Abstract
Inflammation involves a coordinated, sequential, and self limiting sequence of events controlled by positive and negative regulatory mechanisms. Recent studies have shown that microRNAs (miRNAs), an evolutionarily conserved class of endogenous 22-nucleotide noncoding RNAs, contribute to the regulation of inflammation by repressing gene expression at the posttranscriptional level. In this study, we characterize the profile of miRNAs induced by LPS in human polymorphonuclear neutrophils (PMN) and monocytes. In particular, we identify miR-9 as the only miRNA (among 365 analyzed) up-regulated in both cell types after TLR4 activation. miR-9 is also induced by TLR2 and TLR7/8 agonists and by the proinflammatory cytokines TNF-α and IL-1β, but not by IFNγ. Among the 3 different genes encoding miR-9 precursors in humans, we show that LPS selectively induces the transcription of miR-9–1 located in the CROC4 locus, in a MyD88- and NF-κB-dependent manner. In PMN and monocytes, LPS regulates NFKB1 at both the transcriptional and posttranscriptional levels, and a conserved miR-9 seed sustained a miR-9-dependent inhibition of the NFKB1 transcript. Overall, these data suggest that TLR4-activated NF-κB rapidly increases the expression of miR-9 that operates a feedback control of the NF-κB-dependent responses by fine tuning the expression of a key member of the NF-κB family.
Keywords: inflammation, innate immunity, Toll-like receptors, cytokines, NFKB1
The innate immune response is the first line of defense against infectious agents and is mainly exerted by phagocytes, including polymorphonuclear neutrophils (PMN) and monocyte/macrophages. This response is triggered by the recognition of pathogen-associated molecular patterns of invading microorganisms by members of the Toll/IL-1 receptor (TLR) superfamily (1) among others. These receptors signal through similar intracellular pathways that start with the recruitment to the Toll-IL-1R (TIR) domain present in the receptor tail with 1 of 4 possible TIR domain-containing adaptor molecules. The combination of adaptor molecules involved not only depends upon the specific TLR engaged, but also defines the consequent cellular events. In particular, the MyD88 and TIR domain-containing adapter protein/MyD88 adapter-like protein (TIRAP/MAL) mediates the early NF-κB activation, while the TIR domain-containing adapter inducing IFNβ/TIR-containing adapter molecule-1 (TRIF/TICAM-1) and TRIF-related adapter molecule/TIR-containing adapter molecule-2 (TRAM/TICAM-2) mediate the delayed NF-κB and IFN-regulatory factor (IRF) 3 signals (2, 3). As examples, TLR4 induces proinflammatory cytokines via either a MyD88-dependent rapid activation of the transcription factor NF-κB or costimulatory and antiviral proteins through a more delayed activation of both NF-κB and IRF-3 mediated by TRIF. Conversely, TLR3 exclusively signals through the TRIF-dependent pathway and does not activate the MyD88-dependent pathway (2, 3). Importantly, a variety of extracellular and intracellular negative feedback pathways have evolved to prevent an inappropriate inflammatory response following activation of TLRs. These include the regulation of TLR expression, the production of molecules that compete with their ligand binding or signaling activities, and the generation of dominant negative splice variants or posttranslational modifications of signal transducers of the TLR signaling cascade (4, 5).
An emerging class of regulators of gene expression is represented by microRNAs (miRNAs), which act at the posttranscriptional level via an RNA interference mechanism (6, 7). miRNAs biogenesis involves the initial transcription by RNA polymerase II of primary miRNAs (pri-miRNA), which are subsequently cleaved by the RNase III enzyme Drosha and Dicer to produce the 21- to 23-nt double-stranded RNA duplexes (6). The mature miRNA guide strand is then loaded into the miRNA-induced silencing complex, where it guides the recognition and translational repression or degradation of target mRNAs (8). In mammals, a host of genes are processed to produce over 700 miRNA (miRNA registry at www.sanger.ac.uk/Software/Rfam/mirna), which have been implicated in a wide array of biological processes ranging from cellular development and differentiation to tumors (6, 7). Recently, activation of the innate immune response also has been associated with changes in the expression of selected miRNAs [namely miR-146 (9, 10), miR-155 (11, 12), miR-132 (10), and miR-125b (12)]. However, the ability of inflammatory ligands to modulate miRNA expression and, more importantly, the role of regulated miRNAs in the development of an adequate immune response are just beginning to be explored.
Herein, we report the profiles of miRNAs induced by inflammatory stimuli in human PMN and monocytes and identify miR-9 as a previously unrecognized LPS-responsive miRNA induced in a MyD88- and NF-κB-dependent manner in both cell types. We also show that miR-9 takes part of a regulatory circuitry controlling cell activation by means of inhibitory feedback loop acting at the level of NFKB1, a transcriptional regulator with a key role in the inflammatory response.
Results
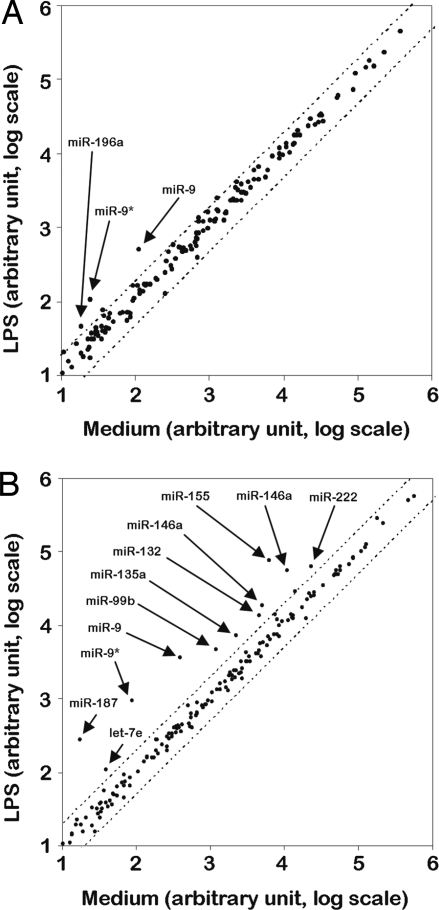
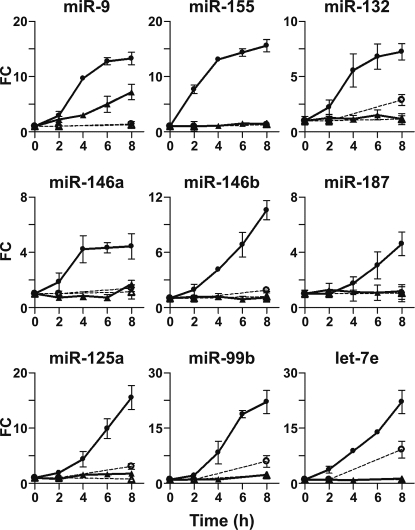
To identify miRNAs potentially involved in the responses of peripheral human PMN and monocytes to stimuli of bacterial origin, the miRNA pattern of expression was investigated in PMN and monocytes stimulated for 8 h with 100 ng/ml LPS using a TaqMan-based Low Density Array. As shown in Fig. 1, LPS induced an up-regulation of 12 miRNAs in PMN and/or monocytes while no miRNA was significantly down-regulated (Fig. 1). LPS-induced miRNAs identified in the array were evaluated in a time-course analysis by RT-qPCR (Fig. 2). Consistent with the array data, the expression of miR-155, miR-132, miR-146a, miR-146b, miR-187, miR-125a, miR-99b, and let-7e rapidly increased in LPS-treated monocytes but not in PMN (Fig. 2). Interestingly, both 3′-end (miR-9) and 5′-end (miR-9*) forms of miR-9 were the only miRNAs consistently induced by LPS in both PMN and monocytes, being already detectable after 2 h and steadily increasing over the time period assessed (Fig. 2 and supporting information (SI) Fig. S1A). Up-regulation of miR-9 expression in PMN and monocytes stimulated with LPS for 24 h was further confirmed in Northern blot analysis (Fig. S1B). Conversely, the induction of miR-222 and miR-196a observed in the array analysis was not confirmed by RT-qPCR analysis (not shown). miR-9 was then chosen for a more detailed analysis, given that it is the only miRNA up-regulated in response to LPS in both cell types and that it has not been previously reported to be involved in the inflammatory response.
Fig. 1.
miRNAs induced by LPS in PMN and monocytes. PMN (A) and autologous monocytes (B) were cultured for 8 h in medium alone or in the presence of 100 ng/ml LPS. The miRNA fraction was purified and changes in miRNA expression levels were determined using a micro fluidic card as described in Materials and Methods. Results are expressed as arbitrary units on a log scale using RNU44 as reference control. The mean values of 2 individual experiments performed are shown. Dotted lines represent the 2 and 0.5 boundary values for fold induction.
Fig. 2.
Kinetics of LPS-induced miRNAs in PMN and monocytes. PMN and monocytes were cultured for the indicated times in medium alone (- -▵- - PMN, - -○- - monocytes) or in the presence of 100 ng/ml LPS (—▴— PMN, —●— monocytes). miRNA fraction was purified and miR-9, miR-155, miR-132, miR-146a, miR-146b, miR-187, miR-125a, miR-99b, and let-7e expression was determined by RT-qPCR and normalized to the let-7a levels, as described in Materials and Methods. The results are expressed as fold change and are representative of 3 individual experiments.
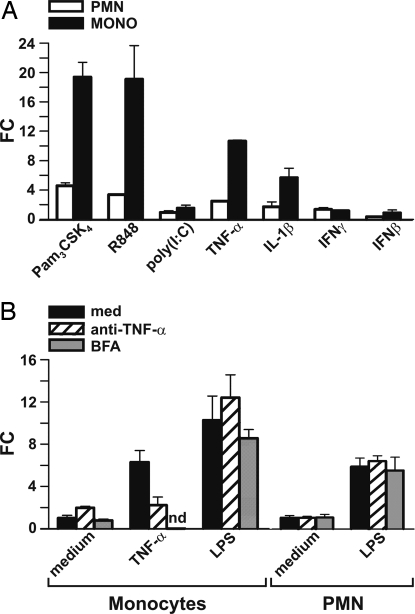
LPS triggers different patterns of responses in PMN and monocytes, partly because of the selective activation of the different MyD88- and TRIF-dependent signaling pathways downstream of the pattern-recognition receptor TLR4 (1). To investigate the requirement of MyD88 and/or TRIF adaptors in the induction of miR-9 expression by LPS and to evaluate miR-9 regulation by other TLRs, PMN and monocytes were stimulated with Pam3CSK4 (100 ng/ml), a synthetic lipoprotein agonist at TLR2 that selectively requires TIRAP/MyD88; Resiquimod (R848, 10 μM), a TRL7/8 ligand signaling through MyD88 only; or polyinosinic:polycytidylic acid [poly(I:C)] (50 μg/ml), a synthetic mimetic of viral double-stranded RNA (dsRNA) that interacts with endosomal TLR3 and utilizes TRIF-mediated signaling (1). As shown in Fig. 3A, activation of TLR2 and TLR7/8 resulted in up-regulation of miR-9 expression in both cell types, while that of TLR3 was ineffective. Conversely, poly(I:C) readily induced miR-155 in monocytes (Fig. S2), as previously reported in other cell types (11), demonstrating that the lack of miR-9 induction was not due to a general failure of monocytes to activate the TRIF-dependent pathway downstream TLR3. In agreement with the lack of TLR3 expression in human PMN (13), poly(I:C) had no effect on miR-9 expression in this cell type (Fig. 3A). Taken together, these data suggest that in human phagocytes, activation of the MyD88-dependent signaling pathway is necessary and sufficient to increase miR-9 expression in response to LPS and that no additional TRIF-dependent signals are required.
Fig. 3.
miR-9 is induced by MyD88-activating TLR agonists and proinflammatory cytokines. (A) PMN and monocytes were cultured for 8 h with 100 ng/ml LPS, 100 ng/ml Pam3CSK4, 10 μM R848 or 50 μg/ml poly(I:C), TNF-α, 20 ng/ml IL-1β, 1000 U/ml IFNγ, or 1000 U/ml IFNβ. miRNA fraction was extracted and analyzed for miR-9 expression by RT-qPCR. (B) monocytes and PMN were pretreated for 30 min with medium (black bars), 10 μg/ml anti-TNF-α MoAbs (hatched bars), or 5 μg/ml brefeldin A (gray bars) before stimulation with TNF-α or LPS. miRNA fraction was extracted after 8 h and analyzed for miR-9 expression by RT-qPCR. miRNA expression is depicted as fold change units after let-7a normalization. Data show one experiment representative of 3. nd: not determined.
To test whether cytokines involved in the response to bacterial and/or viral infection are also effective at inducing miR-9 expression, PMN and monocytes were stimulated with TNF-α (5 ng/ml), IL-1β (20 ng/ml), IFNγ (1000 U/ml), or IFNβ (1000 U/ml). The proinflammatory cytokines TNF-α and IL-1β increased miR-9 levels in both PMN and monocytes, while IFNγ and IFNβ were ineffective (Fig. 3A). Since miR-9 is up-regulated by both LPS and TNF-α, we tested whether TLR4 induction of miR-9 required a TNF-α autocrine signaling as previously reported for miR-155 (11). Anti TNFα MoAbs completely blocked miR-9 induction by TNF-α, but were ineffective when LPS was used (Fig. 3B), indicating that TNF-α is not involved in the induction of miR-9 by LPS. In addition, up-regulation of miR-9 expression was not modified by treatment with brefeldin A before LPS stimulation, ruling out the possible involvement of soluble mediators released in response to LPS for miR-9 up-regulation (Fig. 3B). Taken together, these data candidate miR-9 as a novel miRNA involved in the responses of human phagocytes to selected stimuli of bacterial origin or proinflammatory cytokines.
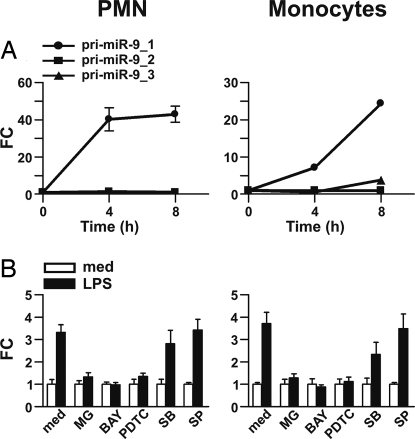
In both mouse and human genomes, miR-9 can be generated by processing of 3 different miR-9 primary transcripts encoded by distinct genes (C1orf61 for pri-miR-9–1, BC036480 for pri-miR-9–2, and CR612213 for pri-miR-9–3, respectively). LPS induced a time-dependent increase in pri-miR-9–1 levels and had no effect on the other 2 miR-9 primary transcripts, both in PMN and monocytes (Fig. 4A). The miR-9–1 primary transcript derives from the C1orf61 locus which encodes for CROC-4 protein, a transcriptional activator for the c-fos proto-oncogene (14). An EST database analysis revealed the existence of an internal product of the C1orf61 locus. Both transcriptional units (here called CROC-4a and CROC-4b: Fig. S3A) generate the miR-9–1 precursor and are activated by LPS in PMN and monocytes (Fig. S3B). Analysis of the C1orf61 locus with the transcription start sites predictor SwitchGear software (available at http://genome.ucsc.edu) supports the existence of an internal transcriptional unit. Inspection of the genomic sequence located 2 kb upstream of the predicted start sites of the 2 transcripts identified putative promoter regions with consensus binding sites for known LPS-sensitive transcription factors, including NF-κB (Fig. S3A). The observation that miR-9 induction by LPS depends on the activation of the MyD88 pathway and the identification of NF-κB consensus binding sites within the 2 putative pri-miR-9–1 promoters suggested that the miR-9 induction by LPS may result from the transcriptional activity of NF-κB. This hypothesis was confirmed by the suppressive effect on LPS-dependent miR-9 induction of NF-κB inhibitors (MG-132, BAY-117082 and PDTC) (Fig. 4B). In contrast, inhibitors of p38 (SB-203580) and JNK (SP-600125) were ineffective (Fig. 4B). Collectively, these data demonstrate that in PMN and monocytes inflammatory stimuli sustain the NF-κB-dependent transactivation of C1orf61 locus and consequent production of pri-miR-9–1.
Fig. 4.
LPS up-regulates pri-miR-9–1 in a NF-κB-dependent manner. (A) PMN and monocytes were cultured in the presence or absence of LPS for the indicated time; total RNA was extracted and pri-miR-9–1 (—●—), pri-miR-9–2 (—▴—), and pri-miR-9–3 (—■—) were analyzed by RT-qPCR and normalized to the 18S RNA as described in Materials and Methods. Results show that only pri-miR-9–1, but not pri-miR-9–2 or pri-miR-9–3, was induced by LPS. (B) PMN and monocytes were pretreated for 30 min with medium, 10 μM MG-132, 10 μM BAY-117082, 300 μM PDTC, 20 μM SP-600125, or 10 μM SB-203580 and subsequently cultured for 8 h with or without LPS. miR-9 expression levels were determined by RT-qPCR and expressed as fold change after let-7a normalization. Data show 1 experiment representative of at least 2 for each panel.
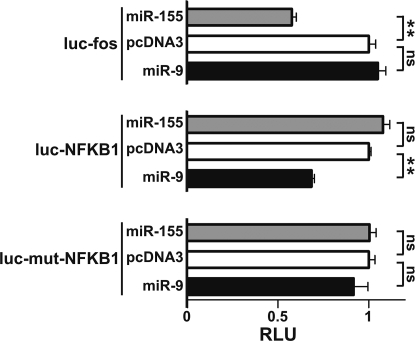
To gain insight on the biological relevance of miR-9 induction under inflammatory conditions, we searched for predicted miR-9 targets, focusing our attention on regulators of transcription, which have been frequently shown to be preferential miRNA targets (15). In agreement with this, the public database of animal miRNA miRGen (available at http://www.diana.pcbi.upenn.edu/miRGen/v3/miRGen.html), which integrates analysis from PicTar (16), MiRanda (17), and TargetScan (18), predicted among high score miR-9 targets the transcriptional regulators Onecut2 and PRDM1/Blimp-1, which have been previously validated as miR-9 targets but are not expressed in PMN and monocytes (data not shown). A miR-9 seed was also predicted in one of few highly conserved regions present in the 3′-UTR of the NFKB1 gene (Fig. S4A). Interestingly, the potential regulatory loop between NF-κB and miR-9 was also confirmed by the miPromotor software. Thus, to test whether miR-9 postranscriptionally affects NFKB1, reporter construct containing the renilla luciferase gene fused to the NFKB1 3′-UTR (luc-NFKB1) was transiently transfected in HEK-293 cells together with expression plasmids encoding miR-9. As shown in Fig. 5, miR-9 significantly reduced luc-NFKB1 luciferase activity, and the introduction of point mutations in the miR-9 seed in the 3′-UTR of NFKB1 (luc-mut-NFKB1) reverted the inhibitory activity of miR-9, demonstrating that the NFKB1 3′-UTR contains an active seed of miR-9. The specificity of miR-9 was also demonstrated by the lack of effect on a c-fos 3′-UTR reporter construct (luc-fos), which does not contain a miR-9 seed. Conversely, the c-fos 3′-UTR presents a miR-155 seed (Fig. S4B) and was significantly inhibited by miR-155, in agreement with previous reports (19) (Fig. 5).
Fig. 5.
The NFKB1 gene is a molecular target of miR-9. The indicated luciferase constructs (luc vectors) were cotransfected with expression vectors encoding miR-155 (gray columns), miR-9 (black columns), or the pcDNA3 empty vector (white columns). Results are expressed as mean (± SD, n = 3) of the ratio between renilla luciferase and firefly control luciferase activities (RLU), adjusted to 1. **: P < 0.01; ns: P > 0.05.
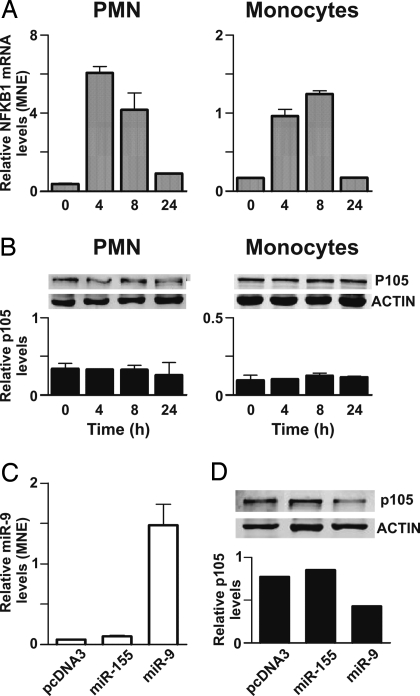
Finally, we analyzed the expression profile of NFKB1 in PMN and monocytes stimulated with LPS at the transcript and protein levels (Fig. 6). Interestingly, LPS rapidly induced a consistent increase in NFKB1 transcripts in PMN and monocytes (Fig. 6A), but the transcript up-regulation was not paralleled by a comparable increase of the corresponding protein, which showed constant expression levels (Fig. 6B). These results indicate that NFKB1 is subjected to a LPS-dependent regulation at the transcriptional level and suggest that a second regulatory mechanism, acting at the posttranscriptional level, is also operative. In agreement with this finding, monocytes overexpressing miR-9 (Fig. 6C) showed a reduced expression of the endogenous NFKB1/p105 protein (Fig. 6D). Conversely, transfection of monocytes with either the pcDNA3 empty vector or with the miR-155-encoding vector (Fig. 6C) did not alter the levels of expression of the endogenous NFKB1/p105 protein (Fig. 6D), confirming the specificity of the effect of miR-9.
Fig. 6.
Analysis of the endogenous miR-9 target: NFKB1 mRNA and protein expression. (A) PMN and monocytes were cultured in the presence of LPS for the indicated times. Total RNA was purified and used to assay NFKB1 mRNA expression by RT-qPCR, as described in Materials and Methods. Relative NFKB1 gene expression is depicted as MNE units after GAPDH normalization. Data reported are representative of 3 independent experiments. (B) Whole-cell extracts (20 μg) were usually loaded on gels and immunoblots were performed by simultaneously using Abs specific for NFKB1 and Abs specific for actin, followed by incubation with Alexa Fluor-680 goat anti-rabbit Abs. One experiment representative of 3 is shown. The relative NFKB1/p105 levels (± SD, n = 3), quantified as described in SI Materials and Methods, are reported below each panel. (C) 6 × 106 monocytes were transfected with 5 μg of pcDNA3 empty vector, miR-155-encoding vector, or miR-9-encoding vector as described in Material and Methods. 48 hours posttransfection total RNA was purified and analyzed for miR-9 expression by RT-qPCR. Relative miR-9 expression is represented as MNE units after let-7a normalization. (D) Fifteen micrograms of whole cell extracts, purified from transfected monocytes cultured in the same conditions as in (C), were usually loaded on gels and NFKB1/p105 protein was detected as described above. The relative NFKB1/p105 levels, normalized for the total actin, are reported below the Western blot. (C and D) One experiment representative of 2.
Discussion
In the last few years, miRNAs have emerged as a class of gene expression regulators involved in a variety of biological processes (7, 20), and recent evidence indicates that they also play a major role in immunity (21). In particular, miRNAs have been involved in myeloid and lymphoid differentiation (22–25), as well as in the induction of an appropriate adaptive immune response (25–27). A rapid increase in the expression of selected miRNAs has also been recently observed during the activation of an innate immune response (10–12), but the relevance of miRNAs in this process is just beginning to be explored.
In this study, we investigated the potential involvement of miRNAs in the regulation of innate immune response by evaluating the profile of miRNAs expressed in human PMN and monocytes after TLR engagement or cytokine activation. An RT-qPCR-based screening on human monocytes exposed to LPS not only confirmed the induction of miRNAs previously described in monocytic cell lines or mouse macrophages, namely of miR-146 (10), miR-155 (11, 12), and miR-132 (10), but also uncovered the induction of previously unrecognized miRNAs, including miR-187, the miR-125a/miR-99b/let-7e cluster, and miR-9/9*. Interestingly, out of the 365 tested, miR-9/9* was the only miRNA induced by LPS also in human PMN.
Initially identified as a brain-specific miRNA, miR-9 has been implicated in mammalian neuronal development and function, notably by inhibiting expression of antineurogenic transcription factor-encoding genes (28, 29). Outside the central nervous system, miR-9 was subsequently found to play a critical role in the control of the secretory function of insulin-producing cells by maintaining appropriate levels of the transcription factor Onecut2 (30), and more recently it was shown to down-regulate the transcription factor PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells, thus interfering with normal B-cell terminal differentiation and contributing to the pathogenesis of Hodgkin lymphoma (31). To our knowledge, the identification of miR-9 as an LPS-responsive miRNA in primary human PMN and monocytes represents the first evidence linking miR-9 to the innate immune response.
Several lines of evidence support the notion that miR-9 is directly induced by LPS via the MyD88/NF-κB-dependent pathway. First, LPS efficiently up-regulated miR-9 expression in human PMN, in which TRL4 engagement does not activate the TRIF-dependent pathway (32). Second, activation of TLR3, the only TLR known to transduce its signal independently from MyD88 (1), does not modulate miR-9 expression. This observation supports the conclusion that in monocytes the induction of miR-9 proceeds in a MyD88-dependent manner, even though in these cells both MyD88- and TRIF-dependent signaling pathways can be simultaneously activated downstream TLR4. By contrast, activation of TRIF-dependent pathway downstream of TLR3 is sufficient to induce miR-155 expression in monocytes as previously reported (11). Finally, activation of TLR2 and TLR7/8, known to engage only the MyD88-dependent pathway, led to increased miR-9 expression in both PMN and monocytes. Taken together, these data rule out any contribution of the TRIF-dependent pathway downstream of TLR4 to LPS-induced miR-9 expression. TLR4 activation also leads to the production of proinflammatory cytokines, such as TNFα and IL-1β, in both PMN and monocytes and to the production of antiviral type I IFN in monocytes. In turn, these mediators can signal through their receptor in an autocrine fashion. However, we provide demonstrations indicating that induction of miR-9 by LPS occurs in a direct manner, without the involvement of endogenous mediators. First, activation of PMN and monocytes with IFNβ does not promote miR-9 expression. Secondly, even though TNFα is able to induce miR-9 expression, its presence is not required for TLR4-mediated production of miR-9 because incubation of PMN and monocytes with anti-TNFα antibodies before LPS stimulation did not suppress the up-regulation of miR-9 expression. Finally, inhibition of protein secretion by brefeldin A does not impair LPS-induced miR-9 up-regulation, ruling out the involvement of any LPS-induced endogenous mediator.
In the human genome, 3 distinct genes encode different pri-miR-9 leading to the generation of identical mature miR-9. Notwithstanding, quantitative analysis of the levels of expression of each pri-miR-9 demonstrated that miR-9 accumulation in response to LPS entirely derived from the pri-miR-9–1. A detailed analysis of EST database revealed the existence in the C1orf61 locus of 2 transcriptional units regulated by independent promoters. Both transcriptional units were activated by LPS, and inspection of the upstream promoter regions of miR-9–1, together with the analysis of miR-9 induction in the presence of different signaling pathway inhibitors, showed that NF-κB activation is involved in the induction of this miRNA.
One model for miRNA-based regulation of gene expression envisages a global effect on the cell transcriptome mediated by a direct effect of the miRNA on a large number of target transcripts, which are diminished to negligible levels. In agreement with this model, an overrepresentation of the seeds of miRNAs induced in a defined experimental condition in the 3′-UTR of genes down-regulated in the same condition has been demonstrated, as in the case of the miR-155 seed in LPS-down-regulated genes (26). An alternative mechanism, also sustained by the frequent observation of a complex cross-talk between miRNAs and transcription factors (19, 22, 24, 26), foresees an indirect effect of miRNAs on the cell transcriptome mediated by the control of transcripts encoding for key transcription regulators. As this mechanism has also been previously reported for miR-9 (30, 31), among predicted miR-9 targets we focused our attention on genes directly involved in the control of transcription under inflammatory conditions. Cotransfection experiments of miR-9 with 3′UTR luciferase reporters identified NFKB1 as miR-9 target. Interestingly, the nfkb1 gene encodes 2 functional proteins (p50 and its precursor p105) with distinct biological activities during LPS responses (33). The active nfkb1 gene product (p50) can form heterodimers with other NF-κB subunits, which function as transcriptional activators. Alternatively, p50 can form homodimers that have been shown to fulfill an anti-inflammatory role by attenuating transcription of proinflammatory cytokines and by activating IL-10 expression (34, 35). Notably, the relative amount of each of the NF-κB family members may affect the outcome of the innate immune response (34, 35). Unbalanced expression of p50 has been observed under pathological conditions in which negative pathways of regulation prevail such as LPS tolerance, chronic inflammatory conditions, and cancer (36, 37). Analysis of the endogenous levels of this miR-9 target showed that NFKB1 mRNA rapidly and transiently increased during the initial hours of the LPS response, while the NFKB1/p105 protein levels remained constant for up to 24 h. This behavior perfectly fits the “micromanaging model,” according to which miRNAs contribute to maintain the optimal level of expression of some genes, particularly regulatory genes, that might have a narrow window of optimal expression (20). Consistent with this hypothesis, we found that miR-9-overexpressing monocytes display decreased NFKB1/p105 levels. Conversely, variations in NFKB1/p105 expression were not observed after transfecting miR-155, indicating that this effect is specific for miR-9. Since NF-κB is a key regulator of inflammation, the NFKB1 levels are likely to be a strictly controlled and timely regulated event of relevance for the proper progression of the inflammatory response. On the basis of our observations it is tempting to propose a model in which the parallel NF-κB-dependent induction of NFKB1 and miR-9 provides a mean to smooth out the fluctuations in gene expression and fine tune the synthesis of this key transcription factor, thus allowing the proinflammatory phase of the LPS response to correctly proceed. It will be important to assess how pathological conditions in which regulatory circuits of inflammation prevail, such as the systemic anti-inflammatory response syndrome associated with LPS tolerance and cancer, affect the miR-9 regulatory axis.
In summary, the results reported here extend to freshly isolated human monocytes and PMN previous observations on the induction of miRNAs by LPS in mouse macrophages and cell lines. In addition to the classic miR-155 and miR-146, a new set of miRNAs (miR-9, miR-187, miR-125a, miR-99b, and let-7e) were found to be LPS-responsive miRNAs in human monocytes. Among these, only miR-9 was also induced in neutrophils. Induction of miR-9 was also mediated by the proinflammatory cytokines IL-1β and TNF-α, but not by IFNγ. Interestingly, NFKB1/p105/p50 was identified in silico and experimentally as a miR-9 target. Given the regulatory function of p50 homodimers these results suggest a model whereby induction of miR-9 acts a tuning mechanism to prevent negative regulation by p50 homodimers as occurs in monocytes in systemic anti-inflammatory response syndrome and in cancer.
Materials and Methods
Materials.
The detailed list of the materials is provided in SI Materials and Methods.
Cell Purification and Culture.
Human PMN and monocytes were purified and cultured as described in SI Materials and Methods. The HEK293 cell line was grown in DMEM (Cambrex) supplemented with 10% FCS (Euroclone), 100 U/ml penicillin/streptomycin (Cambrex), and 2 mM l-glutammine (Cambrex).
Quantification of miRNAs Expression Level.
PMN and monocytes were stimulated with 100 ng/ml LPS for 8 h and the RNA fraction that is highly enriched for small RNA species (≤ 200 bp) was isolated by using the mirVana isolation kit (Ambion, Applied Biosystems), according to the manufacturer's protocol. The small RNA fractions were reverse transcribed and the analysis of the expression level of 365 miRNA was performed using a TaqMan-based Low Density Array. Details on reverse transcription as well as TaqMan Array are presented in SI Materials and Methods. Experimental data were then analyzed by SDS2.2.2 software and the relative miRNA expression values were calculated using RNU44 as endogenous control (38). miRNAs with a threshold cycle <33 that showed a fold change >2 or <0.5 in samples treated with LPS as compared to control samples were considered as differentially expressed.
Real Time RT-PCR (RT-qPCR).
miRNAs differentially expressed were validated using individual TaqMan miRNA Assay (Applied Biosystems) as described in details in SI Materials and Methods. The expression of miR-9 precursors (pri-miR-9–1, pri-miR-9–2 and pri-miR-9–3), CROC-4a, CROC-4b and NFKB1 was quantified by RT-qPCR (see SI Materials and Methods for details). The primers and probes used are described in Table S1.
Northern Blot.
Small RNA fraction was purified from PMN and monocytes and processed for Northern blot analysis as described (39) and detailed in SI Materials and Methods.
Constructs Generation and Luciferase Reporter Assay.
Generation of miR-9-, miR-155-encoding vectors, luc-fos, luc-NFKB1 and luc-mut-NFKB1 together with the luciferase reporter assay are described in SI Materials and Methods. The oligonucleotides used to generate the constructs are listed in Table S1.
Monocytes Transfection.
Freshly purified monocytes (6 × 106) were transfected with 5 μg of plasmid DNA (pcDNA3 empty vector, pcDNA3-miR-155, or pcDNA3-miR-9) using the Amaxa Nuclofector and the Human Monocyte Nucleofector kit (Amaxa), according to the manufacturer's protocol. Cells were then cultured and processed as detailed in SI Materials and Methods.
Western Blot.
Preparation of cell lysates and Western blot analysis were conducted as previously described (40) and described in details in SI Materials and Methods.
Statistical Analysis.
Data are expressed as means ± SD. Statistical changes in luciferase expression were determined using the one-way ANOVA with α set to 0.05 according to the Newman-Keuls test.
Supplementary Material
Acknowledgments.
This work was supported by grants from Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN and FIRB projects), the Fondazione Cariverona, the University of Verona (Joint Project grant), the Fondazione Cariplo (NOBEL project), and the Programma Straordinario di Ricerca Oncologica 2006 from Alleanza Contro il Cancro and Istituto Superiore di Sanità (RNBIO project). The experimental work was conducted in part in the context and with the support of the Fondazione Humanitas per la Ricerca (Rozzano, Milan, Italy). The generous contribution of the Italian Association for Cancer Research is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810909106/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill LA. How Toll-like receptors signal: What we know and what we don't know. Curr Opin Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 5.Mantovani A, Locati M, Polentarutti N, Vecchi A, Garlanda C. Extracellular and intracellular decoys in the tuning of inflammatory cytokines and Toll-like receptors: The new entry TIR8/SIGIRR. J Leukoc Biol. 2004;75:738–742. doi: 10.1189/jlb.1003473. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Perry MM, et al. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 13.Tamassia N, et al. Activation of an immunoregulatory and antiviral gene expression program in poly(I:C)-transfected human neutrophils. J Immunol. 2008;181:6563–6573. doi: 10.4049/jimmunol.181.9.6563. [DOI] [PubMed] [Google Scholar]
- 14.Jeffrey PL, et al. CROC-4: A novel brain specific transcriptional activator of c-fos expressed from proliferation through to maturation of multiple neuronal cell types. Mol Cell Neurosci. 2000;16:185–196. doi: 10.1006/mcne.2000.0866. [DOI] [PubMed] [Google Scholar]
- 15.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 17.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 19.Gottwein EN, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 21.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: Tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Fontana L, et al. MicroRNAs 17–5p-20a–106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 24.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 25.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 28.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leucht C, et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 30.Plaisance V, et al. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 31.Nie K, et al. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamassia N, et al. The MyD88-independent pathway is not mobilized in human neutrophils stimulated via TLR4. J Immunol. 2007;178:7344–7356. doi: 10.4049/jimmunol.178.11.7344. [DOI] [PubMed] [Google Scholar]
- 33.Pereira SG, Oakley F. Nuclear factor-kappaB1: Regulation and function. Int J Biochem Cell Biol. 2008;40:1425–1430. doi: 10.1016/j.biocel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Cao S, Zhang X, Edwards JP, Mosser DM. NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 2006;281:26041–26050. doi: 10.1074/jbc.M602222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 36.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 37.Saccani A, et al. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Varallyay E, Burgyan J, Havelda Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat Protoc. 2008;3:190–196. doi: 10.1038/nprot.2007.528. [DOI] [PubMed] [Google Scholar]
- 40.Crepaldi L, et al. Up-regulation of IL-10R1 expression is required to render human neutrophils fully responsive to IL-10. J Immunol. 2001;167:2312–2322. doi: 10.4049/jimmunol.167.4.2312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.