for the VVC-resistant S + 3FP mutant are shown as gray squares and triangles (error bars, SEM in each case). The theoretical curves derived from the model are shown as continuous lines in the shades of the dataset each is fitted to. (A) Inhibition of the S + 3FP (gray) and S (black) viruses in PBMC from 2 different donor pools. The resistant virus is partially or negligibly inhibited at the highest VVC concentrations. Datasets from other donor pools fall between these profiles when plotted similarly. (B) An example of enhancement of S + 3FP virus infection (gray) at the highest VVC concentration is contrasted with inhibition of S virus (black) on PBMC from different donor pools. Different relative amounts of the CCR5-A and CCR5-B modulate the different levels of inhibition seen at the highest VVC concentrations in the different experiments. (C) Inhibition of TZM-bl cell infection by S + 3FP (gray) and S (black). The model fits the data well when the VVC affinities of CCR5-A and CCR5-B are assumed to differ by >37-fold, a value that was predetermined by measuring the IC50 values for the respective viruses on TZM-bl cells. The small residual infectivity of S + 3FP at the highest VVC concentrations results from an excess of CCR5-B (high-affinity for VVC) over CCR5-A (lower affinity for VVC). This excess would be ≈20-fold if the efficiency of S + 3FP viral entry were the same for free CCR5-B as for VVC-CCR5-A complexes.
As noted above (Fig. 3), the experimental VVC dose-response curves against the resistant S + 3FP mutant in PBMC have complex shapes. Inhibition is maximal at intermediate VVC concentrations, but a plateau arises at higher concentrations that varies from partial inhibition to enhanced infectivity (negative inhibition) in experiments on PBMC from different donor pools (Fig. 5 A and B). Similar data profiles, as well as intermediate forms, arise with PBMC from other donor pools. We have yet to determine whether the various curve shapes are donor-specific, and how they relate to variation in CCR5-expression levels; studies on cells from individual donors, rather than the pooled cells from 2 or 3 donors that we routinely use, will be necessary.
The difference between the plateaus of inhibition at the highest VVC concentrations for the sensitive and resistant viruses is much smaller in the TZM-bl cell assay than with PBMC (Fig. 5C). However, as with the PBMC-based data, the experimental datasets from TZM-bl cells can be closely mimicked by the nonlinear-regression-generated curves based on the model (Fig. 5 and Fig. S6). Thus, in both the experimental data and the theoretical curves, inhibition of the VVC-resistant virus is incomplete at the plateau, but the IC50 is lower than for the wild-type virus; these 2 apparently conflicting observations are otherwise hard to reconcile with each other.
Fitting the function derived from the model to the experimental PBMC data yields values for 3 parameters: the affinities of VVC for CCR5-A and CCR5-B and w, which reflects the relative amount of the form of CCR5 that is infection-permissive when ligated by VVC; also, w can be interpreted as expressing a variable capacity of a test virus to use these VVC-CCR5 complexes (SI Materials and Methods). For the PBMC-derived data, w is close to 0 for the wild-type S virus (0–0.0040; Table S2); for the resistant S + 3FP virus, it varies above and below 1 between experiments on cells from different donor pools; it is <1 (0.34 ± 0.029) for the data showing partial inhibition at the plateau, it approaches 1 (1.0 ± 0.037) when there was no inhibition at the highest VVC concentrations (Fig. 5A, Table S2), but it exceeds 1 (1.8 ± 0.059) when high concentrations of VVC enhance infection (Fig. 5B, Table S2). In each of the 3 cases, the estimated dissociation constant of VVC for CCR5-A was distinct from that for CCR5-B (9.6–50 and 0.30–1.6 nM, respectively, with ratios of 7.5–170; Table S2). The modeled Kd values for S on PBMC from 3 donor pools were in an intermediate range of 1.3–6.4 nM.
Modeling the data for S from the TZM-bl cell assay yielded a value of w close to 0 (w = 0.0095), similar to the one derived from PBMC with the same virus (w = 0.0040) (Table S2). The w value for infection of TZM-bl cells by the resistant S + 3FP virus was 7-fold higher (≈0.06) than for S on those cells, but it was lower than for S + 3FP on PBMC (0.34–1.8). A dissociation constant for VVC binding to CCR5-A on TZM-bl cells could not be precisely estimated, but it must exceed 25 nM; for CCR5-B on the same cells, it could be estimated to be 0.67 ± 0.045 nM. Therefore, the corresponding ratio of >37 may fall within the range derived from the PBMC experiments (7.5–170).
Discussion
Resistance to small molecule CCR5 inhibitors, exemplified by MVC and VVC, is now well documented in vitro, and has arisen in vivo (4, 6–9, 11). Cross-resistance within the class is usually observed (5, 6, 14), but sometimes not (4). Contrary to initial assumptions, the principal resistance pathway does not involve switching to CXCR4; the virus instead acquires the ability to recognize the inhibitor-CCR5 complex while retaining use of the free coreceptor (4, 5). Entry/replication of some resistant viruses can increase when an inhibitor is present, because they recognize the inhibitor-CCR5 complex more efficiently than free CCR5, and/or because more inhibitor-CCR5 complexes are available on the cell (5, 11). The resistant viruses replicate efficiently, and their phenotype is stable, at least in PBMC cultures, exemplified by the lack of reversion of both the CC101.19 (13) and the D1/85.16 isolate (Fig. S1) after prolonged culture in the absence of AD101 and VVC, respectively. Therefore, the altered Env configuration enabling D1/85.16 to use the VVC-CCR5 complex must also recognize free CCR5 approximately as efficiently as Env from the parental CC1/85 isolate. The same applies to the AD101-resistant isolate CC101.19 and clones (5, 6).
How the Env complex adjusts to use the inhibitor-CCR5 complex is under investigation. The generally accepted model of gp120-CCR5 interactions involves the crown of the V3 region contacting the second extracellular loop (ECL2), whereas the bridging sheet and more conserved residues in the V3 stem bind the tyrosine-sulfated amino-terminal domain [N terminus (Nt)] (1, 10, 16). The available evidence suggests that resistant viruses become less dependent on, or even independent of, the V3-ECL2 interaction (8, 17). The genetic pathway typically involves accumulating multiple sequence changes in V3 that, presumably, impede its binding to ECL2 (4, 6–9, 11). However, we described a VVC-resistant clone with no V3 sequence changes (14); its phenotype was comparable with the AD101-resistant (and VVC-cross resistant) viruses derived from the same parental isolate under similar experimental conditions (6, 12). Thus, there is a second genetic pathway to the same phenotype.
Here, we show that this alternative pathway not only does not involve V3, but that the critical changes do not even lie within gp120; instead, they are located within the FP of gp41. This finding was unexpected. The 3 FP changes are collectively compatible with fusion; the resistant viruses are replication-competent, as is the corresponding, inhibitor-sensitive JR-FL mutant. However, no naturally occurring viruses in the HIV Sequence Database contain all 3 changes, although some subtype C strains have 2 (G516V+M518V or M518V+F519I). Of note is that one of the FP changes, G516V, was recently shown to confer CXCR4 usage on a clonal R5 virus (18). Perhaps the coreceptor switching of that virus and the altered CCR5 interactions of the VVC-resistant CC1/85 variants involve similar mechanisms. However, the 3 FP changes combined did not enable CC1/85 variants to use CXCR4 in PBMC, and CXCR4 usage was not responsible for VVC resistance in TZM-bl cells (Fig. S2). We are now investigating whether any of the 3 FP changes individually (or in pairs) affect VVC sensitivity, fusion efficiency, and coreceptor usage. However, the effect of the FP changes is context-dependent. Thus, a JR-FL variant engineered to have the same FP sequence as resistant D1/85.16-derived variants remained VVC-sensitive (Fig. S3); likewise, the V3 changes that confer resistance on CC1/85 have no such effect on JR-FL.
Although the gp120 subunit is not responsible for CCR5 inhibitor resistance, gp120 sequence changes may influence the phenotype of D1/85.16 and some mutants derived from it. We are assessing whether compensatory changes elsewhere in Env influence the stability and fitness of the FP-engineered viruses; they may subtly alter how CCR5 is recognized, particularly at high inhibitor concentrations (compare S + 3FP and R/S + 3FP in Figs. 3 and 4; see Table 1).
How FP changes alter the gp120-CCR5 interaction remains to be understood, but it seems reasonable to assume they act at a distance; there are no grounds to believe that the FP contacts the coreceptor before or during fusion. When unligated by an inhibitor, CCR5 provides a stimulus in a chain of conformational changes in Env that culminate in membrane fusion. This stimulus is abrogated for wild-type Env, but not the resistant variant if an inhibitor is bound. The V3 sequence changes may facilitate an interaction between Env and the inhibitor-CCR5 complex by changing the normal requirements for both V3 and the bridging sheet to contact different regions of CCR5 (ECL2 and the Nt). In principle, the FP substitutions may act analogously, via allosteric effects on Env conformations that functionally mimic the consequences of the V3 changes. An alternative hypothesis is that the FP substitutions lower the threshold for fusion triggering, such that bridging-sheet interactions with the CCR5 Nt now suffice to drive the Env refolding that mediates fusion. Although structural information on the native Env trimer would clarify intersubunit effects, there is now considerable information about how gp41 sequence changes affect gp120 structure and function. The first study of how Env responds to a selection pressure, a serum from a HIV-1-infected individual, involved a point substitution in gp41 (A582T) that created a IIIB-variant resistant to neutralizing antibodies (NAbs) that specifically target CD4-binding site-associated gp120 epitopes (19, 20). Other influences of sequence changes in the gp41 ectodomain and cytoplasmic tail on neutralization by gp120 ligands are now known (21–23). The VVC- and AD101-resistant D1/85.16 and CC101.19 isolates are somewhat more NAb sensitive than their parental virus, CC1/85, further suggesting that the FP and V3 changes affect the overall geometry of the Env complex. The greatest difference in sensitivity between D1/85.16 and CC1/85, ≈58-fold, was to NAb 2G12 against a conformational epitope comprising gp120 N-linked glycans (24). How this difference arises is yet another unknown that will probably require Env trimer structural information. Alterations in gp41, including the cytoplasmic tail, can lead to CD4 independence and reduced CCR5 dependence (25), and secondary resistance substitutions affecting small molecule inhibitors of gp120-CD4 binding were also mapped to gp41 (26).
The manifestation of resistance to CCR5 inhibitors is assay-dependent; the relative efficiency with which the resistant virus uses the inhibitor-bound and inhibitor-free forms of CCR5, the viral inoculum size, and the CCR5 surface density are all relevant variables (4, 5, 7, 27). In PBMC, resistance usually appears as a rightwards shift in the inhibitor dose-response curve (IC50 increase), whereas in CCR5-expressing cell lines such as TZM-bl there is a plateau effect, and resistance is defined by MPI values <100% (4, 5, 7). In general, the engineered FP mutants behaved similarly to the naturally resistant virus, but with some informative exceptions. Thus, replication of the S/R, S + 3FP, and R/S + 3FP viruses in PBMC was partially inhibited (40–60%) by low VVC concentrations (≈0.3–2 nM), but enhanced by higher concentrations (up to 5 μM), and there was considerable variation in the extent of resistance among donor pools (Figs. 1, 3, and 5 A and B). Replication of the engineered viruses in TZM-bl cells was hypersensitive to low VVC concentrations, compared with the parental and naturally resistant clones, but there was a small yet consistent persistent fraction of viruses replicating in the presence of very high VVC concentrations (Fig. 4). The manifestation of resistance of some CXCR4-using viruses to the small molecule inhibitor AMD3100 has also been shown to involve plateau heights that vary with the cell type and among PBMC from different donors (28). The explanation may be similar to what we propose here for CCR5; thus, the derivation of analogous models could be informative.
Our model of CCR5-inhibitor resistance explains the complex curve shapes arising for the VVC-resistant clones in PBMC and TZM-bl cells. Its key feature is the existence of 2 CCR5 forms, CCR5-A and CCR5-B, which bind VVC and related inhibitors with low and high affinity (in reality, there may be >2 forms; see SI Materials and Methods and Fig. S5). The model also assumes that wild-type and resistant HIV-1 variants differentially use free and inhibitor-bound conformations of these CCR5 forms. By varying the proportions of CCR5-A and CCR5-B and their affinities for VVC, we can derive infection-inhibition curves that mimic data generated in both PBMC and TZM-bl cells. The similarities between the experimental results and the model curves support the central hypothesis that free and VVC-ligated forms of different CCR5 subpopulations, respectively, can mediate entry of the resistant S + 3FP variant, whereas the wild-type S virus can use both free CCR5 forms to various degrees. Thus, the model explains the distinct inhibition profiles of the 2 viruses on the 2 cell types.
How reasonable are the model and its underlying assumptions? Antibody-reactivity profiles show that multiple CCR5 conformations are present in proportions that vary between cell types (16). These different conformations may arise because CCR5 can be coupled intracellularly to G proteins, which affects the shape of the ligand-binding sites in the extracellular loops and Nt (29). Also, the CCR5 Nt is tyrosine-sulfated at 3 positions posttranslation, 2 of which (Tyr-10 and Tyr-14) are required for recognition by wild-type HIV-1 (30). Perhaps overexpressing CCR5 in engineered cell lines like TZM-bl and U87.CD4.CCR5 saturates the tyrosine sulfotransferases, creating partially sulfated CCR5 proteins that are antigenically and functionally distinct. The Tyr-sulfation requirements of the various resistant isolates remain to be investigated. Overall, the postulates that different forms of CCR5 exist in different proportions on different cell types and are recognized with different efficiencies by wild-type and resistant HIV-1 variants seem plausible. Quite what forms correspond to our theoretical CCR5-A and CCR5-B remains to be investigated experimentally; based on the curve shapes in Figs. 4 and 5, we argue that the CCR5-B, which has a high affinity for VVC and related inhibitors, will be overexpressed relative to CCR5-A in engineered cells like TZM-bl and, by extrapolation, U87.CD4.CCR5. Also, because CCR5-B may be used less efficiently by wild-type HIV-1 in those cells, it could perhaps be a variant that is under-sulfated on the Nt tyrosines. The relative usage of the CCR5 Nt and ECL2 does appear to alter between wild-type and resistant HIV-1 variants, the latter becoming more dependent on the Nt (8). However, why under-sulfation of the Nt would increase the affinity of CCR5 for small molecule inhibitors, as this interpretation of our model implies, remains to be understood. Perhaps intermolecular interactions involving CCR5 and/or other cellular proteins involve the Nt and indirectly modify the binding site for the small molecule inhibitors in the transmembrane helices. Overall, we hope that our experimental results, and the model they have helped generate, will drive new lines of research into how HIV-1 uses its coreceptors under different conditions, and into the general biology of GPCRs.
Materials and Methods
Viruses and Inhibitors.
The study viruses are listed in Table S1 (5, 6, 14). Infectious stocks were prepared by transient transfection of 293T cells with pNL4-3/env proviral plasmids by using Lipofectamine 2000 (Invitrogen), and titrated before testing. Full-length env genes were sequenced as described (12, 14), and aligned with MacVector 10.0.2. VVC (SCH-D, SCH-417690), and AD101 (SCH-350581) (12) were provided by Julie Strizki, AMD3100 by Gary Bridger.
Construction of Chimeric NL4-3/env Proviruses.
The env genes with interchanged gp120/gp41 subunits from viruses S and R were constructed by using the unique MfeI sites, sequenced, then subcloned into pNL4-3 to produce chimeric infectious viruses (6). Site-directed mutagenesis was performed with QuikChange II (Stratagene), using pBluescript KS(+) plasmids containing EcoRI/XhoI fragments that were then subcloned into pNL4-3.
Infection-Inhibition Assays.
Virus sensitivity to VVC or AD101 was assessed by using PBMC from 2 or 3 random blood donors, as described (6, 14, 27). TZM-bl cells were seeded at 1 × 104 per well in 96-well plates and allowed to adhere overnight; inhibitors were added for 1h before virus infection. After culture for 48h, the supernatant was removed, cells lysed, and luciferase expression quantified as RLU by using the Bright-Glo Luciferase Assay System (Promega). Background values from mock-infected cells, which were also cultured with each VVC or AD101 dose to control for cell viability, were subtracted. A 4-parameter sigmoidal function was fitted to the inhibition data by nonlinear-regression (Prism, Graphpad). In testing theoretical models, we fit different functions as described (SI Materials and Methods). HIV-1 replication (p24 production) in U87.CD4.CCR5 and U87.CD4.CXCR4 cells was measured as described (6, 12).
For modeling CCR5 inhibitor resistance, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Antonia Thomas, Nicole Labutong, and Samson Jacob for technical support; Min Lu, Reem Berro, Rogier Sanders, and Antu Dey for helpful discussions; and past contributions by Andre Marozsan, Pavel Pugach, Julie Strizki, and Shawn Kuhmann to work on VVC-resistant viruses. This work was supported by National Institutes of Health Grant R01 AI41420.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The env sequences of the D1/85.16-derived viruses reported in this paper have been deposited in the GenBank database (accession nos. FJ713453–FJ713458).
This article contains supporting information online at www.pnas.org/cgi/content/full/0811713106/DCSupplemental.
References
- 1.Kuhmann SE, Hartley O. Targeting chemokine receptors in HIV: A status report. Annu Rev Pharmacol Toxicol. 2008;48:425–461. doi: 10.1146/annurev.pharmtox.48.113006.094847. [DOI] [PubMed] [Google Scholar]
- 2.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors-central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 3.Westby M, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westby M, et al. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J Virol. 2007;81:2359–2371. doi: 10.1128/JVI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pugach P, et al. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhmann SE, et al. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78:2790–2807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogert RA, et al. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2–V5 domain of gp120. Virology. 2008;373:387–399. doi: 10.1016/j.virol.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Laakso MM, et al. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 2007;3:e117. doi: 10.1371/journal.ppat.0030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba M, et al. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob Agents Chemother. 2007;51:707–715. doi: 10.1128/AAC.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21:171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 11.Tsibris AM, et al. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J Virol. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trkola A, et al. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci USA. 2002;99:395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anastassopoulou CG, et al. Escape of HIV-1 from a small molecule CCR5 inhibitor is not associated with a fitness loss. PLoS Pathog. 2007;3:e79. doi: 10.1371/journal.ppat.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marozsan AJ, et al. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338:182–199. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 15.Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369:245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B, et al. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 17.Lin G, et al. Replication-competent variants of human immunodeficiency virus type 2 lacking the V3 loop exhibit resistance to chemokine receptor antagonists. J Virol. 2007;81:9956–9966. doi: 10.1128/JVI.00385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, et al. Coreceptor tropism can be influenced by amino acid substitutions in the gp41 transmembrane subunit of human immunodeficiency virus type 1 envelope protein. J Virol. 2008;82:5584–5593. doi: 10.1128/JVI.02676-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klasse PJ, et al. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(→Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology. 1993;196:332–337. doi: 10.1006/viro.1993.1484. [DOI] [PubMed] [Google Scholar]
- 20.Thali M, et al. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol. 1994;68:674–680. doi: 10.1128/jvi.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blish CA, Nguyen MA, Overbaugh J. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 2008;5:e9. doi: 10.1371/journal.pmed.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park EJ, Quinnan GV., Jr Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J Virol. 1999;73:5707–5713. doi: 10.1128/jvi.73.7.5707-5713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Back NK, et al. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J Virol. 1993;67:6897–6902. doi: 10.1128/jvi.67.11.6897-6902.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pugach P, Ketas TJ, Michael E, Moore JP. Neutralizing antibody and anti-retroviral drug sensitivities of HIV-1 isolates resistant to small molecule CCR5 inhibitors. Virology. 2008;377:401–407. doi: 10.1016/j.virol.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor BM, et al. An alteration of human immunodeficiency virus gp41 leads to reduced CCR5 dependence and CD4 independence. J Virol. 2008;82:5460–5471. doi: 10.1128/JVI.01049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin PF, et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci USA. 2003;100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ketas TJ, et al. Cell surface expression of CCR5 and other host factors influence the inhibition of HIV-1 infection of human lymphocytes by CCR5 ligands. Virology. 2007;364:281–290. doi: 10.1016/j.virol.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison JE, et al. Baseline Resistance of Primary Human Immunodeficiency Virus Type 1 Strains to the CXCR4 Inhibitor AMD3100. J Virol. 2008;82:11695–11704. doi: 10.1128/JVI.01303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenakin T. Ligand-selective receptor conformations revisited: The promise and the problem. Trends Pharmacol Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 30.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.