Abstract
Recent genome-wide association studies (GWAS) have identified multiple risk loci for common obesity (FTO, MC4R, TMEM18, GNPDA2, SH2B1, KCTD15, MTCH2, NEGR1 and PCSK1). Here we extend those studies by examining associations with adiposity and type 2 diabetes in Swedish adults. The nine single nucleotide polymorphisms (SNPs) were genotyped in 3885 non-diabetic and 1038 diabetic individuals with available measures of height, weight and body mass index (BMI). Adipose mass and distribution were objectively assessed using dual-energy X-ray absorptiometry in a sub-group of non-diabetics (n = 2206). In models with adipose mass traits, BMI or obesity as outcomes, the most strongly associated SNP was FTO rs1121980 (P < 0.001). Five other SNPs (SH2B1 rs7498665, MTCH2 rs4752856, MC4R rs17782313, NEGR1 rs2815752 and GNPDA2 rs10938397) were significantly associated with obesity. To summarize the overall genetic burden, a weighted risk score comprising a subset of SNPs was constructed; those in the top quintile of the score were heavier (+2.6 kg) and had more total (+2.4 kg), gynoid (+191 g) and abdominal (+136 g) adipose tissue than those in the lowest quintile (all P < 0.001). The genetic burden score significantly increased diabetes risk, with those in the highest quintile (n = 193/594 cases/controls) being at 1.55-fold (95% CI 1.21–1.99; P < 0.0001) greater risk of type 2 diabetes than those in the lowest quintile (n = 130/655 cases/controls). In summary, we have statistically replicated six of the previously associated obese-risk loci and our results suggest that the weight-inducing effects of these variants are explained largely by increased adipose accumulation.
INTRODUCTION
Obesity is a major global health burden with a multitude of co-morbidities including type 2 diabetes, cardiovascular disease, certain cancers, sleep apnea, and osteoarthritis (1). Although the current obesity epidemic is likely related to decreased habitual physical activity levels and changes in dietary nutrient intake, heritability studies indicate that genetic factors also affect the predisposition to obesity (2). While several monogenic causes of obesity have been well-described during the past decade, the progress in defining the genetic basis of common obesity had been frustratingly slow prior to 2007; since then, obesity-predisposing variants in the FTO (3), MC4R (4,5), TMEM18, GNPDA2, SH2B1, KCTD15, MTCH2, NEGR1 (6) and PCSK1 (7) genes have emerged. All of these studies focused primarily on anthropometric measures of obesity, such as body mass index (BMI) or bioimpedance, with none having described associations with objectively assessed adipose distribution.
The purpose of the present study was to attempt to replicate the previously reported genetic associations with obesity in ethnically homogeneous cohorts from northern Sweden. We also sought to extend previous studies by examining genotype associations singly and in combination with measures of adipose mass, adipose distribution and type 2 diabetes.
RESULTS
Table 1 shows participant characteristics for the different cohorts studied here.
Table 1.
Participant characteristics
| Variable | All non-diabetic individuals, n or mean (SD) | Diabetic individuals, n or mean (SD) | Non-diabetic individuals with body scans, n or mean (SD) |
|---|---|---|---|
| Sex (n = M/F) | 1391/2494 | 609/429 | 516/1690 |
| Age (years) | 52.6 (9.5) | 53.8 (7.3) | 52.2 (10.3) |
| Height (m) | 1.68 (0.09) | 1.71 (0.10) | 1.67 (0.09) |
| Weight (kg) | 73.2 (13.5) | 88.4 (14.7) | 71.8 (13.5) |
| Body mass index (kg/m2) | 25.7 (4.0) | 30.4 (4.7) | 25.6 (4.1) |
| Total adipose mass (kg) | – | – | 25.1 (9.2) |
| Abdominal adipose mass (kg) | – | – | 1.52 (0.54) |
| Gynoid adipose mass (kg) | – | – | 2.53 (0.83) |
| Total lean mass (kg) | – | – | 44.2 (9.9) |
Genotype associations with measures of body composition and adipose distribution (continuous trait models)
Table 2 summarizes the adjusted mean level of BMI for each SNP. Six of the nine variants were statistically associated with BMI, with one additional SNP approaching statistical significance. The SNP most strongly associated with BMI localized to the FTO gene (rs1121980) (P < 0.0001). The MC4R rs17782313 SNP statistically interacted with sex (P = 0.02), whereby the minor allele was associated with higher BMI in women (β = 0.41 kg/m2 per copy of the minor allele; P = 0.0034), but no effect was observed in men (β = − 0.03 kg/m2 per copy of the minor allele; P = 0.83). Similar interaction effects were observed for total (P = 0.024) and abdominal (P = 0.046) adipose mass.
Table 2.
Adjusted mean body mass index (BMI) for nine purported obesity-risk variants in non-diabetic individuals (n = 3885)
| Nearest gene | SNP | Risk allele (frequency) | Adjusted means (95% CI) by genotype |
P-value | ||
|---|---|---|---|---|---|---|
| MM | Mm | mm | ||||
| FTO | rs1121980 | A (0.42) | 25.3 (25.1–25.6) | 25.8 (25.6–26.0) | 26.1 (25.8–26.4) | <0.0001 |
| SH2B1 | rs7498665 | C (0.40) | 26.0 (25.7–26.3) | 25.8 (25.6–26.0) | 25.4 (25.2–25.6) | 0.0006 |
| MTCH2 | rs4752856 | T (0.37) | 25.5 (25.3–25.7) | 25.8 (25.6–26.0) | 25.9 (25.5–26.2) | 0.025 |
| MC4R | rs17782313 | C (0.26) | 25.6 (25.4–25.7) | 25.8 (25.6–26.0) | 26.2 (25.7–26.6) | 0.013 |
| NEGR1 | rs2815752 | A (0.58) | 25.5 (25.2–25.8) | 25.6 (25.4–25.8) | 25.9 (25.7–26.1) | 0.023 |
| GNPDA2 | rs10938397 | G (0.37) | 25.6 (25.4–25.8) | 25.7 (25.5–25.9) | 26.0 (25.7–26.4) | 0.068 |
| TMEM18 | rs6548238 | A (0.82) | 25.8 (25.6–25.9) | 25.6 (25.3–25.8) | 25.2 (24.5–25.8) | 0.049 |
| KCTD15 | rs11084753 | G (0.63) | 25.7 (25.5–25.9) | 25.7 (25.5–25.9) | 25.6 (25.2–25.9) | 0.472 |
| PCSK1 | rs6235 | C (0.30) | 25.7 (25.5–25.9) | 25.7 (25.5–25.9) | 25.8 (25.4–26.3) | 0.588 |
M = major allele; m = minor allele; SNP = single nucleotide polymorphism. Means are adjusted for age and sex. P-values are from linear models assuming an additive mode of genetic inheritance.
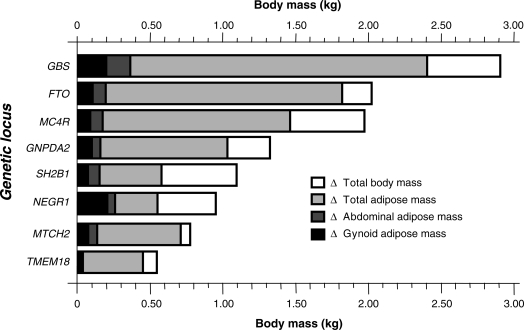
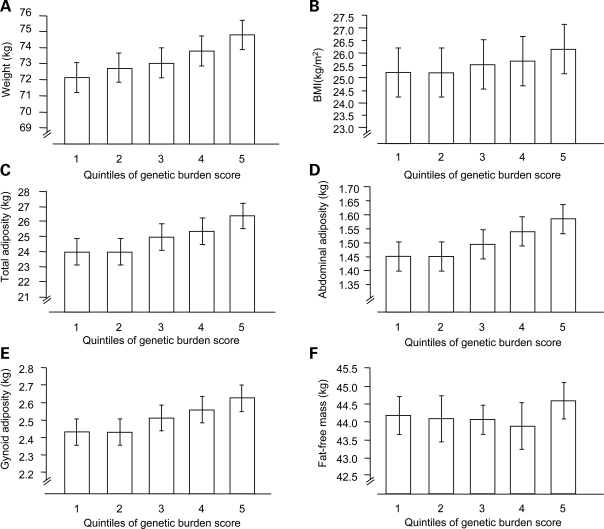
The FTO SNP (rs1121980) was also strongly associated with total, abdominal and gynoid adipose mass (Table 3). As in earlier studies (3), the per allele increase in weight for FTO rs1121980 was 1.03 kg (P = 0.005), of which 0.91 kg was attributable to increased total adipose mass (P = 0.0007). As shown in Table 3, although trends for association with measures of adiposity were evident for several of the remaining SNPs in directions consistent with the associations with obesity, only three reached a formal level of statistical significance (FTO, MTCH2 and GNPDA2). Figure 1 shows the absolute weight difference between major and minor allele homozygotes at each of the obesity-associated loci and between the first and fifth quintiles of the genetic burden score (GBS). The figure also illustrates how much of the total weight difference between genotypes at a given locus is attributable to adipose accumulation. When the overall contribution of these SNPs was assessed using the GBS, strong associations were evident for all anthropometric (Fig. 2A and B) and dual-energy X-ray absorptiometry (DEXA) measures of obesity (Fig. 2C–E). For example, when stratified into quintiles, those with the highest level of the GBS (Q5) weighed approximately 2.56 kg more than those with the lowest level of the GBS (Q1) (β = 0.87 kg per SD increase in score, P < 0.0001). Consistently, BMI was 0.97 kg/m2 higher (β = 0.32 kg/m2 per SD increase in the score, P < 0.0001), total adipose mass was 2.41 kg higher (β = 0.76 kg per SD increase in the score, P = 0.0001), abdominal adipose mass was 0.14 kg higher (β = 0.04 kg per SD increase in the score, P = 0.0004), and gynoid adipose mass was 0.19 kg higher (β = 0.06 kg per SD increase in the score, P = 0.0002) in the top versus bottom quintiles of the GBS.
Table 3.
Effect estimates and P-values for tests of association between nine purported obesity-risk variants and adipose traits measured using DEXA (n = 2206)
| Nearest gene | SNP | Total adipose mass (kg) |
Abdominal adipose mass (kg) |
Gynoid adipose mass (kg) |
|||
|---|---|---|---|---|---|---|---|
| β | P-value | β | P-value | β | P-value | ||
| FTO | rs1121980 | 0.91 | 0.0007 | 0.05 | 0.0008 | 0.07 | 0.004 |
| SH2B1 | rs7498665 | 0.32 | 0.24 | 0.02 | 0.17 | 0.03 | 0.28 |
| MTCH2 | rs4752856 | 0.38 | 0.16 | 0.03 | 0.04 | 0.04 | 0.12 |
| MC4R | rs17782313 | 0.46 | 0.15 | 0.02 | 0.20 | 0.02 | 0.36 |
| NEGR1 | rs2815752 | 0.23 | 0.41 | 0.02 | 0.14 | 0.03 | 0.18 |
| GNPDA2 | rs10938397 | 0.44 | 0.12 | 0.03 | 0.06 | 0.05 | 0.05 |
| TMEM18 | rs6548238 | 0.30 | 0.38 | 0.00 | 0.91 | 0.03 | 0.32 |
| KCTD15 | rs11084753 | 0.07 | 0.80 | 0.00 | 0.92 | 0.02 | 0.51 |
| PCSK1 | rs6235 | 0.15 | 0.62 | 0.00 | 0.95 | 0.01 | 0.62 |
| GBS (SD units) | – | 0.76 | 0.0001 | 0.04 | 0.0004 | 0.06 | 0.0002 |
β Indicates the difference in adipose mass per copy of the risk allele at each locus. Data are from separate generalized linear models, assuming an additive mode of genetic inheritance and adjusted for age and sex. GBS = genetic burden score.
Figure 1.
Differences in weight (kg) and total, abdominal and gynoid adipose mass (kg) between major and minor allele homozygotes at each of the seven nominally associated loci (P < 0.1) and between the first and fifth quintiles of the genetic burden score. Data are adjusted for age and sex (n = 2206). As described in the results, nominal gene–sex interactions were observed for total and abdominal adipose mass at the MC4R locus.
Figure 2.
Associations between the genetic burden score (expressed in quintiles) and obesity indices. Error bars are 95% confidence intervals. P-values are derived from linear regression models adjusted for age and sex. (A) P = 2.32 × 10−5; (B) P = 3.69 × 10−6; (C) P = 0.0001; (D) P = 0.0004; (E) P = 0.0002; (F) P = 0.099).
Genotype associations with obesity (categorical trait models)
The following results are derived from the cohort of non-diabetic individuals only (n = 3885). Table 4 shows obesity risk estimates (odds ratios; OR) for each of the genetic predictor variables ranked by magnitude of effect in non-diabetics. As with the BMI models reported above, the FTO rs1121980 variant was the most strongly associated SNP, with a per allele OR of 1.15 (95% CI 1.05–1.25; P = 0.0016). Five of the remaining SNPs (SH2B1, MTCH2, MC4R, NEGR1 and GNPDA2) were significantly associated with obesity in directions consistent with prior reports of association with BMI (6–8). For the GBS, the risk of obesity increased by 1.13 (95% CI 1.06–1.21; P = 0.0002) per SD unit increase, with a 1.40-fold (95% CI 1.14–1.72; P = 0.002) increased risk of being obese for those in the top compared with the bottom quintiles of the score. One SNP (MC4R rs17782313) statistically interacted with sex (P = 0.02), whereby the minor allele was associated with increased obesity risk in women (OR = 1.20; 95% CI 1.06–1.36; P = 0.0034), but no effect was observed in men (OR = 0.94; 95% CI 0.79–1.12; P = 0.48).
Table 4.
Estimates of obesity risk (odds ratios) and discriminative power (ROCAUCs) for nine purported obesity-risk variants singly and in combination in non-diabetic individuals (n = 3885)
| Nearest gene | SNP | Risk allele (frequency) | OR | 95% CI | P | ROCAUC | PROCAUC |
|---|---|---|---|---|---|---|---|
| FTO | rs1121980 | A (0.42) | 1.15 | (1.05–1.25) | 0.002 | 0.5435 | 2.00 × 10−4 |
| SH2B1 | rs7498665 | C (0.40) | 1.13 | (1.04–1.24) | 0.007 | 0.5348 | 1.72 × 10−6 |
| MTCH2 | rs4752856 | T (0.37) | 1.12 | (1.03–1.23) | 0.009 | 0.5257 | 1.10 × 10−7 |
| MC4R | rs17782313 | C (0.26) | 1.11 | (1.00–1.22) | 0.042 | 0.5293 | 1.04 × 10−6 |
| NEGR1 | rs2815752 | A (0.58) | 1.10 | (1.01–1.20) | 0.028 | 0.5263 | 2.21 × 10−7 |
| GNPDA2 | rs10938397 | G (0.37) | 1.10 | (1.00–1.20) | 0.045 | 0.5157 | 2.45 × 10−8 |
| TMEM18 | rs6548238 | A (0.82) | 1.10 | (0.98–1.22) | 0.095 | 0.5278 | 7.00 × 10−8 |
| KCTD15 | rs11084753 | G (0.63) | 1.05 | (0.96–1.14) | 0.33 | 0.5087 | 3.57 × 10−7 |
| PCSK1 | rs6235 | C (0.30) | 1.05 | (0.96–1.16) | 0.30 | 0.5221 | 1.20 × 10−6 |
| GBS (SD units) | – | – | 1.13 | (1.06–1.21) | <0.001 | 0.5752 | 2.00 × 10−5 |
| GBS (5th versus 1st quintile) | – | – | 1.40 | (1.14–1.72) | 0.002* | – | – |
For the calculation of odds ratios (OR), obesity is defined according to WHO (13,14). The area under the receiver operator characteristic curves (ROCAUC) reported in the table are unadjusted. PROCAUC is the significance level of the test for difference between the ROCAUC for each single nucleotide polymorphism (SNP) or the genetic burden score (GBS) versus the ROCAUC for all SNPs (0.6147). These comparisons test the ability of each genetic predictor to discriminate between 353 obese (BMI >30 kg/m2) and 1370 normal weight (BMI 18.5–24.9 kg/m2) individuals. As described in the results, gene–sex interactions were observed for obesity (P = 0.02) at the MC4R locus. *P-value obtained by direct comparison of fifth versus first GBS quintiles, adjusted for age and sex (n = 1325).
Comparison of genetic effects on obesity receiver-operating characteristic curves (ROCAUC models)
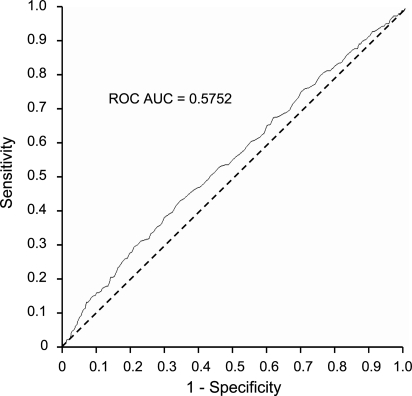
The power of each of the SNPs to discriminate between normal weight and obese individuals [i.e. the receiver-operating characteristic curves (ROCAUC)] was compared against the discriminative power of the GBS as the criterion variable (Table 4). The crude ROCAUC for the GBS was 0.5752 (Fig. 3) (P < 0.0001). This compared with a ROCAUC of 0.5881 (P < 0.0001) when all eight SNPs were entered simultaneously into the ROC model. The combination of all SNPs was a significantly more powerful discriminator than any single SNP, with or without adjustment for age and sex (P < 0.05). The FTO variant was the most discriminative of the individual SNPs (crude ROCAUC = 0.5435; P < 0.0001), with the crude ROCAUCs ranging from 0.5087 to 0.5348 for the remaining SNPs.
Figure 3.
Unadjusted receiver-operating characteristic curve showing the combined ability of the genetic burden score to discriminate between 353 obese (BMI >30 kg/m2) and 1370 normal weight (BMI 18.5–24.9 kg/m2) individuals.
Genetic effects on risk of type 2 diabetes
Table 5 shows the associations between each of the nine obesity-risk variants and type 2 diabetes. In models adjusted for age and sex, four variants were significantly associated with type 2 diabetes risk (FTO rs1121980: P = 0.013; MC4R rs17782313: P = 0.009; GNPDA2 rs10938397: P = 0.028; TMEM18 rs6548238: P = 0.007). A tendency for association with diabetes was observed for the SH2B1 rs7498665 variant (P = 0.06). After additional adjustment for BMI, none remained statistically significant. Although PCSK1 rs6235 was not significantly associated with diabetes in the age- and sex-adjusted models (P = 0.21), additional adjustment for BMI rendered a nominally significant protective effect for this variant (P = 0.04).
Table 5.
Estimates of type 2 diabetes risk (odds ratios) with and without adjustment for body mass index (BMI) for nine purported obesity-risk variants (n=1038 diabetes case and 3885 non-diabetic controls)
| Nearest gene | SNP | Risk allele (frequency) | Adjusted age, sex |
Adjusted age, sex, BMI |
||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |||
| FTO | rs1121980 | A (0.42) | 1.14 | (1.03–1.26) | 0.013 | 1.02 | (0.91–1.15) | 0.70 |
| SH2B1 | rs7498665 | C (0.40) | 1.11 | (1.00–1.23) | 0.06 | 1.04 | (0.92–1.17) | 0.57 |
| MTCH2 | rs4752856 | T (0.37) | 1.02 | (0.92–1.13) | 0.70 | 0.96 | (0.86–1.08) | 0.54 |
| MC4R | rs17782313 | C (0.26) | 1.17 | (1.04–1.31) | 0.009 | 1.10 | (0.97–1.26) | 0.15 |
| NEGR1 | rs2815752 | A (0.58) | 1.07 | (0.97–1.19) | 0.18 | 1.05 | (0.93–1.18) | 0.41 |
| GNPDA2 | rs10938397 | G (0.37) | 1.13 | (1.01–1.26) | 0.028 | 1.17 | (1.02–1.35) | 0.35 |
| TMEM18 | rs6548238 | A (0.82) | 1.21 | (1.05–1.39) | 0.007 | 1.14 | (0.97–1.33) | 0.11 |
| KCTD15 | rs11084753 | G (0.63) | 1.00 | (0.90–1.12) | 0.94 | 1.01 | (0.90–1.14) | 0.82 |
| PCSK1 | rs6235 | C (0.30) | 0.93 | (0.83–1.04) | 0.21 | 0.87 | (0.77–0.99) | 0.04 |
The risk of type 2 diabetes was significantly increased across the spectrum of the GBS following adjustment for age and sex (linear P < 0.0001). For each SD unit increase in the score, the risk of diabetes increased significantly (OR = 1.17; 95% CI 1.09–1.27), with those in the highest quintile of the GBS (n = 193/594 cases/controls) being at 1.55 (95% CI 1.21–1.99) greater risk of type 2 diabetes than those in the lowest quintile (n = 130/655 cases/controls). When all SNPs were simultaneously entered into the model, the unadjusted ROCAUC for type 2 diabetes was 0.5643 (P < 0.0001), which was significantly larger than any of the individual SNP ROCAUCs. The ROCAUC for the GBS was 0.5524, which did not differ from the model containing all SNPs (P difference = 0.14). Additional adjustment for BMI attenuated the association, with those in the highest quintile of the score no longer being at significantly greater risk of diabetes relative to those in the lowest quintile (OR 1.22; 95% CI 0.92–1.62). The ROCAUC for the model including age and sex (ROCAUCs = 0.6382) improved significantly when the nine SNPs were added to the model (ROCAUCs = 0.6654) (P < 0.0001). However, BMI explained all the predictive power attributable to the SNPs (ROCAUCs: age, sex and BMI = 0.8236 versus age, sex, BMI and SNPs = 0.8236).
DISCUSSION
In this study we examined the associations of nine recently discovered obesity loci with anthropometric and DEXA-derived measures of adipose mass, adipose distribution and type 2 diabetes risk in adults from northern Sweden. The previously reported genetic associations for these variants with obesity replicated for six loci. To assess the overall burden conveyed by the variants, we composed a weighted genetic risk score and show that this is strongly associated with obesity and the accumulation of total, abdominal and gynoid adipose tissue. Because obesity is an established risk factor for type 2 diabetes, it is important to determine whether the increased genetic predisposition to obesity translates to increased type 2 diabetes risk, which we were able to confirm for several variants. All associated variants clearly conveyed their effects via obesity.
During the past two decades, a great number of studies have been published purporting to have identified gene variants that predispose to obesity [as summarized in (9)].The credibility and relevance of these studies is indicated by the success of replication attempts within and beyond the geographic and ethnic groups from which the original reports hailed. Unfortunately, very few of these earlier reports have been adequately replicated. Thus, the identification of the FTO locus in 2007 (3) and the subsequent discovery of several other genetic risk factors for obesity (4–7) marked an important breakthrough in comprehending the genetic basis of common obesity. However, the extension of the initial studies' findings to more detailed and complex disease phenotypes is necessary if the clinical relevance of genetic variation is to be ascertained. Figure 1 illustrates the total difference in body mass between major and minor allele homozygotes at each of the obesity-associated loci, with the weight difference attributable to total, abdominal and gynoid adipose tissue overlaid. While these data illustrate that the weight differences observed for all of the variants are largely related to increased adiposity, for some loci fairly large proportions of the weight difference between homozygotes is attributable to lean mass. For example, the FTO, MCTH2 and TMEM18 variants were predominantly associated with adipose accumulation, whereas the variances in weight for the SH2B1 and NEGR1 variants appear to be explained to a larger extent by the accumulation of non-adipose tissue (e.g. bone, muscle and organ tissue). These characteristics may be of relevance when considering the importance of these variants in cardiovascular and metabolic disease etiology. This is because the health risks associated with weight gain vary depending on whether fat or lean tissue is the source of the accumulated weight.
For one variant (MC4R rs17782313), nominally significant interactions with sex were observed, where the associations between the genotypes and obesity measures were consistent with prior reports in females, but were inconclusive in males. None of the interaction P-values was robust to correction for multiple statistical comparisons, suggesting that these observations may be false-positive. Furthermore, no prior evidence of sex-effects at this locus has been reported. Nonetheless, it is important to highlight that some individuals in this cohort who underwent DEXA scans did so because they were athletes, had sustained recent injuries, or were concerned about skeletal health (10). Whether differences in the distributions of such factors by sex could underlie the interaction effects observed here is difficult to quantify owing to a lack of detailed information on these parameters.
Although during recent years the prevalence of obesity in northern Sweden has been rising at a similar rate to its European neighbors, the incidence of type 2 diabetes has remained relatively stable in Sweden during this time (11). However, the present study suggests that the genetic basis of obesity in people from Västerbotten is similar to other geographic populations. Thus, studies which seek to determine the mechanisms that protect this population against the diabetes-inducing effects of obesity might yield important insights into diabetes prevention that are relevant to other populations. In this context, studies focusing on gene–environment interactions may help elucidate why the relationship between obesity and diabetes differs between populations.
In conclusion, we have replicated the genotype associations with obesity for six (FTO rs1121980, SH2B1 rs7498665, MTCH2 rs4752856, MC4R rs17782313, NEGR1 rs2815752, GNPDA2 rs10938397) of nine previously associated loci. We also quantified the combined effect of these variants on obesity risk and described the extent to which the associations with obesity are attributable to increased total and regional adipose mass. Furthermore, we illustrate the degree to which these variants increase the risk of type 2 diabetes, showing that this risk is largely dependent on BMI. These extensions of the original findings provide further insight into the role common genetic variation plays in human obesity and the associated risk of metabolic disease. However, it is also evident that the effect of these variants on obesity and type 2 diabetes risk is fairly modest. Thus, it is unlikely that in its present form this information will add substantially to current risk prediction algorithms for these diseases.
MATERIALS AND METHODS
Participants
The 4923 individuals included in this report were white Swedish adults living within the county of Västerbotten in northern Sweden who had participated in the Northern Swedish Health and Disease Study (NSHED), a prospective cohort study of common diseases of later life (12). Participant characteristics are shown in Table 1. All living participants provided written informed consent. Permission to undertake genetic analyses in these materials was obtained from the research ethics committee of Umeå University.
Measures of obesity
Anthropometric measures were collected as part of NSHED by trained research nurses using a protocol standardized across study centers (12). Height and weight were measured using a calibrated wall-mounted stadiometer and scales, respectively. BMI was calculated as weight in kilograms divided by height in meters-squared (kg/m2). Obesity is defined according to WHO (13). In a subgroup of 2206 individuals, DEXA scans were performed for the assessment of total and regional adipose distribution (Lunar DPX-L, IQ, or Prodigy IV, GE Lunar, WI, USA), the methods for which have been described in detail previously (10). Briefly, participants dressed in lightweight clothing free of metallic parts, were instructed to lie flat on the DEXA table as the scanning arm passed over their body. DEXA scanners were calibrated daily with body mass phantoms according to the manufacturer's protocols.
Diabetes diagnosis
To test whether the obesity variants were associated with type 2 diabetes, we genotyped 1038 people with clinically manifest type 2 diabetes. The type 2 diabetes ascertainment procedures included clinical chart review and an independent oral glucose tolerance test as described in detail previously (15). Participant characteristics are shown in Table 1.
Genetic analyses
DNA was extracted at the Medical Biobank in Umeå from peripheral white blood cells. Genomic DNA samples were subsequently diluted to 4 ng/µl. Taqman MGB chemistry (Applied Biosystems, Foster City, CA, USA) was used for all genotyping assays in accordance with the recommended protocol (16). Genotyping success and concordance rates were >94% and >99% for all single nucleotide polymorphisms (SNPs), respectively.
Genetic burden score
A score summarizing the overall obesity burden conveyed by each of the eight variants replicate by the GIANT consortium was computed by weighting each variant [i.e. by multiplying the genotype at each locus by its per allele effect on BMI reported in the original meta-analysis; see Table 4 for reference (6)] and then summing-up these values.
Statistical analysis
All statistical analyses were undertaken using the SAS software (v9.1, SAS Institute, Cary, NC, USA). Haploview v4.0 (http://www.broad.mit.edu/mpg/haploview) was used to determine Hardy–Weinberg equilibrium (HWE). All genotypes fulfilled HWE expectations (P > 0.01). Generalized linear models were used to test genotype associations for continuous outcome traits (i.e. height, weight, BMI and DEXA measures of adiposity). Logistic regression was used to assess the association between each variant and obesity or type 2 diabetes as binary variables defined according to the definitions of the World Health Organization and ADA (13,14). All models were adjusted for age and sex and assumed an additive mode of inheritance. For anthropometric outcome variables, interaction tests were performed to determine whether the genetic effects differed by sex. This was achieved by fitting a term for genotype (0,1,2) × sex (0,1) to the regression models. For the models where obesity or type 2 diabetes were the outcomes, the area under the ROCAUC for each variant was calculated and compared with the ROCAUC derived from a model including all genotypes simultaneously. These analyses were performed using the methods described by DeLong et al. (17).
SUPPLEMENTARY MATERIAL
FUNDING
Novo Nordisk (370579201 to P.W.F.); the Swedish Heart-Lung Foundation (20070633 to P.W.F.); the Swedish Diabetes Association (DIA2006-013 to P.W.F.) and the Wellcome Trust (F.P. and I.B.). E.C.B. was supported by a studentship from the Portuguese Foundation for Science and Technology and P.W.F. in part by funding from Västerbotten's Health Authority (ALF strategic appointment 2006–2009). Funding to Pay the Open Access Charge was provided by Swedish Diabetes Association.
Supplementary Material
ACKNOWLEDGEMENTS
We thank those who participated in this study, Kerstin Enquist and Thore Johansson for their expert technical assistance with DNA extraction, and Kathleen Jablonski for statistics advice. We also thank the staff of the Sports Medicine Unit at Umeå University for undertaking the DEXA scans, the staff of the Västerbotten Intervention Program for additional data collection, and members of the Genetic Investigation of ANthropometric Traits (GIANT) consortium for providing pre-publication access to locus information.
Conflict of Interest statement: I.B. owns stock in Glaxo-Smith-Kline (GSK) and Incyte (INCY), in values greater than 10,000 USD. None of the other authors have anything to declare.
REFERENCES
- 1.Must A., Spadano J., Coakley E.H., Field A.E., Colditz G., Dietz W.H. The disease burden associated with overweight and obesity. JAMA. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard C., Tremblay A., Despres J.P., Nadeau A., Lupien P.J., Theriault G., Dussault J., Moorjani S., Pinault S., Fournier G. The response to long-term overfeeding in identical twins. N. Engl. J. Med. 1990;322:1477–1482. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- 3.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 5.Loos R.J., Lindgren C.M., Shengxu L., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. for the Genetic Investigation of ANthropometric Traits (GIANT) consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benzinou M., Creemers J.W., Choquet H., Lobbens S., Dina C., Durand E., Guerardel A., Boutin P., Jouret B., Heude B., et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 8.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 9.Rankinen T., Zuberi A., Chagnon Y.C., Weisnagel S.J., Argyropoulos G., Walts B., Perusse L., Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 10.Wiklund P., Toss F., Weinehall L., Hallmans G., Franks P.W., Nordstrom A., Nordstrom P. Abdominal and Gynoid Fat Mass Are Associated with Cardiovascular Risk Factors in Men and Women. J. Clin. Endocrinol. Metab. 2008;93:4360–4366. doi: 10.1210/jc.2008-0804. [DOI] [PubMed] [Google Scholar]
- 11.Eliasson M., Lindahl B., Lundberg V., Stegmayr B. No increase in the prevalence of known diabetes between 1986 and 1999 in subjects 25–64 years of age in northern Sweden. Diabet. Med. 2002;19:874–880. doi: 10.1046/j.1464-5491.2002.00789.x. [DOI] [PubMed] [Google Scholar]
- 12.Hallmans G., Agren A., Johansson G., Johansson A., Stegmayr B., Jansson J.H., Lindahl B., Rolandsson O., Soderberg S., Nilsson M., et al. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand. J. Public Health Suppl. 2003;61:18–24. doi: 10.1080/14034950310001432. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization. Obesity: Preventing and Managing the Global Epidemic. Geneva: World Health Organization; 2000. Technical report series 894. [PubMed] [Google Scholar]
- 14.American Diabetes Association. Clinical practice recommendations 2002. Diabetes Care. 2002;25:S1–S147. doi: 10.2337/diacare.25.2007.s1. [DOI] [PubMed] [Google Scholar]
- 15.Franks P.W., Rolandsson O., Debenham S.L., Fawcett K.A., Payne F., Dina C., Froguel P., Mohlke K.L., Willer C., Olsson T., et al. Replication of the association between variants in WFS1 and risk of type 2 diabetes in European populations. Diabetologia. 2008;51:458–463. doi: 10.1007/s00125-007-0887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranade K., Chang M.S., Ting C.T., Pei D., Hsiao C.F., Olivier M., Pesich R., Hebert J., Chen Y.D., Dzau V.J., et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.