Abstract
Arsenic is the most common toxic substance in the environment, ranking first on the Superfund list of hazardous substances. It is introduced primarily from geochemical sources and is acted on biologically, creating an arsenic biogeocycle. Geothermal environments are known for their elevated arsenic content and thus provide an excellent setting in which to study microbial redox transformations of arsenic. To date, most studies of microbial communities in geothermal environments have focused on Bacteria and Archaea, with little attention to eukaryotic microorganisms. Here, we show the potential of an extremophilic eukaryotic alga of the order Cyanidiales to influence arsenic cycling at elevated temperatures. Cyanidioschyzon sp. isolate 5508 oxidized arsenite [As(III)] to arsenate [As(V)], reduced As(V) to As(III), and methylated As(III) to form trimethylarsine oxide (TMAO) and dimethylarsenate [DMAs(V)]. Two arsenic methyltransferase genes, CmarsM7 and CmarsM8, were cloned from this organism and demonstrated to confer resistance to As(III) in an arsenite hypersensitive strain of Escherichia coli. The 2 recombinant CmArsMs were purified and shown to transform As(III) into monomethylarsenite, DMAs(V), TMAO, and trimethylarsine gas, with a Topt of 60–70 °C. These studies illustrate the importance of eukaryotic microorganisms to the biogeochemical cycling of arsenic in geothermal systems, offer a molecular explanation for how these algae tolerate arsenic in their environment, and provide the characterization of algal methyltransferases.
Keywords: arsenic detoxification, arsenic methylation, As(III) S-adenosylmethyltransferase, thermophile
Arsenic is the most common toxic substance in the environment, ranking first on the Superfund list of hazardous substances (http://www.atsdr.cdc.gov/cercla/07list.html). It is introduced to the environment primarily from geologic sources and is acted on biologically, creating an arsenic biogeocycle (1). Geothermal environments are well known for their elevated arsenic content (2) and thus provide an excellent setting in which to study microbe–arsenic interactions. Thus far, studies conducted in the Alvord Desert Basin (3), Mono Lake (4, 5), and Yellowstone National Park (YNP) (6–9) have focused on microbial arsenite [As(III)] oxidation. However, studies aimed at identifying the organisms participating in these and other As transformations have focused almost entirely on microorganisms belonging to the domains Archaea and Bacteria (3, 9–12). By contrast, comparatively little attention has been paid to the Eukarya that inhabit these extreme environments, much less their potential contribution to biogeochemical cycles in these extreme habitats.
The structurally simple unicellular eukaryotic red algae Cyanidioschyzon, Cyanidium, and Galdieria (order Cyanidiales) are acidophilic, moderate thermophiles that visually dominate the biomass of microbial communities inhabiting the cooler (38–57 °C) reaches of outflow channels of acidic (pH 0.2–3.5) geothermal features in YNP (13) (Fig. 1) and are the only identified photoautotrophs in thermo-acidic environments (14). Variations in 18S rRNA and microsatellite markers (13, 15) and rbcL (16) genes cloned from geothermal environments suggest that significant population-level diversity occurs within the Cyanidiales, and recent cultivation efforts have uncovered interesting evidence of biogeographical groups in New Zealand, Japan, and YNP (15). Although these primitive plants are commonly observed growing in high arsenic environments [e.g., adjacent to realgar (AsS); Fig. 1C], little is known about how they tolerate arsenic or about their potential contribution to the biogeocycling of this environmental toxicant. Algae phylogenetically related to Cyanidioschyzon merolae (18S rRNA, 99%; rbcL, 99%) have been isolated from various acidic geothermal locations in YNP (13, 15), and a representative Cyanidioschyzon strain that oxidizes As(III) to As(V) in pure culture has been identified (13). In this article, we show that this isolate oxidizes As(III), reduces As(V), and methylates arsenic, converting As(III) to several organic forms. As part of these investigations, we also characterize the methyltransferases involved and offer a molecular explanation for how these algae tolerate arsenic in their environment and provide the characterization of algal methyltransferases.
Fig. 1.
Cyanidiales mats dominate the microbial biomass in cooler reaches of acidic geothermal outflow channels in YNP. The images were taken on September 15, 2008 (A and B) and October 30, 2008 (C). (A) East Fork, Tantalus Creek drains the Norris Geyser Basin. The algal mat is recovering from UV-associated mat decline (15). (B) Nymph Creek, located in the Norris-Mammoth corridor, is heavily shaded and does not experience mat decline. (C) Cyanidiales algae associated with realgar-like mineral phases and geothermal-derived waters containing 0.76 mM total arsenic. The orange-yellow deposits were determined by scanning electron microscope energy-dispersive X-ray spectroscopy to be realgar-like, with an As/S ratio range of 1.1 to 1.3 (Fig. S1).
Results
Bioassays.
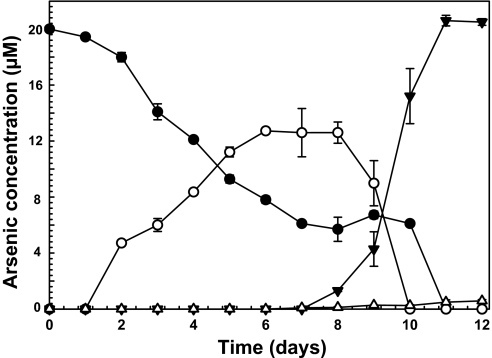
Assays with pure cultures of strain 5508 were quite reproducible. There was an initial period of As(III) oxidation to As(V) (Fig. 2), followed by a rapid decline in As(V) levels concomitant with accumulation of trimethylarsine oxide (TMAO) (Fig. 2). Although trimethylarsine [TMAs(III)] gas might be expected to be the final product, only minor amounts were detected by GC-MS analysis of the headspace gases. Because the algal cultures were sealed to monitor gas production, photosynthesis would fill the headspace with oxygen, which spontaneously oxidizes TMAs(III) to TMAO (17), so it is not surprising that TMAO was the major methylated species accumulating in the algal culture.
Fig. 2.
Arsenic transformations by Cyanidioschyzon sp. isolate 5508. Algal cultures were incubated with 20 μM arsenite for the indicated times, and the products were analyzed as described in Methods: ●, As(III); ○, As(V); ▼, TMAO; △, DMAs(V). The data represent the average of 2 replicate cultures. The experiment was repeated 4 times with equivalent results.
Cloning of the arsM Genes.
PCR cloning using total DNA extracts from the alga yielded 2 arsM genes, CmarsM7 (GenBank accession no. FJ476310) and CmarsM8 (GenBank accession no. FJ463403), with each encoding proteins of 400 residues that differ at 3 residues: K279R, G379D, and A383E (Fig. S2). Sequencing of multiple clones suggested the sequence deviations were not caused by PCR or sequencing errors. CmArsM7 and CmArsM8 are 99.5% identical to the putative product of gene CMT 024C (GenBank accession no. AP006502), annotated as an arsenic methyltransferase in C. merolae strain 10D chromosome 20, and 89.7–90.0% identical to CME 010C (GenBank accession no. AP006487), annotated as a probable arsenic methyltransferase in chromosome 5. The 2 CmArsMs share 48% identity and 66% similarity with a predicted protein in the marine eukaryotic alga Ostreococcus lucimarinus (GenBank accession no. ABP00263.1), 34% identity, 56% similarity (across 180 aa) to an annotated arsenic methyltransferase in Aspergillus fumigatus (GenBank accession no. XM_748062), and to the AS3MT arsenic methyltransferases of rat and human (44% identity, 60% similarity to ArsM7) (18). They also exhibit significant, but lower, similarity to the prokaryotic Rhodopseudomonas palustris ArsM (38% identity, 51% similarity to CmArsM7) (19). The greatest similarity is within the N-terminal regions of the predicted proteins. There is little similarity in the C-terminal regions except for the presence of 1 or 2 cysteine pairs at the C terminus of each. With glycine residues on each side, the cysteine pairs likely stick out from the surface of the protein. Vicinal cysteines are known to form strong As(III) binding sites, so these might serve as the initial As(III) binding site, similar to the cysteine-rich N termini of heavy metal translocating P-type ATPases (20).
CmArsM Enzymes Function to Detoxify As(III).
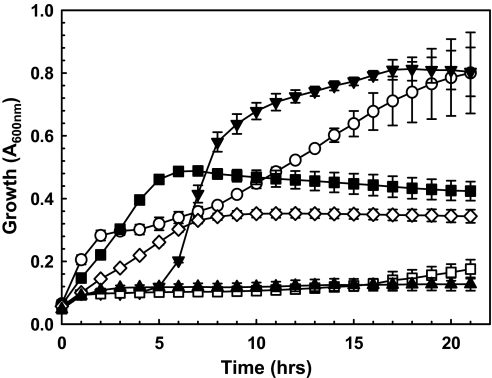
Fewer than 0.5% of the nuclear genes in C. merolae contain introns (21), and thus expression studies with cloned genes are a straightforward exercise. The CmarsM7 and CmarsM8 genes were cloned into an Escherichia coli vector and expressed in E. coli strain AW3110, which has no orthologous arsM gene and is As(III)-hypersensitive resulting from the deletion of the chromosomal arsRBC operon (22). Cells expressing either CmarsM gene could grow in As(III) concentrations as high as 0.10 mM (Fig. 3 shows results with CmarsM7), demonstrating the ability of the gene products to confer arsenic tolerance, consistent with the idea that these enzymes have a physiological role in arsenic detoxification.
Fig. 3.
Expression of algal CmarsM7 confers arsenite resistance in E. coli. Cultures of E. coli ΔarsRBC bearing the indicated plasmids were assayed with the indicated concentrations of sodium arsenite. ▼, vector plasmid pET28a+, no arsenite; ○, pET28a-alg-arsM7, no arsenite; □, vector plasmid pET28a+, 50 μM As(III); ■, pET28a-alg-arsM7, 50 μM As(III); ▲, vector plasmid pET28a+, 100 μM As(III); ◇, pET28a-alg-arsM7, 100 μM As(III). The data represent the average of 3 replicate cultures.
Reaction Products of CmArsM.
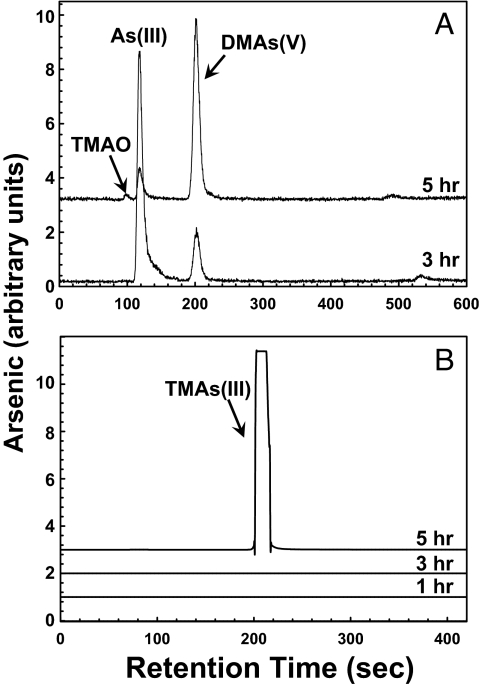
Analysis of the aqueous fraction of sealed AW3110 cultures expressing CmarsM7 (or CmarsM8) revealed formation of dimethylarsenate [DMAs(V)] and small amounts of TMAO (Fig. 4 and Table S1). Cold-trapping gas chromatography coupled with inductively coupled plasma MS (ICP-MS) was used to quantify and speciate the gaseous products. The major product was determined to be TMAs(III). At 3 h, DMAs(V) began to accumulate in the medium, and at 5 h the amount of DMAs(V) in the medium increased, with a concomitant increase in TMAs(III) gas, consistent with DMAs(III) being an intermediate in the pathway of TMAs(III) production as proposed by Challenger (23), who suggested an alternating series of oxidative methylations followed by reductions of As(V) to As(III). Presumably other intermediates in the Challenger pathway such as DMAs(III) also form transiently. Approximately 5% of the arsenic was volatilized after 5 h in vivo at 37 °C, the temperature optimum for growth of E. coli. The amount of both TMAO and TMAs(III) gas produced increased with higher concentrations of medium As(III). The accumulation of volatile arsenic in E. coli heterologously expressing the CmarsM7 gene reflects stability of TMAs(III) in the sealed cultures of the oxygen-consuming E. coli cultures, which rapidly become anaerobic.
Fig. 4.
Biomethylation of arsenite in E. coli expressing algal CmarsM7. (A) HPLC-ICP-MS analysis of the soluble As(III) transformation products generated by E. coli ΔarsRBC expressing pET28a-alg-arsM7 after 3 and 5 h of incubation at 37 °C. (B) The volatile products produced after 1, 3, and 5 h were analyzed from trapped headspace gas. Curves are displaced on the y axis for clarity.
Enzymatic Mechanism.
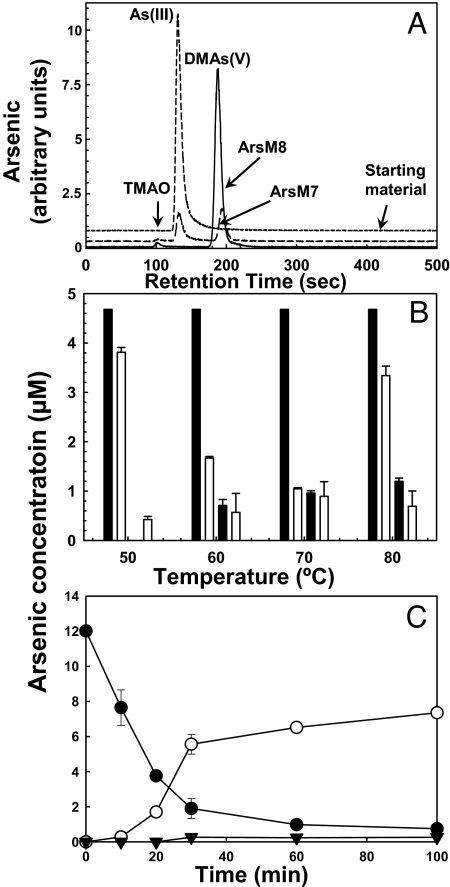
To elucidate the mechanism of arsenic methylation by the algal enzymes, recombinant CmArsM7 and CmArsM8 were purified from E. coli cytosol and assayed for As(III)-S-adenosylmethionine methyltransferase activity. At 37 °C, both enzymes converted As(III) to DMAs(V) after 6 h, with small amounts of TMAO appearing after 8 h (Fig. 5A and Table S2). When assayed at temperatures ranging from 50 °C to 80 °C, purified CmArsM7 converted As(III) to monomethylarsenite [MMAs(III)] and DMAs(V), with a temperature optimum at 60–70 °C (Fig. 5B). There was an initial small loss of total arsenic, apparently caused by binding to the column material, but no volatilization was observed at early time points. At 70 °C, the pH optimum for CmArsM7 was 7.5 measured over the range 2 to 8.5. The rate of arsenic transformation was considerably accelerated at higher temperatures compared with 37 °C, with nearly all of the As(III) converted to DMAs(V) within 30 min at 70 °C (Fig. 5C). All of the As(III) was converted to TMAO and TMAs(III) after 12 h. Although the ratios of MMAs(III) to DMAs(V) varied somewhat from assay to assay with the purified enzyme, at 50°C the concentration of MMAs(III) tended to be lower than DMAs(V), suggesting that at temperatures more environmentally relevant to Cyanidioschyzon the conversion of MMAs(III) to DMAs(V) is faster than the next step, the conversion of DMA(V) to DMA(III), which may be rate limiting in the overall reaction under these conditions.
Fig. 5.
Purified CmArsM is a thermophylic arsenite S-adenosylmethionine methyltransferase. (A) The products of methylation by 2.2 μM purified CmArsM7 and CmArsM8 at 37 °C were analyzed by HPLC-ICP-MS, with instrumental output in arbitrary units. (B) The products of methylation by 3 μM CmArsM7 were examined as a function of assay temperature after 30 min. From left to right the 4 bars at each temperature are: starting amount of As(III), As(III), MMAs(III), and DMAs(V). (C) Arsenic methylation was assayed with 1.5 μM CmArsM7 and analyzed by HPLC-ICP-MS: ●, As(III); ▼, MMAs(III); ○, DMAs(V). The error bars indicate the standard error of 3 assays. Curves are displaced on the y axis for clarity.
Discussion
The studies described herein demonstrate As(III) methylation by the ecologically relevant Cyanidioschyzon sp. pure culture isolate 5508 and by the enzymes encoded by genes cloned from this alga. The optimal in vitro enzymatic rates at elevated temperatures (Fig. 5C) are consistent with the thermophilic properties of the alga, and when considered with the considerable biomass of this organism in these springs (Fig. 1), it would seem to appear that the Cyanidiales play a significant role in the As biogeochemical cycle in the geothermal environment. Indeed, As(III) oxidation rates in the outflow channel dominated by these algae are significant (7).
The ability of these unicellular plants to transform arsenic is arguably a major mechanism for tolerance to high concentrations of arsenic in acidic geothermal settings and contributes to their ecological fitness and ability to flourish in such surroundings. Isolate 5508 may use at least 2 different arsenic detoxification strategies that may be coupled and compartmentalized in a fashion that is well suited with their environment. Because As(III) is the dominant form of arsenic in acidic geothermal waters (2), the cell may first attempt to detoxify its immediate environment by converting As(III) to the less toxic As(V) oxyanion. However, inorganic phosphate is typically below detection in such environments (2, 7), and thus the resulting As(V) would likely be readily taken up by the cell via phosphate permeases (24). Consequently, As(III) methylation could represent an additional mechanism to rid the cell of the accumulated As(V), with the expectation that, under in situ conditions, the eventual final product would be TMA(III), a volatile gas that would leave the cell, presumably by a passive mechanism. It should be noted that the rat enzyme also synthesizes TMA(III) (25), so the entire methylation pathway is well conserved across the Eukarya.
The observed time course of As(V) accumulation (Fig. 2) is consistent with As(III) oxidation occurring outside of the cytoplasm compartment, which is well documented for bacteria (26) and would represent an interesting addition to algal extracellular enzymes [e.g., carbonic anhydrase (27) and extracellular phosphatase (28)]. The subsequent disappearance of As(V) (Fig. 2) likely reflects As(V) uptake by the phosphate transporter when the As(V)/phosphate ratio significantly exceeded unity. As(V) uptake would occur when phosphate levels become depleted at latter stages of the culture cycle due to cellular P consumption. The As(V) is then rereduced in the cytosol to As(III), the substrate for methylation. We examined this scenario in experiments where As(V) was added to the medium in place of As(III) and at the same concentrations; the time course and amount of TMAO formed were essentially identical with that observed with As(III), and thus argues that the level of extracellular As(V) per se is not a determining factor to arsenic uptake and initiation of As(III) methylation. In still further experiments where the cells were suspended in phosphate-free medium containing As(III), the onset and rate of TMAO formation reflected enhanced access of arsenic to the cytoplasm; i.e., >50% of the arsenic was methylated within 66 h. In summary, the data indicate that As(V) uptake is the rate-limiting factor for intracellular reduction to As(III) and subsequent methylation.
In vitro characterization of the purified recombinant enzymes generated data that are also internally consistent with the balance of the study in that the pH optimum of 7.5 in an acidophilic organism is consistent with a cytosolic location, and Topt of 60–70 °C is clearly consistent with the thermophilic nature of the alga. Further, these studies also offer clarification of our mechanistic understanding of an important step in the biotransformation of arsenic; i.e., the MMAs(III) intermediate has been predicted (23) but never observed.
Regarding the ecological significance of these findings, identification and characterization of arsenic methylation in this thermoacidophilic Yellowstone alga correlates well with the observation that methylated arsenic in the Yellowstone geothermal complex is found at highest concentrations in acidic features (29, 30). Thus it would appear that algae play a significant role in arsenic cycling in the geothermal environment as also found in a range of marine and freshwater environments (1, 31). These observations indicate that arsenic methylation forms an important component of the global arsenic biogeocycle.
Methods
Alga and Bacterial Cultures.
The isolation and description of Cyanidiales isolate 5508 has been described (32). Unless otherwise noted, Allen's medium (32) was used for routine culturing of this organism and in all experiments where arsenic speciation was examined. For in vivo As methylation and oxidation assays with isolate 5508, 20 mM NaH2AsO3 was added to media in duplicate sterile 70-mL capped (gray rubber septa) serum bottles with a 100% CO2 headspace (replenished on day 7). Starting culture optical density was adjusted to A595 nm = 0.050, and flasks were incubated at 42 °C under constant illumination (80 mE m−2·s−2). For As(III) resistance assays with E. coli strains AW3110(DE3) (ΔarsRBC) or AW3110(DE3) pET28-alg-arsM were used. Resistance assays with E. coli were performed as described (19).
DNA Manipulations, PCRs, and Protein Purification.
DNA was extracted from 5508 cultures as described (34). For amplifying both CmarsM genes, the forward primer 5′-TTAGCAGCATCGTCGCCCTGTCGC-3′ and reverse primer 5′-ATGCCGTGCAGCTGTGCGTCTG-3′ were designed based on arsM sequences found in the genome sequence of C. merolae strain 10D. Amplicons were cloned into pCR2.1-Topo (Invitrogen) as 1.2-kb fragments and then sequenced at the Ohio State University Plant Microbe Genomics Facility (Columbus).
For construction of plasmids for CmarsM expression and purification of ArsM proteins, the 1.2-kb fragments containing the ATG start codon but excluding the stop codon were PCR- amplified by using the forward primer (5-GTGCTCGAGGCGGCCGCGCAGCATCGTCGCCCTGTCGC-3′) (NotI site underlined) and reverse primer (5′-GATATACCATGGCGTGCAGCTGTGCGTCTGG-3′) (NcoI site underlined), then cloned into pET28a(+) as a NcoI/NotI digest, generating plasmid pET28-alg-arsM, in which the arsM gene is under the control of the T7 promoter. All of the sequences were verified by DNA sequencing. His-tagged ArsM proteins were purified as described (19).
Arsenic Speciation Analysis.
Reaction samples were processed as described (19). Protein was removed from the reaction samples by centrifugation using a 10-kDa cut-off Amicon Ultrafilter. The filtrate was speciated by HPLC (PerkinElmer Series 200) ICP-MS (PerkinElmer ELAN 9000) using an anion exchange column (Hamilton PRP-X100) eluted with a step gradient composed of 9 mL of mobile phase A (20 mM ammonium bicarbonate, pH 8.5), and 18 mL of mobile phase B (20 mM ammonium sulfate, pH 7.0) at a flow rate of 1.5 mL/min. For in vivo assays with the cultured isolate 5508, diluted culture fluids were speciated by using HPLC-ICP-MS (Agilent 7500ce) with an anion exchange column (Agilent G3154A). A 2.0 mM KH2PO4/0.2 mM EDTA/5% methanol (pH 6.0) buffer was used as the mobile phase at a flow rate of 1.0 mL/min.
As(III) methylation was assayed both in vivo and in vitro. Algal culture samples (0.1 mL) were withdrawn daily for arsenic analysis. Culture samples were centrifuged (10 min, 7,500 × g), and the supernatant was removed, diluted appropriately, and kept frozen at −20 °C until analysis by HPLC-ICP-MS. Duplicate controls containing arsenic but no algal cells and duplicate killed-cell controls, where the media was inoculated with autoclaved algal cells, were prepared, incubated, and sampled in the same way.
For in vivo assays with E. coli strain AW3110(DE3) (ΔarsRBC) bearing either vector plasmid pET28a+ or pET28a-alg-arsM7, cultures were first grown overnight at 37 °C in LB media containing 25 μg/mL kanamycin and 0.3 mM IPTG, and then diluted 50-fold into fresh, prewarmed LB media containing 25 μg/mL kanamycin, 0.3 mM IPTG, and 25 μM sodium arsenite. The cultures were divided either into 4-mL aliquots in capped vials or 5-mL capped chambers, as described above, and grown with gentle shaking at 37 °C. At the indicated times, arsenic species in the reaction solution and filters were analyzed in triplicate by HPLC-ICP-MS. In vitro assays with purified ArsM were performed in a buffer containing 50 mM K2HPO2, 8 mM reduced glutathione, and 0.3 mM S-adenosylmethionine chloride, pH 7.4, unless otherwise indicated.
The gaseous arsenic compounds evolved from the cell cultures were analyzed by cryogenic trapping of the evolved gaseous arsenic species followed by GC and ICP-MS analysis of these arsenic species (35). Cell cultures (5 mL) were incubated with As(III) in capped 29.4-mL chambers. After incubation, the chambers were taken out from an incubator and connected to a custom-built cold trapping-GC-ICP-MS system. The headspace in the chambers was purged with helium for 6 min, and the volatile arsenic species were trapped on a GC column immersed in liquid nitrogen. After trapping, liquid nitrogen was removed, and the gaseous arsines trapped on the GC column were separated and carried by helium to the ICP-MS for analysis. Individual gaseous arsenic species were quantified against calibrations of the respective arsine species generated from the corresponding arsenic standards by using hydride generation reactions.
Scanning Electron Microscope Energy-Dispersive X-Ray Analysis.
Samples corresponding to the orange-yellow solid phase observed in the thermal spring and associated with the Cyanidiales algae were taken with sterile spatulas, placed into sterile 15-mL tubes, covered with corresponding spring water, and transported to the laboratory. Portions of the solid phase were air-dried, mounted and sputter-coated with carbon, and imaged with a scanning electron microscope (JEOL 6100), and elemental spectra were acquired from representative spots by using energy dispersive X-rays (NORAN detector with Roentec software). The quantitative analysis from each spectrum was run to determine the ratio of the As (as the L-alpha line) to S (K-alpha line) in the coating.
Supplementary Material
Acknowledgments.
T.R.M. thanks Nancy Equall for assistance with the scanning electron microscope energy-dispersive x-ray analysis. This work was supported by National Institutes of Health Grants GM55425 and AI043428 (to B.P.R.), National Aeronautics and Space Administration Grants NAG 5-8807 and NSF MCB0702212 (to T.R.M.), Montana Agricultural Experiment Station Grant 911310 (to T.R.M.), and grants from Alberta Water Research Institute, Metals in the Human Environment Strategic Network, and Persistent Toxic Substances Innovative Consortium (to X.C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. FJ476310 and FJ463403).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900238106/DCSupplemental.
References
- 1.Bhattacharjee H, Rosen BP. Arsenic metabolism in prokaryotic and eukaryotic microbes. In: Nies DHS, editor. Molecular Microbiology of Heavy Metals. Vol 6. Heidelberg: Springer; 2007. pp. 371–406. Microbiology Monographs. [Google Scholar]
- 2.Nordstrom DK, Ball JW, McCleskey RB. Ground water to surface water: Chemistry of thermal outflows in Yellowstone National Park. In: Inskeep W, McDermott TR, editors. Geothermal Biology and Geochemistry in Yellowstone National Park. Bozeman: Montana State University; 2005. pp. 73–94. [Google Scholar]
- 3.Connon SA, Koski AK, Neal AL, Wood SA, Magnuson TS. Ecophysiology and geochemistry of microbial arsenic oxidation within a high arsenic, circumneutral hot spring system of the Alvord Desert. FEMS Microbiol Ecol. 2008;64:117–128. doi: 10.1111/j.1574-6941.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 4.Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 5.Kulp TR, et al. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science. 2008;321:967–970. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- 6.Gihring TM, Banfield JF. Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol Lett. 2001;204:335–340. doi: 10.1111/j.1574-6968.2001.tb10907.x. [DOI] [PubMed] [Google Scholar]
- 7.Langner HW, Jackson CR, McDermott TR, Inskeep WP. Rapid oxidation of arsenite in a hot spring ecosystem, Yellowstone National Park. Environ Sci Technol. 2001;35:3302–3309. doi: 10.1021/es0105562. [DOI] [PubMed] [Google Scholar]
- 8.Macur RE, Jackson CR, Botero LM, McDermott TR, Inskeep WP. Bacterial populations associated with the oxidation and reduction of arsenic in an unsaturated soil. Environ Sci Technol. 2004;38:104–111. doi: 10.1021/es034455a. [DOI] [PubMed] [Google Scholar]
- 9.D'Imperio S, Lehr CR, Breary M, McDermott TR. Autecology of an arsenite chemolithotroph: Sulfide constraints on function and distribution in a geothermal spring. Appl Environ Microbiol. 2007;73:7067–7074. doi: 10.1128/AEM.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahoe-Christiansen J, D'Imperio S, Jackson CR, Inskeep WP, McDermott TR. Arsenite-oxidizing Hydrogenobaculum strain isolated from an acid-sulfate-chloride geothermal spring in Yellowstone National Park. Appl Environ Microbiol. 2004;70:1865–1868. doi: 10.1128/AEM.70.3.1865-1868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oremland RS, et al. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph, strain MLHE-1. Appl Environ Microbiol. 2002;68:4795–4802. doi: 10.1128/AEM.68.10.4795-4802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oremland RS, et al. A microbial arsenic cycle in a salt-saturated, extreme environment. Science. 2005;308:1305–1308. doi: 10.1126/science.1110832. [DOI] [PubMed] [Google Scholar]
- 13.Lehr CR, et al. Cyanidia (Cyanidiales) population diversity and dynamics in an acid-sulfate-chloride spring in Yellowstone National Park. J Phycol. 2007;43:3–14. [Google Scholar]
- 14.Reysenbach AL, Cady SL. Microbiology of ancient and modern hydrothermal systems. Trends Microbiol. 2001;9:79–86. doi: 10.1016/s0966-842x(00)01921-1. [DOI] [PubMed] [Google Scholar]
- 15.Toplin JA, Norris TB, Lehr CR, McDermott TR, Castenholz RW. Biogeographic and phylogenetic diversity of thermoacidophilic cyanidiales in Yellowstone National Park, Japan, and New Zealand. Appl Environ Microbiol. 2008;74:2822–2833. doi: 10.1128/AEM.02741-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciniglia C, Yoon HS, Pollio A, Pinto G, Bhattacharya D. Hidden biodiversity of the extremophilic Cyanidiales red algae. Mol Ecol. 2004;13:1827–1838. doi: 10.1111/j.1365-294X.2004.02180.x. [DOI] [PubMed] [Google Scholar]
- 17.Parris GE, Brinckman FE. Reactions which relate to environmental mobility of arsenic and antimony. II. Oxidation of trimethylarsine and trimethylstibine. Environ Sci Technol. 1976;10:1128–1134. doi: 10.1021/es60122a010. [DOI] [PubMed] [Google Scholar]
- 18.Lin S, et al. A novel S-adenosyl-l-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- 19.Qin J, et al. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arguello JM, Eren E, Gonzalez-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 22.Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177:981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challenger F. Biological methylation. Adv Enzymol Related Subjects Biochem. 1951;12:429–491. doi: 10.1002/9780470122570.ch8. [DOI] [PubMed] [Google Scholar]
- 24.Rosen BP, Liu Z. Transport pathways for arsenic and selenium: A mini-review. Environ Int. 2008 doi: 10.1016/j.envint.2008.07.023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters SB, et al. Glutathione modulates recombinant rat arsenic (+3 oxidation state) methyltransferase-catalyzed formation of trimethylarsine oxide and trimethylarsine. Chem Res Toxicol. 2004;17:1621–1629. doi: 10.1021/tx0497853. [DOI] [PubMed] [Google Scholar]
- 26.Silver S, Phung le T. A bacterial view of the periodic table: Genes and proteins for toxic inorganic ions. J Ind Microbiol Biotechnol. 2005;32:587–605. doi: 10.1007/s10295-005-0019-6. [DOI] [PubMed] [Google Scholar]
- 27.Nimer NA, Brownlee C, Merrett MJ. Extracellular carbonic anhydrase facilitates carbon dioxide availability for photosynthesis in the marine dinoflagellate prorocentrum micans. Plant Physiol. 1999;120:105–112. doi: 10.1104/pp.120.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallmann A. Enzymes in the extracellular matrix of Volvox: An inducible, calcium-dependent phosphatase with a modular composition. J Biol Chem. 1999;274:1691–1697. doi: 10.1074/jbc.274.3.1691. [DOI] [PubMed] [Google Scholar]
- 29.Planer-Friedrich B, et al. Speciation of volatile arsenic at geothermal features in Yellowstone National Park. Geochim Cosmochim Acta. 2006;70:2480–2491. [Google Scholar]
- 30.Planer-Friedrich B, Merkel BJ. Volatile metals and metalloids in hydrothermal gases. Environ Sci Technol. 2006;40:3181–3187. doi: 10.1021/es051476r. [DOI] [PubMed] [Google Scholar]
- 31.Bentley R, Chasteen TG. Microbial methylation of metalloids: Arsenic, antimony, and bismuth. Microbiol Mol Biol Rev. 2002;66:250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehr CR, Kashyap DR, McDermott TR. New insights into microbial oxidation of antimony and arsenic. Appl Environ Microbiol. 2007;73:2386–2389. doi: 10.1128/AEM.02789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen MB. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Archiv Mikrobiol. 1959;32:270–277. doi: 10.1007/BF00409348. [DOI] [PubMed] [Google Scholar]
- 34.Botero LM, et al. Poly(A) polymerase modification and reverse transcriptase PCR amplification of environmental RNA. Appl Environ Microbiol. 2005;71:1267–1275. doi: 10.1128/AEM.71.3.1267-1275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan C, Lu X, Qin J, Rosen BP, Le XC. Volatile arsenic species released from Escherichia coli expressing the AsIII S-adenosylmethionine methyltransferase gene. Environ Sci Technol. 2008;42:3201–3206. doi: 10.1021/es702910g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.