Summary
Growth and expansion of ventricular chambers is essential during heart development and is achieved by proliferation of cardiac progenitors. Adult cardiomyocytes, by contrast, achieve growth through hypertrophy rather than hyperplasia. Although epicardial-derived signals may contribute to the proliferative process in myocytes, the factors and cell types responsible for development of the ventricular myocardial thickness are unclear. Using a co-culture system, we found that embryonic cardiac fibroblasts induced proliferation of cardiomyocytes, in contrast to adult cardiac fibroblasts that promoted myocyte hypertrophy. We identified fibronectin, collagen and heparin-binding EGF-like growth factor as embryonic cardiac fibroblast-specific signals that collaboratively promoted cardiomyocyte proliferation in a paracrine fashion. β1-integrin was required for this proliferative response, and ventricular cardiomyocyte-specific deletion of β1-integrin in mice resulted in reduced myocardial proliferation and impaired ventricular compaction. These findings reveal a previously unrecognized paracrine function of embryonic cardiac fibroblasts in regulating cardiomyocyte proliferation.
Keywords: Heart, fibroblast, integrin, ventricular formation
Introduction
Tissue morphogenesis is an intricate process in which cells derived from many embryonic sources coalesce, interact and develop into mature organs. In this process, cell proliferation and differentiation are tightly regulated spatiotemporally by cell-cell interactions to insure the tissue attains the necessary size, shape, structure and function. The molecular and cellular mechanisms involved in these cell-cell interactions are complex but are still largely unknown.
Cardiac ventricular formation involves growth of the heart muscle by proliferation of cardiomyocytes and the transmural subdivision into two distinct tissue architectures: trabecular myocardium on the inside and compact myocardium to the outside (Pennisi et al., 2003; Srivastava, 2006). Trabecular myocardium, which contains highly organized muscular ridges, forms as a result of interactions between cardiomyocytes and endocardium from embryonic day (E) 9.5 to E13.5 in mouse hearts (Grego-Bessa et al., 2007; Kang and Sucov, 2005; Smith and Bader, 2007). Compact myocardium is highly mitotic and distributed along the outer ventricular wall. Compact myocardium becomes morphologically recognizable around E11.5 and continues to proliferate throughout gestation. How this proliferative activity of cardiac progenitors is regulated and subsequently terminated after birth remains unknown (Toyoda et al., 2003). In particular, the regulation of cardiac growth in late embryonic stages is unclear, but signals from the endocardium and epicardium may influence cardiomyocytes in a paracrine fashion (Lavine et al., 2005; Smith and Bader, 2007). However, signals from these two layers may not be sufficient for compact myocardium expansion transmurally given the thickness achieved late in development.
Along with cardiomyocytes, cardiac fibroblasts are found throughout cardiac tissue and account for up to two-thirds of the cells in the adult heart (Baudino et al., 2006; Camelliti et al., 2005). They are embedded within the extracellular matrix (ECM) of the connective tissue and are, to a large extent, responsible for its synthesis. Under physiological conditions, fibroblasts provide a mechanical scaffold for cardiomyocytes and coordinate pump function of the heart (Miragoli et al., 2006). In diseased hearts, fibroblasts have central and dynamic roles in modulating cardiac function. The ECM and growth factors secreted from fibroblasts promote cardiomyocyte hypertrophy, leading to myocardial remodeling (Sano et al., 2000; Weber and Brilla, 1991). Although these findings demonstrate that cardiac fibroblasts are critical in the adult heart, little is known about their development and roles in the embryonic heart. Moreover, it has not been determined if the ECM affects myocardial proliferation and ventricular compaction.
We examined the development and function of cardiac fibroblasts in mouse embryonic hearts. In particular, we explored the embryonic cardiac fibroblast-specific signals which regulated cardiomyocyte proliferation and ventricular chamber formation. Our findings suggest that embryonic, but not adult, cardiac fibroblasts secrete high levels of fibronectin, collagen, and heparin-binding EGF-like growth factor (HBEGF), which collaboratively interact and regulate mitotic activity in cardiomyocytes through β1 integrin signaling.
Results
Cardiac fibroblasts develop coincident with ventricular compaction
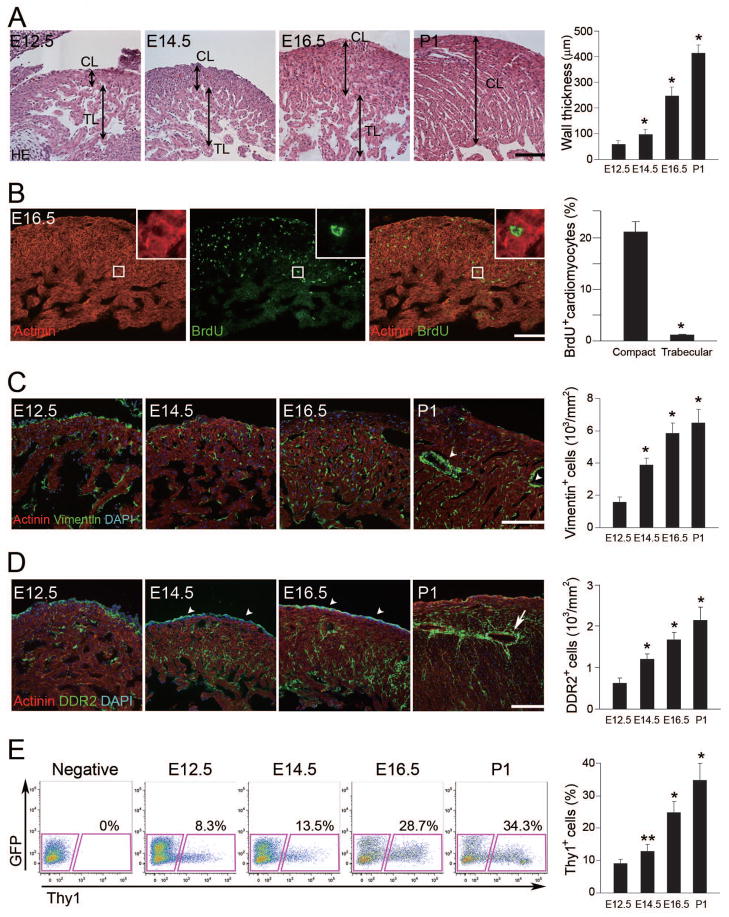
We analyzed the time course of myocardial growth in the ventricles of developing mice from E12.5 to postnatal day (P) 1. The wall thickness gradually increased with development, and growth of the compact layer was greater than that of the trabecular layer (Figure 1A), consistent with previous reports (Toyoda et al., 2003). We labeled E16.5 mouse hearts with BrdU and determined proliferating cardiomyocytes by immunostaining for actinin (cardiomyocyte marker) and BrdU (proliferation marker). Surprisingly, BrdU+ cardiomyocytes were abundant not only in the outer compact layer, but also transmurally. As expected, proliferation was barely detectable in the trabecular layer (Figure 1B).
Figure 1. Cardiac Fibroblasts Develop Concordantly with Ventricular Compaction.
(A) Sections of left ventricles stained with HE. TL, trabecular layer; CL, compact layer. Quantitative analyses of wall thickness in the compact layer (n = 4). (B) Immunofluorescent staining for actinin (red) and BrdU (green). BrdU+ cells were more abundant in the compact layer than in the trabecular layer. Inset is a high-magnification view. Quantitative analyses were shown (n = 4). (C) Immunofluorescent staining for actinin (red), vimentin (green) and DAPI (blue, nuclei). Vimentin+ cells appeared in the myocardium by E12.5 and gradually increased over time. Note that vimentin+ cells existed around vessels in P1 heart (arrowheads), indicating perivascular fibroblasts. Vimentin+ cell numbers in the left ventricles (n = 4). (D) Development of DDR2+ cells (green) throughout the myocardium. Arrowheads indicate DDR2+ epicardium, and an arrow indicates perivascular fibroblasts. DDR2+ cell numbers in the left ventricles (n = 4). (E) FACS analyses for Thy1+ cells. Thy1+ cells were increased during development (n = 3). Representative data are shown in each panel. All data are presented as means ± SEM. *, P<0.01; **, P<0.05 vs relative control. Scale bars, 100 μm.
We next analyzed the distribution of cardiac fibroblasts in E16.5 hearts by immunostaining with an antibody to vimentin. Vimentin+ cells did not express actinin and existed abundantly in the compact layer along with cardiomyocytes (Figure S1A). Vimentin+ cells appeared in the compact myocardium at E12.5, and increased in number throughout development (Figure 1C). We also analyzed development of cardiac fibroblasts using a more fibroblast-specific marker, Discoidin Domain Receptor 2 (DDR2) (Baudino et al., 2006; Camelliti et al., 2005; Goldsmith et al., 2004). DDR2 was expressed in some vimentin+ cells and in the epicardium, but not in CD31+ (endothelial cell marker) or smooth muscle-myosin heavy chain+ (SM-MHC, smooth muscle cell marker) cells, consistent with previous reports (Figure S1B and Figure 1D). The number of DDR2+ cells within the myocardium (excluding those within the epicardium) also increased during development (Figure 1D). To analyze fibroblast cell number more quantitatively, fluorescence-activated cell sorting (FACS) for Thy-1 (fibroblast, T-lymphocyte, and neuron marker) was performed (Hudon-David et al., 2007). We found that the fibroblast markers, vimentin, DDR2, periostin (Postn), and fibroblast-specific protein 1 (Fsp1) were more highly expressed in Thy-1+ cells, but the cardiomyocyte markers, Nkx2.5 and actinin, were more abundant in Thy-1− cells (Figure S1C). FACS analysis revealed that Thy-1+ cells were increased in number with development (Figure 1E). Thus, multiple markers indicated that cardiac fibroblasts developed coincident with growth of the compact layer.
Embryonic cardiac fibroblasts promote cardiomyocyte proliferation
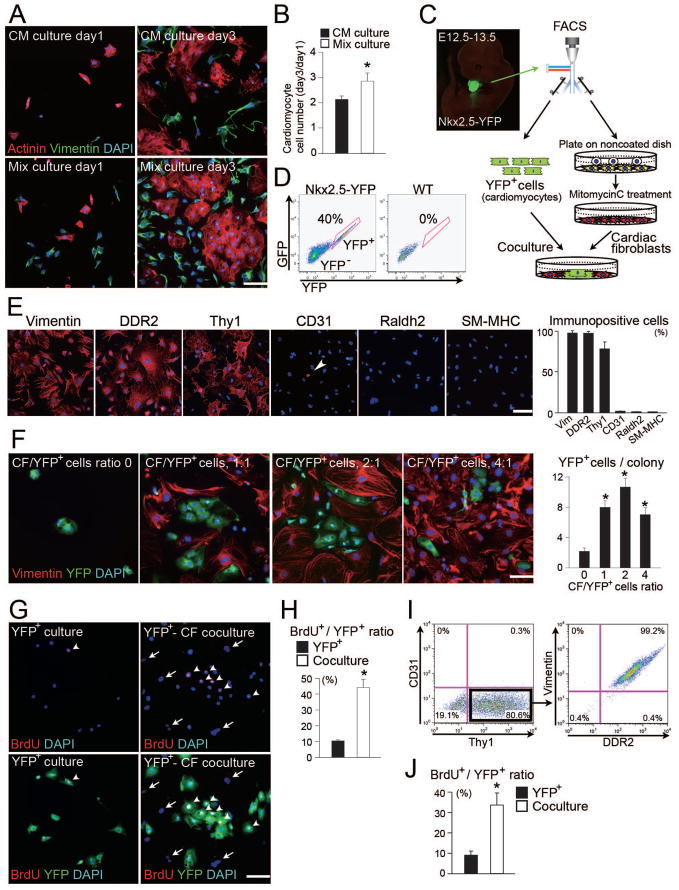
To determine the roles of cardiac fibroblasts in cardiogenesis, we performed primary culture experiments with E12.5–13.5 mice hearts. We compared a whole-heart cell (mixed population) culture with a cardiomyocyte-enriched culture, which was obtained by the conventional preplating method (Engel et al., 1999; Ieda et al., 2007). In either culture, cells were incubated for 3 days and analyzed by immunostaining for actinin, vimentin, and DAPI. The number of cardiomyocytes increased more in the mixed population culture than in the cardiomyocyte-enriched culture (Figure 2A and B). To determine the effect of cell density on cardiomyocyte proliferation, the cell number plated in cardiomyocyte-enriched culture was varied, and compared with the mixed population culture by BrdU labeling (Figure S2A). Non-myocytes supported cardiomyocyte proliferation, but higher density of cardiomyocytes did not show significant effects. The number of TUNEL+ cardiomyocytes (apoptosis marker) was not different among these cultures (Figure S2B), suggesting non-myocytes supported cardiomyocyte increase by augmenting cell proliferation. The percentage of non-myocytes was around 10% at day 1, but increased up to 30–40% by day 3 in the cardiomyocyte-enriched culture; in the mixed population cultures, non-myocytes were 40% of cells at day 1, and increased up to 50–60% by day 3. Due to the high contamination and proliferation of non-myocytes, even in the cardiomyocyte-enriched culture, it was difficult to determine the role of non-myocytes using this conventional cardiomyocyte isolation method; we also could not rule out the possibility that proliferating non-myocyte progenitors might transdifferentiate into cardiomyocytes (Laugwitz et al., 2005).
Figure 2. Cardiac Fibroblasts Promote Cardiomyocyte Proliferation in Co-culture.
(A) Cardiomyocyte-enriched culture (CM culture) and mixed population culture (Mix culture) at days 1 and 3. Immunofluorescent staining for actinin (red), vimentin (green) and DAPI (blue). (B) Increase of actinin+ cell number during a 3-day culture (n = 3). (C) Schematic representation of the co-culture strategy and Nkx2.5-YFP mouse embryo. (D) FACS was used to compare E12.5 Nkx2.5-YFP and wild-type mouse heart cells: 40% of the transgenic heart cells were YFP+. (E) Immunofluorescent staining for vimentin, DDR2, Thy-1, CD31, Raldh2, and SM-MHC in cardiac fibroblast culture. The majority of cells were positive for fibroblast markers (n = 4). (F) YFP+ cells were co-cultured with varying numbers of cardiac fibroblasts. The ratios of cardiac fibroblasts to YFP+ cells varied from 0:1 to 4:1. CF, cardiac fibroblasts. Note that YFP+ cells formed colonies in the co-culture with cardiac fibroblasts. YFP+ cell numbers per colony in the co-cultures (n = 3). (G) Immunofluorescent staining for BrdU (red), YFP (green) and DAPI (blue) in the pure cardiomyocyte culture (YFP+ culture) or the co-culture of cardiomyocytes with fibroblasts (YFP+-CF coculture). Arrowheads indicate BrdU+ cells, and arrows indicate YFP− fibroblasts. Note that fibroblasts were all BrdU−. (H) Ratios of BrdU+ cells to YFP+ cells indicate the percentage of proliferating cardiomyocytes (n = 5). (I) Thy-1+CD31− cells were FACS sorted from fibroblast culture, and analyzed for vimentin and DDR2 expressions. (J) Ratios of BrdU+ cells to YFP+ cells in the coculture with Thy-1+CD31− cells (n = 3). Representative data are shown in each panel. All data are presented as means ± SEM. *, P < 0.01 vs relative control. Scale bars, 100 μm.
To overcome these issues, we developed a co-culture system using E12.5–13.5 Nkx2.5-YFP mice, which were obtained by crossing the Nkx2.5 cardiac ventricular enhancer–Cre (ventricular cardiomyocyte lineage) transgenic mice with R26R-EYFP (reporter) mice (McFadden et al., 2005; Srinivas et al., 2001). Nkx-YFP+ cells co-stained with actinin, but not with vimentin (Figure S2C). These YFP+ cells were purified from Nkx2.5-YFP embryos by FACS and co-cultured with putative mitomycin-treated embryonic cardiac fibroblasts (Figure 2C and D). To determine the purity of the embryonic cardiac fibroblast population, we assessed the expression of several independent markers. We confirmed that the majority of the cells expressed vimentin (nearly 100%), DDR2 (nearly 100%), and Thy-1 (80%), while rare cells expressed CD31, retinaldehyde dehydrogenase 2 (Raldh2, epicardium marker) and SM-MHC (Figure 2E), consistent with previous reports (Goldsmith et al., 2004). To explore the dose dependency of cardiac fibroblasts in this co-culture system, we varied the ratios of cardiac fibroblasts to cardiomyocytes from 0 to 4:1 and cultured them for 3 days. Nkx-YFP+ cells were significantly increased in number and formed colonies in the co-culture with cardiac fibroblasts in a dose-dependent manner (Figure 2F). To determine if the increase of Nkx-YFP+ cells was due to cell proliferation, BrdU labeling was performed. Approximate 40% of the YFP+ cells, co-cultured with fibroblasts, were labeled with BrdU, in contrast to only 10% in the pure cardiomyocyte culture (Figure 2G and H). To purify a more homogeneous fibroblast population, we FACS sorted Thy-1+CD31− cells from fibroblast culture (Figure 2I). 80% of the cells were Thy-1+, while only 0.3% were CD31+. We confirmed that Thy-1+CD31− cells were double positive for vimentin and DDR2 by FACS, consistent with the immunocytochemistry results. BrdU labeling showed that Thy-1+CD31− fibroblasts also promoted YFP+ cell proliferation (Figure 2J). These results suggested that embryonic cardiac fibroblasts promote cardiomyocyte proliferation.
Embryonic cardiac fibroblasts promote cardiomyocyte proliferation through fibronectin and collagen
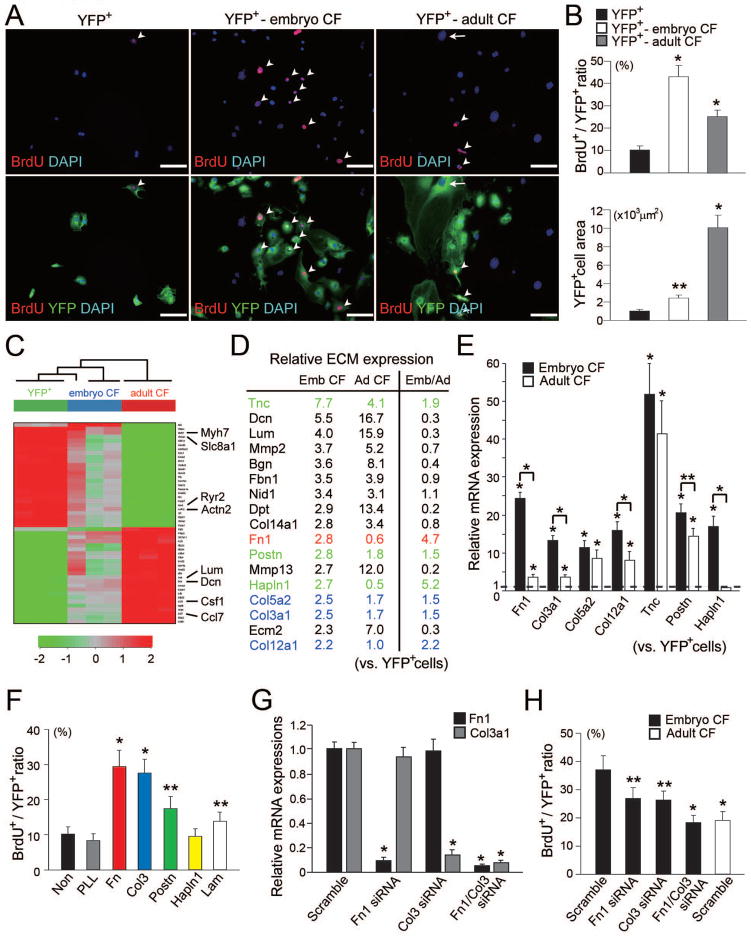
Cardiomyocytes proliferate during embryogenesis, but after birth, they lose this capacity and switch to hypertrophic growth (Soonpaa et al., 1996). To determine whether the proliferative effect of embryonic cardiac fibroblasts on cardiomyocytes differed from that of adult cardiac fibroblasts, we co-cultured Nkx-YFP+ cells with adult cardiac fibroblasts. Adult cardiac fibroblasts, prepared by standard methods (Smolenski et al., 2004), induced cardiomyocyte proliferation but to a significantly lesser degree than embryonic cardiac fibroblasts (Figure 3A and B). In contrast, features of hypertrophy, including sarcomeric organization and increased cell size, were observed in the co-culture with adult cardiac fibroblasts (Figure S3A). These results are consistent with physiological changes in the heart and suggest that signals derived from embryonic or adult cardiac fibroblasts contribute to cardiomyocyte proliferation or hypertrophy, respectively.
Figure 3. Embryonic Cardiac Fibroblasts Promote Cardiomyocyte Proliferation through Fibronectin and Collagen Synthesis.
(A) Immunofluorescent staining for BrdU (red), YFP (green) and DAPI (blue) in the pure cardiomyocyte culture (YFP+), and co-cultures of cardiomyocytes with embryonic (YFP+-embryo CF) or adult cardiac fibroblasts (YFP+-adult CF). Arrowheads indicate BrdU+ cells, and arrow indicates a hypertrophied cardiomyocyte. YFP− cells are fibroblasts. (B) Ratios of BrdU+ cells to YFP+ cells (upper panel) and the YFP+ cell areas (lower panel) (n = 4). (C) The heatmap image of hierarchical clustering based on the most variable genes among Nkx-YFP+ cells, embryonic cardiac fibroblasts, and adult cardiac fibroblasts (n = 3 in each group). The scale extends from 0.25- to 4-fold over mean (−2 to +2 in log2 scale) as indicated on the bottom. (D) Profiling of ECM gene expression in embryonic (Emb CF) and adult cardiac fibroblasts (Ad CF). The genes upregulated at least twofold in the embryonic or adult cardiac fibroblasts compared to Nkx-YFP+ cells by microarray analyses are listed with their fold enrichment (n = 3). The ratios of gene expressions in embryonic fibroblasts to those in adult fibroblasts are also shown (Emb/Ad). Fibronectin (red), collagen family (blue) and other ECM genes (green) upregulated in embryonic cardiac fibroblasts are highlighted. (E) qRT-PCR showing enrichment of ECM genes in embryonic cardiac fibroblasts (n = 3). (F) Ratios of BrdU+ cells to YFP+ cells in the pure cardiomyocytes cultured on the different ECMs (n = 4). (G) qRT-PCR confirmed that Fn1 and Col3a1 gene expression was downregulated by Fn1, Col3a1, or both Fn1 and Col3a1 siRNA knockdown (Fn1, Co13, Fn1/Col3 siRNA, respectively) (n = 4). (H) Ratios of BrdU+ cells to YFP+ cells in the co-culture of cardiomyocytes with siRNA-knockdown embryonic and scramble siRNA-treated adult cardiac fibroblasts (n = 4). All data are presented as means ± SEM. *, P<0.01; **, P<0.05 vs relative control. Scale bars, 100 μm.
To identify candidate fibroblast-derived factors that promote myocyte proliferation, we isolated RNA from Nkx-YFP+ cardiomyocytes, embryonic cardiac fibroblasts, and adult cardiac fibroblasts and profiled mRNA expressions by microarray analyses. The heatmap image of hierarchical clustering based on the most variable genes revealed a striking grouping of the gene expression patterns among the three cell types (Figure 3C). Known genes that are upregulated in cardiomyocytes (Myh7, Slc8a1, Ryr2 and Actn2) were highly expressed in Nkx-YFP+ cells. In contrast, growth factors, cytokines, and ECM were more highly expressed in cardiac fibroblasts (Figure 3D and S3B). Embryonic fibroblasts expressed higher levels of growth factors, Hbegf and Ptn (pleiotrophin), while adult fibroblasts were enriched for more growth-related cytokines (e.g., Il-6, Il-1a) (Figure S3B). Among the ECM genes, we found that Fn1 (Fibronectin1), several members of the collagen family, TnC (tenascin C), Postn (Periostin), and Hapln1 (Hyaluronan and proteoglycan link protein 1) were expressed more abundantly in the embryonic cardiac fibroblasts than in adult fibroblasts and cardiomyocytes (Figure 3D). Interestingly, the expression of laminin genes, encoding another prototypic family of ECM, was not significantly different among cell types (Figure S3C). Quantitative RT-PCR (qRT-PCR) confirmed most of the microarray results (Figure 3E and S3D).
To evaluate the effects of different ECM proteins on cardiomyocyte proliferation, Nkx-YFP+ cells were cultured on non-, poly-L-lysine (PLL)-, Fibronectin-, Collagen III-, Periostin-, Hapln1-, or Laminin-coated plates, followed by BrdU labeling (Figure 3F and S3E). Fibronectin-, Collagen-, and to a lesser extent Periostin- and Laminin-coated plates augmented cardiomyocyte proliferation compared to non-coated plates. Intriguingly, PLL and Hapln1 which are not ligands for integrin, enhanced cell attachment, but did not affect cell proliferation. To analyze cell proliferation on different ECM more quantitatively, we used a mouse cardiomyocyte cell line (HL-1 cells). The increase in HL-1 cell number was significantly higher on fibronectin/gelatin than on laminin, and was due to an increase of cells in the S and G2/M phase and enhanced cell proliferation (Figure S4A–E). These results suggested that interaction with specific ECM is critical for cardiomyocyte proliferation.
To determine if fibronectin and collagen secreted from embryonic cardiac fibroblasts were critical for cardiomyocyte proliferation, we co-cultured Nkx-YFP+ cells with embryonic cardiac fibroblasts in which Fn1, Col3a1, or both Fn1 and Col3a1 were downregulated by siRNA knockdown. We confirmed by qRT-PCR that Fn1 and Col3a1 mRNA were significantly and specifically reduced in the fibroblasts by siRNA knockdown (Figure 3G). We found that knockdown of fibronectin or collagen in embryonic cardiac fibroblasts resulted in decreased cardiomyocyte proliferation, and moreover, knockdown of both had an additive effect (Figure 3H and Figure S4F), reducing the proliferation to levels seen with adult fibroblasts. These results indicated that the enhanced cardiomyocyte proliferative effects of embryonic cardiac fibroblasts require fibronectin and collagen secretion.
Extracellular matrix/β1 integrin signaling is required for cardiomyocyte proliferation in response to growth factors
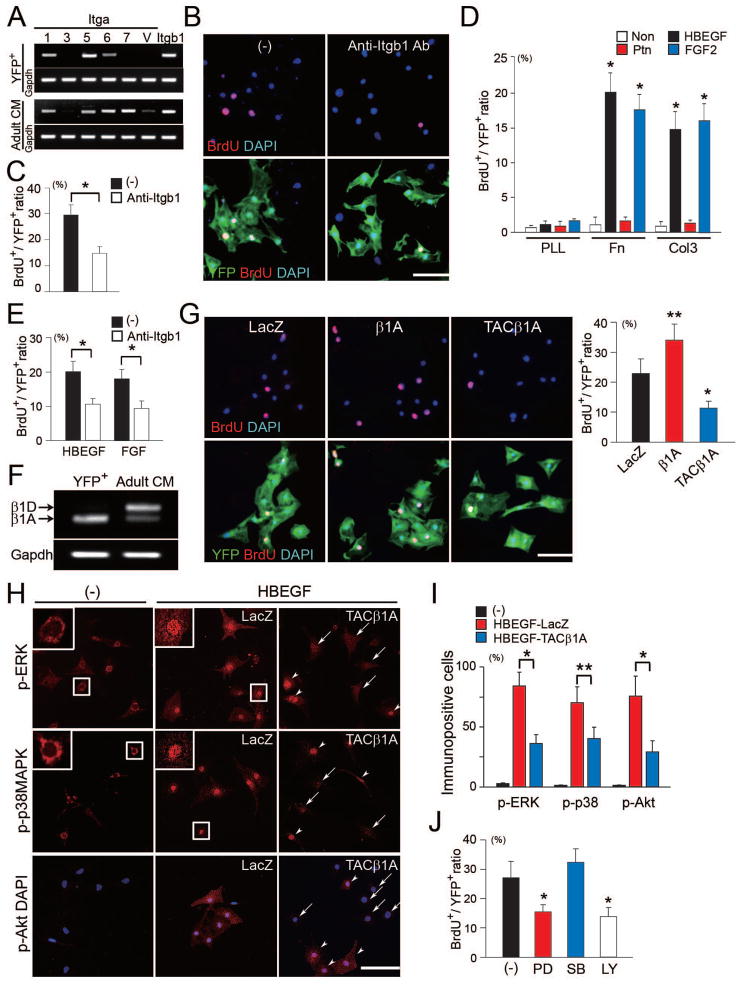
Next, we investigated whether integrins, including the β1 and α subunits that form fibronectin and collagen receptors, were required for cardiomyocyte proliferation (Ross and Borg, 2001). We first analyzed the expressions of integrin subunits in embryonic and adult cardiomyocytes by semiquantitative RT-PCR (Figure 4A). Embryonic cardiomyocytes (Nkx-YFP+ cells) expressed abundant Itga1 (collagen and laminin receptor), Itga5 (fibronectin-specific receptor) and Itgb1, but less Itga6 (laminin-specific receptor). In contrast, adult cardiomyocytes expressed more Itga6, Itga7 (laminin-specific receptor), and Itgav (periostin and tenascin C receptor) than did embryonic cardiomyocytes. To investigate whether β1 integrin was required for cardiomyocyte proliferation in response to embryonic cardiac fibroblasts, we incubated Nkx-YFP+ cells with an anti-β1 integrin-blocking antibody in the co-culture system. To ensure that potential proliferative defects were not caused by cell detachment, cells were cultured on PLL-coated plates. Pretreatment with an anti-β1 integrin-blocking antibody did not affect cell attachment or spreading, but resulted in fewer BrdU+ cardiomyocytes, suggesting that the ECM secreted from embryonic cardiac fibroblasts promoted cardiomyocyte proliferation through β1 integrin signaling (Figure 4B and C).
Figure 4. β1 Integrin Is Required for Cardiomyocyte Proliferation upon Co-culture with Cardiac Fibroblasts.
(A) Semiquantitative RT-PCR of integrin α subunits and β1 in embryonic (YFP+) and adult cardiomyocytes (Adult CM). (B) Immunofluorescent staining for BrdU (red), YFP (green) and DAPI (blue) in co-culture of cardiomyocytes with embryonic cardiac fibroblasts on PLL-coated plates. Cells were cultured in the presence of mock (−) or anti-β1 integrin blocking antibody (Anti-Itgb1 Ab). Cell morphology and attachment were not affected in the presence of anti-β1 integrin blocking antibody. (C) Quantitative data of the ratios of BrdU+ cells to YFP+ cells in (B) (n = 5). (D) HBEGF and FGF2 but not Ptn, augmented cardiomyocyte proliferation on fibronectin or collagen III-coated plates (n = 3). (E) BrdU+ cardiomyocytes induced with HBEGF or FGF were reduced by pretreatment with an anti-β1 integrin-blocking antibody (n = 4). (F) Semiquantitative RT-PCR of integrin β1A and D isoforms in embryonic (YFP+) and adult cardiomyocytes (Adult CM). (G) BrdU+ staining in cardiomyocytes treated with HBEGF after modification of β1A integrin signaling by adenoviral infection with control (LacZ), β1A or TACβ1A (n = 5). (H, I) Activation of phospho-ERK1/2 (p-ERK), phospho-p38MAPK (p-p38MAPK) and phospho-Akt (p-Akt) were determined by immunocytochemistry using phospho-specific antibodies (n = 3). Nkx-YFP+ cells were infected with LacZ or TACβ1A adenovirus, and treated with HBEGF. Note that nuclear translocation of p-ERK and p-p38MAPK after HBEGF stimulation, in contrast to perinuclear localization of MAPK before stimulation (insets). Arrowheads indicate activated cells, and arrows indicate non-activated cells. (J) Nkx-YFP+ cells were pretreated with PD98059 (PD), SB203580 (SB) or LY294002 (LY), and stimulated with HBEGF (n = 4). BrdU+ cardiomyocyes were reduced with PD or LY compared with mock treated cells (−). All data are presented as means ± SEM. *, P<0.01; **, P<0.05 vs. relative control. Scale bars, 100 μm.
To investigate the mechanism underlying ECM/β1 integrin-induced cardiomyocyte proliferation in the co-culture system, we analyzed the interaction between ECM/β1 integrin and growth factors specifically expressed in the embryonic cardiac fibroblasts (Figure S3B and D). Nkx-YFP+ cells were plated on PLL, fibronectin or collagen III, serum starved, then treated with HBEGF or Ptn. Strikingly, HBEGF, but not Ptn, augmented cardiomyocyte proliferation, but only when cells were attached to fibronectin or collagen III (Figure 4D). To investigate whether β1 integrin signaling was involved in this HBEGF-induced cardiomyocyte proliferation, Nkx-YFP+ cells were plated on fibronectin/PLL mixed coated plates (to prevent cell detachment), and treated with HBEGF in the absence or presence of anti-β1 integrin-blocking antibody. Although cell attachment and morphology were not affected, BrdU+ cells were reduced by anti-β1 integrin-blocking antibody in the presence of HBEGF (Figure 4E). Attachment to fibronectin or collagen III through β1 integrin was also required for myocyte proliferation in response to the potent myocardial mitogen, FGF2, which was also highly expressed in the cardiac fibroblasts (17-fold higher in embryonic cardiac fibroblasts than in cardiomyocytes by qRT-PCR) (Figure 4D and E). These results suggested that ECM/β1 integrin signaling is required for cardiomyocyte proliferation in response to the growth factors secreted from cardiac fibroblasts.
β1 integrin consists of two isoforms, β1A and β1D. Although β1D is known to promote the cardiac hypertrophic response, the role of β1A in heart development is not clear. Semiquantitative RT-PCR showed that E12.5–13.5 embryonic cardiomyocytes expressed β1A isoform exclusively, while adult cardiomyocytes expressed more β1D (Figure 4F), consistent with previous reports (Ross and Borg, 2001). To determine the role of β1A, we used adenovirus expressing the full-length β1A integrin (β1A), or a dominant negative version containing the cytoplasmic tail domain of the β1A integrin fused to the extracellular/transmembrane domain of the interleukin-2 receptor (TACβ1A); adenovirus encoding β–galactosidase (LacZ) was used as a control. Previous studies have shown that low-level of TACβ1A expression reduces integrin signaling, but does not inhibit cell adhesion or cytoskeletal organization (Ross et al., 1998). We cultured Nkx-YFP+ cells on fibronectin-coated plates and treated with HBEGF in the presence of each adenoviral vector. We found that overexpression of β1A augmented, but expression of TACβ1A inhibited, cardiomyocyte proliferation, while cell morphology and attachment were maintained (Figure 4G). These results indicated that the β1 integrin, specifically β1A isoform, promotes cardiomyocyte proliferation independent of cell attachment.
Integrins activate mitogen-activated protein kinases (MAPK) and phosphatidylinositol-3-OH kinase (PI3K)/Akt pathways (Giancotti and Ruoslahti, 1999). To investigate the signaling pathways involved in response to the interaction between ECM and growth factors, we analyzed activation of MAPK and Akt by immunocytochemistry (Figure 4H and I). Nkx-YFP+ cells stimulated with HBEGF and cultured on fibronectin-coated plates revealed robust nuclear translocation of phospho-ERK1/2 and phospho-p38MAPK. Activation of ERK and p38MAPK was attenuated by TACβ1A expression. Upregulation of phospho-Akt, induced with HBEGF, was also repressed by inhibition of β1A integrin. To determine which pathways were involved in cardiomyocyte proliferation, Nkx-YFP+ cells were pretreated with specific inhibitors of these pathways then stimulated with HBEGF. We found that PD98059 (MEK inhibitor) and LY294002 (PI3K inhibitor), but not SB203580 (p38MAPK inhibitor), reduced the number of BrdU+ cells (Figure 4J), suggesting that cardiomyocyte proliferation in response to interactions between ECM/β1integrin and HBEGF is mediated by MEK/ERK1/2 and PI3K/Akt, but not p38MAPK.
Cardiomyocyte-specific β1 integrin deletion results in peri-natal lethality
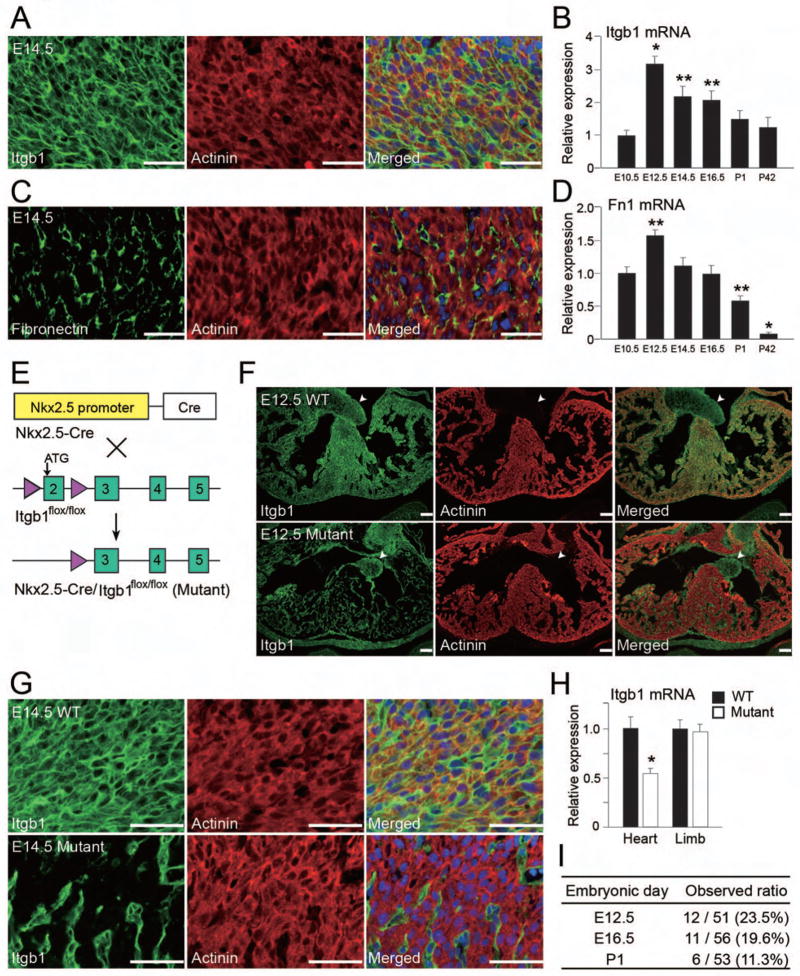
To investigate whether the ECM/β1 integrin signaling is important for myocyte proliferation in vivo, we analyzed the function of β1 integrin and ECM in developing hearts. Immunostaining for β1 integrin and actinin revealed that β1 integrin was abundantly expressed in both cardiomyocytes and non-myocytes in E14.5 mouse hearts (Figure 5A). qRT-PCR for Itgb1, which detected both β1A and β1D, revealed Itgb1 expression in E10.5 hearts. It peaked at E12.5 and was greater in embryonic hearts than in adult hearts (Figure 5B). Consistent with in vitro data, fibronectin was exclusively expressed in non-myocytes (Figure 5C). Interestingly, cardiac Fn1 expression also peaked at E12.5 and changed largely in parallel with Itgb1 expression (Figure 5D). We also found that Col3a1 mRNA was more highly expressed in embryonic hearts than in adult hearts (data not shown).
Figure 5. Generation of Cardiomyocyte-Specific β1 Integrin Knockout Mice.
(A) Immunofluorescent staining for β1 integrin (green), α-actinin (red) and DAPI (blue) in E14.5 mouse hearts. Note that a large proportion of β1 integrin+ cells were colabeled with α-actinin. (B) The time course of Itgb1 mRNA expression determined by qRT-PCR (n = 5). (C) Immunofluorescent staining for fibronectin (green), α-actinin (red) and DAPI (blue) in E14.5 mouse hearts. Note that fibronectin and actinin immunoreactivities were not colocalized. (D) The time course of Fn1 mRNA expression determined by qRT-PCR (n = 5). (E) Scheme of the experiments to generate Nkx2.5-Cre/Itgb1flox/flox mice (Mutant) with the Cre-loxP system. (F) Immunofluorescent staining for β1 integrin (green) and actinin (red) in E12.5 wild-type (WT) and mutant hearts. Note that β1 integrin and actinin were colocalized in WT, but not in mutant hearts. Arrowheads indicate valves, immunopositive for β1 integrin in either heart. (G) Immunofluorescent staining for β1 integrin (green), actinin (red) and DAPI in E14.5 wild-type and mutant hearts, exhibited in higher magnification. β1 integrin is specifically deleted in mutant ventricular cardiomyocytes. (H) Itgb1 mRNA expression in wild-type and mutant hearts and limbs, determined by qRT-PCR (n = 4). (I) Ratios of observed mutant mice at different embryonic stages. Absolute numbers are shown with percentages in parenthesis. Note that expected ratio is 25%. Representative data are shown in each panel. All data are presented as means ± SEM. *, P<0.01; **, P<0.05 vs. relative control. Scale bars, 50 μm (A, G); 100 μm (F).
To determine whether the crosstalk between cardiac fibroblasts and cardiomyocytes through ECM/β1 integrin signaling contributes to cardiogenesis, we specifically deleted β1 integrin in cardiomyocytes by crossing the Nkx2.5 cardiac ventricular enhancer–Cre transgenic mice with Itgb1flox/flox mice (Figure 5E). Immunostaining for β1 integrin and actinin demonstrated that β1 integrin was specifically deleted in mutant ventricular cardiomyocytes, but not in cardiac fibroblasts, endocardium, epicardium or atrial cardiomyocytes (Figure 5F, G and S5). At E12.5, overall Itgb1 mRNA expression levels were lower by about 50% in mutant hearts than in wild-type littermates, but were not decreased in mutant limbs (Figure 5H). Genotyping of offspring revealed 50% lethality of mutants by weaning. Mendelian ratios of mutant embryos were observed until E12.5, but thereafter, death occurred at varying times, ranging from E16.5 to just after birth (Figure 5I).
β1 integrin is required for myocardial proliferation and ventricular compaction in vivo
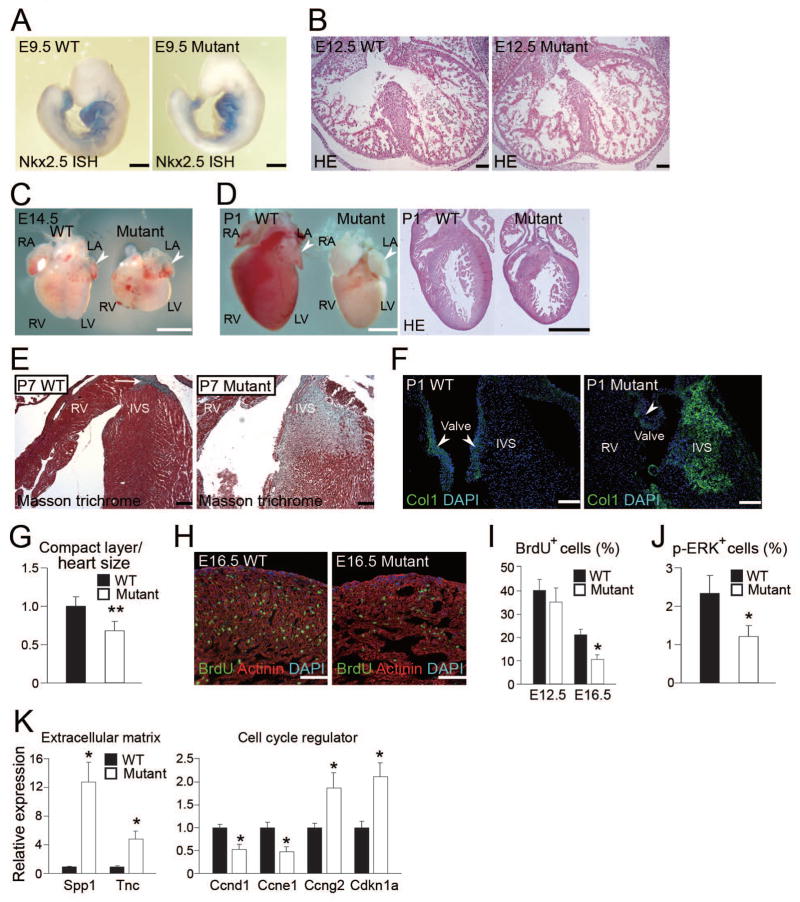
Although Nkx2.5 enhancer–Cre mice express Cre recombinase by E8.5 (McFadden et al., 2005), initial cardiac differentiation and patterning were normal in mutant mice, evidenced by Nkx2.5 and Mlc2v expression at E9.5 and 10.0, respectively (Figure 6A and S6A). H&E staining and immunostaining for actinin revealed that the trabecular layer was well organized at E12.5, and no significant change was detected in mutant hearts (Figure 5F and 6B). However, beginning at E14.5, mutant ventricles were smaller compared with wild type, and the difference became increasingly apparent by P1 (Figure 6C and D). The atrial and the body size were maintained in mutant mice, in accordance with the Cre activity (McFadden et al., 2005). We found that some atria were enlarged in mutant mice suggesting cardiac dysfunction during embryogenesis. Although H&E staining showed that mutant hearts had normal patterning of the ventricle, septum, valves, and outflow tract, Masson-trichrome staining and immunostaining for collagen1 exhibited an accumulation of interstitial fibrosis in mutant hearts after birth (Figure 6D–F).
Figure 6. β1 Integrin Is Required for Myocardial Proliferation, Muscle Integrity and Ventricular Chamber Formation in the Compaction Stage.
(A) Whole-mount in situ hybridization (ISH) for Nkx2.5 in wild-type and mutant E9.5 embryos. (B) H&E staining of wild type and mutant E12.5 hearts. (C) Wild-type and mutant hearts at E14.5. Mutant ventricles are smaller than wild-type, while atria are same size (arrowheads). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. (D) Wild-type and mutant hearts at P1. The right panel shows H&E staining. Note that mutant ventricles were hypoplastic, while the atria were preserved (arrowheads). (E) Masson-trichrome staining in P7 wild-type and mutant hearts. Large amounts of fibrosis were detected in the mutant interventricular septum and right ventricular subendocardium. Arrow indicates His bundle (positive control). (F) Immunofluorescent staining for collagen1 (Col1, green) and DAPI (blue) in the wild-type and mutant P1 hearts. Collagen deposits were observed in mutant hearts. Arrowheads indicate valves (positive control). (G) The ratio of wall thickness in the compact layer to heart size (longest diameter) in wild-type or mutant hearts at P1 (n = 5). (H) Immunofluorescent staining for actinin (red), BrdU (green) and DAPI (blue) in the wild-type and mutant E16.5 left ventricles. BrdU+ cells were reduced in the mutant heart. (I) Percentage of BrdU+ cells in the wild-type and mutant hearts at E12.5 and E16.5. (J) Percentage of p-ERK+ cells in wild-type or mutant hearts at E16.5. (K) qRT-PCR of Spp1 and TnC mRNA (left panel), and Ccnd1, Ccne1, Ccng2, and Cdkn1a mRNA (right panel) in E12.5 wild-type and mutant hearts. Representative data are shown in each panel. All data are presented as means ± SEM. *, P<0.01; **, P<0.05 vs. relative control. Scale bars, 100 μm (B, E, F, H); 500 μm (A); 1 mm (C and D).
The compact layer was thinner and the proportion of the compact layer to the heart size was also smaller in mutant hearts than wild-type littermates, suggesting ventricular hypoplasia was mainly due to growth retardation of the compact myocardium (Figure 6D, G and S6B). The BrdU labeling index was not different between wild-type and mutant hearts at E12.5, but was reduced by 50% in E16.5 mutant hearts compared with wild-type littermates (Figure 6H, I and S6C). We also found that the percent of p-ERK+ cells were significantly less in mutant hearts at E16.5 (Figure 6J and S6D).
To investigate the mechanisms responsible for the abnormalities in β1 integrin mutant mice, we performed mRNA expression microarray analyses of E12.5 wild-type and mutant hearts, well before any obvious dysfunction. 82 genes were upregulated more than twofold, and two were downregulated in mutant hearts (Figure S6E). Among the upregulated genes, nine were inflammatory and immune response genes, and six were ECM genes (Figure S6F). We confirmed that dysregulated ECM genes such as Spp1 and Tnc were upregulated by qRT-PCR (Figure 6K). When microarray analysis was extended to include genes that were upregulated more than 1.2-fold, and downregulated less than 0.8-fold in mutant hearts (p<0.05), many cell cycle-related genes were identified (Figure S6G). We confirmed that Ccnd1 and Ccne1 (cell-cycle promoters) were downregulated, and Ccng2 and Cdkn1a (cell-cycle inhibitors) were significantly upregulated in mutant hearts by qRT-PCR, consistent with proliferative defects in mutant hearts (Figure 6K). These results indicated that myocardial β1 integrin is required for cardiomyocyte proliferation and compact layer growth during mid to late embryogenesis, coincident with the development of cardiac fibroblasts.
Discussion
Here we demonstrated that embryonic cardiac fibroblasts develop coincident with expansion of ventricular compact layer and regulate proliferation of cardiac progenitors. We provide evidence that this occurs via secretion of growth factors and ECM that function through β1 integrin, which we show is essential for normal ventricular expansion during cardiogenesis. In contrast, adult cardiac fibroblasts had a unique function in promoting hypertrophy rather than proliferation, suggesting that a fundamental switch in the cardiac fibroblast gene program may contribute to some of the physiological differences between embryonic and adult heart.
Embryonic vs. adult cardiac fibroblasts
We developed a novel co-culture system to identify the effects of cardiac fibroblasts on cardiomyocytes. Using this system, together with microarray, siRNA knockdown, anti-β1 integrin blocking antibody, and adenovirus experiments, we provide evidence that embryonic cardiac fibroblasts secrete high levels of fibronectin, collagen, and HBEGF, which collaboratively promote cardiomyocyte proliferation through β1 integrin signaling. Although the function and cell source of HBEGF in developing hearts has not been determined, mice carrying a constitutive active form of HBEGF showed cardiac hyperplasia (Yamazaki et al., 2003). This is consistent with our result that HBEGF is a potent mitogen for cardiomyocytes. We also revealed that adult cardiac fibroblasts induced more cardiomyocyte hypertrophy and less proliferation than did embryonic cardiac fibroblasts. The gene expression profiles were very different for embryonic and adult cardiac fibroblasts, and Il6 expression, a potent cardiac hypertrophic factor, was 58-fold higher in the adult fibroblasts than in the embryonic fibroblasts. To our knowledge, this is the first report demonstrating the difference in functions and gene expressions between embryonic and adult cardiac fibroblasts. While there are intrinsic differences between embryonic and adult cardiomyocytes, our data would suggest that the non-cell autonomous signals arising from cardiac fibroblasts contribute to unique aspects of the proliferative vs. hypertrophic response. These findings suggest that the new co-culture system and gene profiling will reveal additional signals that regulate cardiomyocyte proliferation, differentiation and hypertrophy.
Integrin and ECM function in cardiac proliferation
The ECM is important in the growth, division, and differentiation of many cell types, but its role in cardiomyocyte proliferation during embryogenesis has not been determined (Giancotti and Ruoslahti, 1999; Nikolova et al., 2006; Xu et al., 2001). We found that cardiac progenitors proliferated to a greater extent on fibronectin or collagen than on periositn, laminin, hapln1, or poly-lysine-coated plates. Fibronectin or collagen promoted HBEGF-induced cardiomyocyte mitotic activity through β1 integrin. In agreement with our results, α5β1 integrin (fibronectin-specific receptor) directly associates with EGF receptor (HBEGF receptor), making receptor complexes on plasma membrane, which is necessary for optimal activation of growth signaling (Moro et al., 1998). Skeletal myoblasts proliferate on fibronectin, but stop growing and differentiate into myotubes on laminin (Sastry et al., 1999). Ectopic expression of α5 integrin maintained skeletal myoblasts in the proliferative phase, while ectopic expression of the α6 integrin promoted differentiation, indicating the ratio of expression levels of the α integrins is critical for the control of myoblast proliferation. Embryonic cardiomyocytes mainly expressed α1 and α5 integrins, while adult cardiomyocytes expressed more α6 and α7 integrins, consistent with the observation that adult cardiomyocytes specifically attach to laminin-coated plates and undergo hypertrophy (data not shown). Intriguingly, the expression of fibronectin and collagen was higher in the embryonic hearts than in the adult hearts, corresponding to the developmental change of α integrin expressions. These results suggested that the expression of α integrin in cardiomyocytes might be responsible for cardiomyocyte proliferation on fibronectin and collagen.
The roles of endocardium- or epicardium-derived signals in ventricular chamber expansion during mid gestation have been extensively investigated (Grego-Bessa et al., 2007; Smith and Bader, 2007). However, the regulation of ventricular formation in mid-late gestation, particularly the contribution of other cell types, including cardiac fibroblasts, has been less clear. While the origin of cardiac fibroblasts is still ambiguous, some are derived from the epicardium through epithelial-mesenchymal transformation at around E11.5–12.5 in mouse hearts (Cai et al., 2008; Camelliti et al., 2005; Dettman et al., 1998; Zhou et al., 2008). Consistent with this, we found that cardiac fibroblasts appeared in the myocardium by E12.5 and gradually increased in accord with growth of the compact myocardium. The abundant cardiac fibroblasts in the compact myocardium synthesize ECM and growth factors within the embryonic heart. Strikingly, β1 integrin expression, specifically β1A isoform, was also upregulated concordant with the appearance of cardiac fibroblasts in the heart at E12.5, and its expression was maintained highly throughout gestation. β1 integrin mutant mice revealed reduced myocardial proliferation and disruption of muscle integrity, leading to prenatal death. These results, along with the in vitro data, suggested that cardiac fibroblasts regulate cardiomyocyte proliferation during mid-late gestation through β1 integrin signaling.
ECM/β1 integrin signaling is also important in the adult heart. Shai reported that adult cardiomyocte-specific β1 integrin knockout mice (MLC2V-Cre/Itgb1flox/flox) had myocardial fibrosis and congestive heart failure by 6 months of age (Shai et al., 2002). We found that proliferation of cardiac progenitors is regulated by interaction between ECM/β1 integrin and growth factors mediated through ERK1/2 and PI3K/Akt pathways. Activation of ERK1/2 and the expression of cyclinD1 (Ccnd1) and cyclinE1 (Ccne1) were downregulated, while cyclinG2 (Ccng2) and p21 (Cdkn1a) were upregulated in β1 integrin mutant hearts. In agreement with these results, the cyclinD1 promoter and its expression are regulated by ERK1/2 and JNK activities (Giancotti and Ruoslahti, 1999). Anchorage to the ECM is necessary for the down-regulation of the Cdk2 inhibitors, p21 and p27, and conditional deletion of β1 integrin in the mammary gland also revealed reduced mammary cell proliferation due to upregulation of p21 (Giancotti and Ruoslahti, 1999; Li et al., 2005). The recent report of proliferative defects upon cardiac deletion of focal adhesion kinase (FAK) (Peng et al., 2008), an important mediator between β1 integrin and ERK1/2 or PI3K/Akt, is consistent with our findings. However, the heart size of Nkx2.5 enhancer–Cre/Itgb1flox/flox mice were generally smaller than wild type littermates, but those of FAK knock-out mice were not. FGF signaling, also downstream of β1 integrin, is critical for heart size (Lavine et al., 2005) and therefore may account for this difference.
In conclusion, our findings demonstrate that cardiac fibroblasts express specific ECM and growth factors, which collaboratively promote proliferation of myocardial progenitors through β1 integrin signaling. Fibroblasts also play roles in various other processes, including skin epidermis formation and maintenance (Szabowski et al., 2000), mammary morphogenesis (Niranjan et al., 1995), lung epithelial cell proliferation and differentiation (Demayo et al., 2002), and cancer growth and metastasis (Kalluri and Zeisberg, 2006). Therefore, identification of the molecular mechanisms involved in the interaction between cardiomyocytes and fibroblasts may have a general impact on our understanding of tissue development, function, and disease.
Experimental Procedures
Generation of Nkx2.5-YFP and Nkx2.5 enhancer–Cre/Itgb1flox/flox mice
Nkx2.5-YFP mice were obtained by crossing Nkx2.5 cardiac enhancer–Cre mice and R26R-EYFP mice (McFadden et al., 2005; Srinivas et al., 2001). Nkx2.5 enhancer–Cre/Itgb1flox/flox mice were generated by crossing Nkx2.5 cardiac enhancer–Cre mice and Itgb1flox/flox mice (Graus-Porta et al., 2001).
Isolation of embryonic and adult cardiomyocytes
Cardiomyocytes were prepared from E12.5–13.5 mouse embryos by the conventional preplating method (Ieda et al., 2007). Briefly, hearts were minced and digested with collagenase type II (Worthington) solution. To enrich for cardiomyocytes, the cells were preplated for 2 h to remove nonmyocytes. For mixed population culture, cells were directly plated on culture dish without preplating. By preplating, the percentage of nonmyocytes was reduced from 40% to 10% as reported (Engel et al., 1999). In either case, cells were cultured in DMEM/M199 medium containing 10% FBS at a density of 104/cm2.
Adult cardiomyocytes were isolated from 8- to 12-week-old mice by Langendorff perfusion and Thompson’s procedure (O’Connell et al., 2007). For isolation of adult cardiac fibroblasts, hearts were digested with collagenase/dispase (Roche) solution and plated for 2 h. Attached fibroblasts were cultured for 7 days, treated with mitomycin C, and stored in liquid nitrogen.
Isolation of Nkx-YFP+ cells
To isolate Nkx-YFP+ cells, we began with 20–30, E12.5–13.5 Nkx2.5-YFP embryos. YFP+ ventricles were cut into small pieces and digested with collagenase type II solution. A single-cell suspension was obtained by gentle triturating and passing through a 40-μm cell strainer. Nkx-YFP+ live cells (as defined by the lack of propidium iodine staining) were isolated by FACS Diva flow cytometer and cell sorter (BD Biosciences). Embryonic cardiac fibroblasts (YFP− cells) were plated onto plastic dishes for 2 h, cultured for 7 days, and treated with mitomycin C. FACS-sorted Thy1+CD31− cells were treated with mitomycin C. Fibroblasts were stored in liquid nitrogen for the co-culture experiment.
Culture of Nkx-YFP+ cells
Nkx-YFP+ cells were cultured in DMEM/M199 medium containing 10% FBS at a density of 2×104/cm2 on noncoated, PLL-, fibronectin-, laminin-(Sigma Aldrich), collagen III-(BD Biosciences), periostin, or hapln1-(R&D) coated dishes according to the manufacturer’s protocols. In some experiments, cells were maintained in 0.1% FBS media either with or without viral infection for 24h, and stimulated with HBEGF (50 ng/ml), Ptn (1 μg/ml), or FGF2 (50 ng/ml, R&D). For co-culture experiments, mitomycin-treated cardiac fibroblasts or Thy1+CD31− cells were plated at 104/cm2, and Nkx-YFP+ cells (104/cm2) were added 24 h later and cultured for 3 days. An anti-β1 integrin-blocking antibody (10 μg/ml, BD Biosciences), PD98059 (5 μM), SB203580 (5 μM), or LY294002 (10 μM, Calbiochem) was added after a 1-day incubation in some experiments. Unless otherwise stated, cells were cultured on noncoated plates.
FACS analyses and sorting
For Thy1+ cell expression analyses and sorting, single cells were isolated from ICR mouse ventricles, incubated with APC-conjugated anti-Thy1 antibody (eBioscience), analyzed and sorted by FACS Diva with FlowJo software. For sorting of Thy1+CD31− cells from cardiac fibroblasts, APC-conjugated anti-Thy1 antibody and FITC-conjugated anti-CD31 antibody (eBioscience) were used. For vimentin and DDR2 expression analyses, Thy1+CD31− cells were fixed with BD Cytofix/Cytoperm Kit (BD Bioscience), stained with anti-vimentin and DDR2 antibodies, followed by secondary antibodies conjugated with Alexa 488 and 647, and analyzed on a FACS Calibur (BD Biosciences) with FlowJo software.
Histology
H&E and Masson-trichrome staining were performed on paraffin-embedded sections, according to standard practices. The hearts were sectioned longitudinally in 5-μm thickness near the central conduction system to show the 4 chambers. Trabecular and compact layers were determined by their morphology. For each slide stained with H&E, we defined trabecular myocardium as bundles of cardiomyocytes surrounded by endocardium that project across the lumen of the ventricular chamber, and compact myocardium as concentrically organized layers of tightly adherent cardiomyocytes not surrounded by endocardium that constitute the outer ventricular chamber wall. Five parts per section were randomly selected for measuring wall thickness. The proportion of the compact layer to the heart size was the ratio between the length of the compact myocardium and the longest diameter of the ventricle, each measured by Image J software. Data for each mouse were calculated from 10 serial sections, and we observed 4–5 mice in each group. For immunohistochemical studies, hearts or whole embryos were fixed in 4% paraformaldehyde overnight, and then embedded in OCT compound and frozen in liquid nitrogen. Hearts were cut longitudinally in 7-μm sections in the middle to show the four chambers. Sections were stained with primary antibodies against actinin, vimentin, BrdU, β1 integirn (Chemicon), fibronectin (Thermo Scientific), p-ERK, with secondary antibodies conjugated with Alexa 488 or 546, and DAPI. For BrdU labeling, pregnant mice were injected intraperitoneally with BrdU (100 μg/g body weight) 1 h before sacrifice. The numbers of BrdU+ and p-ERK+ cells relative to the total number of total nuclei were counted in randomly selected three fields per section. The datafor each mouse were calculated for 5–10 sections. The numbers of vimentin+ or DDR2+ cells per area were calculated as the ratio between the number of vimentin+ or DDR2+ cells and the myocardial area, measured by Image J software. The data for each mouse were calculated from 10 sections.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 15 min at room temperature, blocked, and incubated with primary antibody against α-actinin (Sigma Aldrich), vimentin (Progen), BrdU (Accurate), GFP (Invitrogen), Ki67 (Novocastra), DDR2, Raldh2 (Santa Cruz), Thy-1, CD31 (BD Biosciences), SM-MHC (Biomedical Technologies), p-ERK, p-p38MAPK or p-Akt (Cell Signaling), with secondary antibodies conjugated with Alexa 488 or 546 (Molecular Probes), and DAPI (Invitrogen). TUNEL staining was performed according to the manufacturer’s protocol (Roche). To determine cell number, actinin+ cardiomyocytes were counted in six randomly selected fields in triplicate. YFP+ cell area was measured by image J software. For BrdU labeling, cells were incubated with 10 μM BrdU (Roche) for the last 2 day. To determine p-ERK+ and p-p38MAPK+ cells, the percentage of cells with nuclear p-ERK and p-p38MAPK were counted. In each experiment, the numbers of immunopositve cells were counted in six randomly selected fields, and 500–1000 embryonic cardiomyocytes or fibroblasts were counted in total.
siRNA knockdown
Mitomycin-treated cardiac fibroblasts were transfected with siRNA against mouse Fn1, Col3a1, Fn1/Col3a1 or scramble siRNA (Dharmacon) with Lipofectamine 2000 (Invitrogen), according to the standard protocol. After a 1-day incubation, cells were used for co-culture experiment. qRT-PCR was performed after a 2-day incubation. We confirmed that the transfection efficiency was more than 90% by FACS Calibur with BLOCK-iT Fluorescent Oligo (Invitrogen).
Adenovirus infection
Adenoviruses were generated as described (Ross et al., 1998). YFP+ cells were infected with adenovirus expressing LacZ, β1A, or TAC-β1 for 24 h prior to the experiments. Viral titer was determined on all stocks using the Adeno-X titer kit (Clontech), and cells were infected at matched multiplicities of infection of 10 to maximize expressed protein, but prevent viral toxicity and detachment of cells. LacZ infection resulted in >90% transfection efficiency.
Microarray Analyses
Mouse genome-wide gene expression analyses were performed using Affymetrix Mouse Gene 1.0 ST Array. YFP+ cells were collected just after sorting, and embryonic, and adult cardiac fibroblasts were collected after plating for 2 h without further incubation. RNA was extracted using Trizol (Invitrogen). Microarray analyses were performed in triplicate from independent biologic samples, according to the standard Affymetrix Genechip protocol. Data were analyzed using the Affymetrix Power Tool (APT, version 1.8.5). Linear models were fitted for each gene on the sample group to derive estimated group effects and their associated significance using the limma package (Smyth, 2004) in R/Bioconductor. Moderated t-statistics and the associated p-values were calculated. P-values were adjusted for multiple testing by controlling for false-discovery rate using the Benjamini-Hochberg method. Gene annotations were retrieved from Affymetrix (version Nov 12, 2007). Differential gene expression was defined using the statistics/threshold combination. Genes differentially expressed >2 fold and p<0.05 were shown in Figure 3, Supplementary figure 3 and Supplementary figure 6F, and genes differentially expressed >1.2 fold or <0.8 fold, and p<0.05 were shown in Supplementary figure 6G.
Statistical analyses
Differences between groups were examined for statistical significance using Student’s t-test or ANOVA. P values of <0.05 were regarded as significant.
Other Experimental Procedures
Other Experimental Procedures are listed in the Supplementary files.
Supplementary Material
Acknowledgments
We are grateful to members of the Srivastava lab and to B.G. Bruneau for critical discussions and comments on the manuscript; to B. Taylor and G. Howard for editorial assistance and manuscript preparation; to C. Barker in the Gladstone genomics core, and to C. Miller, J. Wong, and J. Fish in the histology core; to V. Stepps and M. Bigos in the flow cytometry core for technical assistance; to R. Yeh for bioinformatic support for array data; and to L.F. Reichardt for providing Itgb1flox/flox mice and E.N. Olson for Nkx2.5 enhancer-Cre transgenic mice. D.S. is supported by grants from NHLBI/NIH, March of Dimes Birth Defects Foundation, and the California Institute of Regenerative Medicine and is an Established Investigator of the American Heart Association. M.I. is supported by a grant from Banyu Life Science Foundation International, and R.S.R. is supported by grants from NHLBI/NIH and the Veterans Administration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291:H1015–1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Demayo F, Minoo P, Plopper CG, Schuger L, Shannon J, Torday JS. Mesenchymal-epithelial interactions in lung development and repair: are modeling and remodeling the same process? Am J Physiol Lung Cell Mol Physiol. 2002;283:L510–517. doi: 10.1152/ajplung.00144.2002. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Engel FB, Hauck L, Cardoso MC, Leonhardt H, Dietz R, von Harsdorf R. A mammalian myocardial cell-free system to study cell cycle reentry in terminally differentiated cardiomyocytes. Circ Res. 1999;85:294–301. doi: 10.1161/01.res.85.3.294. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- Goldsmith EC, Hoffman A, Morales MO, Potts JD, Price RL, McFadden A, Rice M, Borg TK. Organization of fibroblasts in the heart. Dev Dyn. 2004;230:787–794. doi: 10.1002/dvdy.20095. [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Muller U. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudon-David F, Bouzeghrane F, Couture P, Thibault G. Thy-1 expression by cardiac fibroblasts: lack of association with myofibroblast contractile markers. J Mol Cell Cardiol. 2007;42:991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Ieda M, Kanazawa H, Kimura K, Hattori F, Ieda Y, Taniguchi M, Lee JK, Matsumura K, Tomita Y, Miyoshi S, et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med. 2007;13:604–612. doi: 10.1038/nm1570. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Kang JO, Sucov HM. Convergent proliferative response and divergent morphogenic pathways induced by epicardial and endocardial signaling in fetal heart development. Mech Dev. 2005;122:57–65. doi: 10.1016/j.mod.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang Y, Naylor MJ, Schatzmann F, Maurer F, Wintermantel T, Schuetz G, Mueller U, Streuli CH, Hynes NE. Beta1 integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. Embo J. 2005;24:1942–1953. doi: 10.1038/sj.emboj.7600674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Miragoli M, Gaudesius G, Rohr S. Electrotonic modulation of cardiac impulse conduction by myofibroblasts. Circ Res. 2006;98:801–810. doi: 10.1161/01.RES.0000214537.44195.a3. [DOI] [PubMed] [Google Scholar]
- Moro L, Venturino M, Bozzo C, Silengo L, Altruda F, Beguinot L, Tarone G, Defilippi P. Integrins induce activation of EGF receptor: role in MAP kinase induction and adhesion-dependent cell survival. Embo J. 1998;17:6622–6632. doi: 10.1093/emboj/17.22.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova G, Jabs N, Konstantinova I, Domogatskaya A, Tryggvason K, Sorokin L, Fassler R, Gu G, Gerber HP, Ferrara N, et al. The vascular basement membrane: a niche for insulin gene expression and Beta cell proliferation. Dev Cell. 2006;10:397–405. doi: 10.1016/j.devcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Niranjan B, Buluwela L, Yant J, Perusinghe N, Atherton A, Phippard D, Dale T, Gusterson B, Kamalati T. HGF/SF: a potent cytokine for mammary growth, morphogenesis and development. Development. 1995;121:2897–2908. doi: 10.1242/dev.121.9.2897. [DOI] [PubMed] [Google Scholar]
- O’Connell TD, Rodrigo MC, Simpson PC. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol. 2007;357:271–296. doi: 10.1385/1-59745-214-9:271. [DOI] [PubMed] [Google Scholar]
- Peng X, Wu X, Druso JE, Wei H, Park AY, Kraus MS, Alcaraz A, Chen J, Chien S, Cerione RA, Guan JL. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proc Natl Acad Sci U S A. 2008;105:6638–6643. doi: 10.1073/pnas.0802319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi DJ, Ballard VL, Mikawa T. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev Dyn. 2003;228:161–172. doi: 10.1002/dvdy.10360. [DOI] [PubMed] [Google Scholar]
- Ross RS, Borg TK. Integrins and the myocardium. Circ Res. 2001;88:1112–1119. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- Ross RS, Pham C, Shai SY, Goldhaber JI, Fenczik C, Glembotski CC, Ginsberg MH, Loftus JC. Beta1 integrins participate in the hypertrophic response of rat ventricular myocytes. Circ Res. 1998;82:1160–1172. doi: 10.1161/01.res.82.11.1160. [DOI] [PubMed] [Google Scholar]
- Sano M, Fukuda K, Kodama H, Pan J, Saito M, Matsuzaki J, Takahashi T, Makino S, Kato T, Ogawa S. Interleukin-6 family of cytokines mediate angiotensin II-induced cardiac hypertrophy in rodent cardiomyocytes. J Biol Chem. 2000;275:29717–29723. doi: 10.1074/jbc.M003128200. [DOI] [PubMed] [Google Scholar]
- Sastry SK, Lakonishok M, Wu S, Truong TQ, Huttenlocher A, Turner CE, Horwitz AF. Quantitative changes in integrin and focal adhesion signaling regulate myoblast cell cycle withdrawal. J Cell Biol. 1999;144:1295–1309. doi: 10.1083/jcb.144.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai SY, Harpf AE, Babbitt CJ, Jordan MC, Fishbein MC, Chen J, Omura M, Leil TA, Becker KD, Jiang M, et al. Cardiac myocyte-specific excision of the beta1 integrin gene results in myocardial fibrosis and cardiac failure. Circ Res. 2002;90:458–464. doi: 10.1161/hh0402.105790. [DOI] [PubMed] [Google Scholar]
- Smith TK, Bader DM. Signals from both sides: Control of cardiac development by the endocardium and epicardium. Semin Cell Dev Biol. 2007;18:84–89. doi: 10.1016/j.semcdb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolenski A, Schultess J, Danielewski O, Garcia Arguinzonis MI, Thalheimer P, Kneitz S, Walter U, Lohmann SM. Quantitative analysis of the cardiac fibroblast transcriptome-implications for NO/cGMP signaling. Genomics. 2004;83:577–587. doi: 10.1016/j.ygeno.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, Angel P. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 2000;103:745–755. doi: 10.1016/s0092-8674(00)00178-1. [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Weber KT, Brilla CG. Pathological hypertrophy and cardiac interstitium. Fibrosis and renin-angiotensin-aldosterone system. Circulation. 1991;83:1849–1865. doi: 10.1161/01.cir.83.6.1849. [DOI] [PubMed] [Google Scholar]
- Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Iwamoto R, Saeki K, Asakura M, Takashima S, Yamazaki A, Kimura R, Mizushima H, Moribe H, Higashiyama S, et al. Mice with defects in HB-EGF ectodomain shedding show severe developmental abnormalities. J Cell Biol. 2003;163:469–475. doi: 10.1083/jcb.200307035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [0–9] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.