Abstract
We investigated the involvement of rare (<1%) copy number variants (CNVs) in 471 cases of schizophrenia and 2792 controls that had been genotyped using the Affymetrix GeneChip® 500K Mapping Array. Large CNVs >1 Mb were 2.26 times more common in cases (P = 0.00027), with the effect coming mostly from deletions (odds ratio, OR = 4.53, P = 0.00013) although duplications were also more common (OR = 1.71, P = 0.04). Two large deletions were found in two cases each, but in no controls: a deletion at 22q11.2 known to be a susceptibility factor for schizophrenia and a deletion on 17p12, at 14.0–15.4 Mb. The latter is known to cause hereditary neuropathy with liability to pressure palsies. The same deletion was found in 6 of 4618 (0.13%) cases and 6 of 36 092 (0.017%) controls in the re-analysed data of two recent large CNV studies of schizophrenia (OR = 7.82, P = 0.001), with the combined significance level for all three studies achieving P = 5 × 10−5. One large duplication on 16p13.1, which has been previously implicated as a susceptibility factor for autism, was found in three cases and six controls (0.6% versus 0.2%, OR = 2.98, P = 0.13). We also provide the first support for a recently reported association between deletions at 15q11.2 and schizophrenia (P = 0.026). This study confirms the involvement of rare CNVs in the pathogenesis of schizophrenia and contributes to the growing list of specific CNVs that are implicated.
INTRODUCTION
Schizophrenia (SZ, MIM 181500) is a chronic mental disorder with a lifetime risk of ∼1% and a strong genetic component, indicated by heritability estimates of up to 85% and a ∼10-fold elevated recurrence risk in first-degree relatives (1). The best-established molecular genetic finding in schizophrenia is association with the 22q11.2 deletion syndrome, with 20–30% of adult carriers of this copy number variant (CNV) having schizophrenia (2). Reciprocally, the 22q11.2 deletion is found in 0.2–0.6% of patients with schizophrenia, a rate significantly higher than in the general population (3–5). Recently, a number of studies have shown that other rare CNVs contribute to schizophrenia (4–8), with the two largest studies (4,5) being adequately powered to identify two specific CNVs that are significantly associated with risk: deletions at 1q21.1 and 15q13.3.
CNVs have also been confirmed to be pathogenic in other neurodevelopmental disorders, most notably in autism, where an increase in the rate of de novo CNVs was established (9) and several recurrent CNVs were shown to increase risk (10–13).
In this study, we sought to identify novel schizophrenia risk CNVs in a sample of 520 patients with schizophrenia or schizoaffective disorder and in the 2998 controls used in the Wellcome Trust Case Control Consortium (WTCCC) study (14). We used Affymetrix 250K NspI and StyI arrays that provide signal intensity data on ∼500 000 single-nucleotide polymorphisms (SNPs). To reduce the false-positive discovery rate, we restricted the analysis to CNVs of >100 kb size, which were independently verified on both arrays with at least 10 SNPs on each array (see Methods), and validated all relevant CNVs with a second platform: Agilent 4×44K arrays.
RESULTS
Of the 520 patients and 2998 controls that had been analysed with Affymetrix microarrays, 471 cases and 2792 controls survived our filtering for high interquartile of the log2ratio (IQR) and for being outliers for high numbers of CNVs, that are likely to be false-positives (see Materials and Methods).
The overall CNV rate per person was similar in cases (1.73 CNVs) and controls (1.76 CNVs), of which 0.56 CNVs/case and 0.60 CNVs/control were deletions. In keeping with previous studies (4), we filtered out common CNVs (defined as present in >1% in the sample) including CNVs that overlapped >50% of their length with those CNVs. This was achieved using PLINK (15). The list of CNVs meeting these criteria is available in the Supplementary Material, which can be uploaded to UCSC as ‘custom tracks’ and visualized against the annotated genome in Build 36 (hg18).
In Table 1, we present the numbers of rare CNVs in cases and controls, stratified by size. Only very large CNVs >1 Mb in size showed a significant excess in cases (odds ratio, OR = 2.26, P = 0.00027), a finding mainly driven by a highly significant excess of deletions (OR = 4.53, P = 0.00013) although there was also a nominally significant excess of large duplications (OR = 1.71, P = 0.04). The association to the large deletion category was also significant after correction for the eight independent secondary tests (four size intervals for each of deletion and duplication; P = 0.001).
Table 1.
Deletions and duplications in cases and controls in different size ranges
| CNV type, size (kb) | Cases | Controls | CNVs per case/control | Case/control ratio | Permutation P–value |
|---|---|---|---|---|---|
| All deletions | 97 | 573 | 0.21/0.21 | 1 | 0.5 |
| All duplications | 153 | 832 | 0.32/0.30 | 1.09 | 0.18 |
| All deletions+duplications | 250 | 1405 | 0.53/0.50 | 1.05 | 0.23 |
| Deletions <200 | 32 | 267 | 0.068/0.096 | 0.71 | 0.98 |
| Duplications <200 | 38 | 244 | 0.081/0.087 | 0.92 | 0.67 |
| Total <200 | 70 | 511 | 0.15/0.18 | 0.81 | 0.95 |
| Deletions 200–500 | 47 | 257 | 0.1/0.092 | 1.08 | 0.32 |
| Duplications 200–500 | 73 | 386 | 0.16/0.14 | 1.12 | 0.21 |
| Total 200–500 | 120 | 643 | 0.25/0.23 | 1.11 | 0.17 |
| Deletions 500–1000 | 5 | 32 | 0.011/0.011 | 0.93 | 0.63 |
| Duplications 500–1000 | 23 | 135 | 0.049/0.048 | 1.01 | 0.52 |
| Total 500–1000 | 28 | 167 | 0.059/0.060 | 0.99 | 0.54 |
| Deletions >1000 | 13 | 17 | 0.028/0.0061 | 4.53 | 0.00013 |
| Duplications >1000 | 19 | 67 | 0.04/0.024 | 1.68 | 0.04 |
| Total >1000 | 32 | 84 | 0.068/0.03 | 2.26 | 0.00027 |
P-values are one-sided on comparing the rate of CNVs in cases versus controls and are based on 10 000 permutations (or 1 000 000, for the significant ones, which are shown in bold).
As CNVs >1 Mb constitute the only group that is significantly enriched in schizophrenia in this study, a number of them could be pathogenic. We present their details in Table 2. The genes within these intervals are listed in Table S1, Supplementary Material.
Table 2.
Positions of rare CNVs >1 Mb in cases
| Sample ID | Chr | Start (bp) | End (bp) | Size (kb) | N genes | Times found in controls |
|---|---|---|---|---|---|---|
| Deletions | ||||||
| 19336A5 | 2 | 139150475 | 140198728 | 1048 | 2 | |
| 20577A5 | 3 | 17179123 | 20586352 | 3407 | 7 | |
| 19326C3 | 5 | 165712 | 1196298 | 1031 | 16 | |
| 19338E3 | 6 | 48110720 | 49298238 | 1188 | 1 | |
| 19339E4 | 7 | 137845291 | 139317586 | 1472 | 15 | |
| 19328D4a | 7 | 157744094 | 158798338 | 1054 | 5 | |
| 19338A2 | 10 | 12673372 | 14643305 | 1970 | 11 | |
| 19325A4 | 17 | 14048304 | 15357533 | 1309 | 8 | |
| 19338F6 | 17 | 14048304 | 15357533 | 1309 | 8 | |
| 19338B6 | 22 | 17275227 | 19791017 | 2516 | 43 | |
| 19326F4 | 22 | 17275227 | 19791017 | 2516 | 43 | |
| 20579B3 | X | 323881 | 2726346 | 2402 | 13 | |
| 19339D3 | X | 6505282 | 8051350 | 1546 | 4 | 3 |
| Duplications | ||||||
| 19338A5 | 1 | 17118758 | 18187208 | 1068 | 12 | |
| 19326C3b | 1 | 144964403 | 146281318 | 1317 | 11 | Two deletionsb |
| 20577F4 | 2 | 106275670 | 107792241 | 1517 | 1 | |
| 20579B4 | 3 | 58527369 | 60146377 | 1619 | 4 | 2 |
| 19336D3 | 7 | 47255074 | 49037822 | 1783 | 9 | |
| 19338B1a | 7 | 157623546 | 158798338 | 1175 | 5 | |
| 19337A5 | 8 | 136873012 | 138138546 | 1266 | – | |
| 19325E5 | 12 | 33576567 | 34650850 | 1074 | 1 | 4 |
| 19337F4 | 13 | 22466197 | 23791694 | 1325 | 8 | 2 |
| 19339B1 | 14 | 80656427 | 82414847 | 1758 | 4 | |
| 19326C3 | 14 | 104812400 | 106318151 | 1506 | 8 | |
| 19339G4 | 15 | 21246055 | 26196279 | 4950 | 13 | |
| 19335E2c | 15 | 28748073 | 30231488 | 1483 | 6 | |
| 19327C3 | 16 | 15032942 | 16189808 | 1157 | 11 | 6 |
| 19327F2 | 16 | 15032942 | 16189808 | 1157 | 11 | 6 |
| 20577B1 | 16 | 15032942 | 16189808 | 1157 | 11 | 6 |
| 19338A5 | 18 | 26180575 | 27565032 | 1384 | 9 | |
| 19326C1 | X | 281199 | 2726346 | 2445 | 13 | 4 |
| 19325H5 | X | 6505282 | 8051350 | 1546 | 4 | 6 |
The column ‘N genes’ shows the number of genes intersecting them, according to the UCSC table browser for all RefSeq genes, as made available in PLINK (15) (downloaded on 24 July 2008). A full list of the genes within the CNVs are given in Table S1, Supplementary Material. The last column shows the number of controls who had identical CNVs.
aThis CNV is present once as a duplication and once as a deletion in cases, but not in any controls.
bThis CNV overlaps the previously identified schizophrenia susceptibility locus 1q21.1 (see Table 3), which was found as a deletion in two controls, whereas one case had the reciprocal duplication.
cThis duplication overlaps the previously identified schizophrenia susceptibility locus 15q13.3 (see Table 3). We find it only as a duplication in one case.
Of the large deletions observed in cases, most of them were single observations making it difficult to draw conclusions concerning their involvement in pathogenesis, even if they were not seen in controls. However, two were observed twice in patients and not in controls, making them nominally significantly associated with schizophrenia (Fisher's exact test, P = 0.02). The first one is the 22q11.2 deletion, which is already known to be associated with schizophrenia (2,3,4,5,16). The second one is a deletion of 17p12 at 14.0–15.4 Mb. One of the probands with the 17p12 deletion was part of a multiply affected family, in which one parent and three siblings (including the proband) had schizophrenia and another sib had major depressive disorder. CNV analysis of all available family members (performed with Agilent arrays; see Materials and Methods) showed that the affected parent, and three of the four sibs with a major psychiatric disorder, also carried the deletion. The absence of the deletion in one schizophrenic member in this family is not compatible with the hypothesis that most risk in this family is attributable to a fairly highly penetrant CNV, as is the existence of a sibling of the unaffected parent with a possible diagnosis of schizophrenia.
Of the large duplications in Table 2, only one was found in more than one patient. It is on 16p13.1, between 15.0 and 16.2 Mb, and was present in three cases and six controls. In addition, one control was deleted for this region, giving a combined burden of CNVs in this region of 0.6% in cases and 0.25% in controls, a non-significant trend (Fisher's exact test, P = 0.17, OR = 2.55). This region has been reported as a susceptibility factor for autism when duplicated (12) and for mental retardation when deleted (12,17).
Another intriguing duplication in a schizoaffective patient spanned 5 Mb, between 21.2 and 26.2 Mb. This covers the Prader–Willi/Angelman Syndrome (PWS/AS) critical region. No controls had deletions or duplications of this interval (Fisher's exact test, P = 0.15).
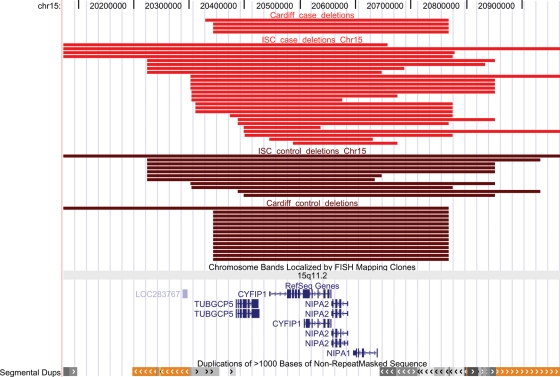
Of the susceptibility CNVs reported in the recent large studies (4,5), two were found in both of these studies (at 1q21.1, between 144.9 and 146.3 Mb, and at 15q13.3, between 28.7 and 30.3 Mb), whereas one at 15q11.2 was reported only in the study of Stefansson et al. (5) and has not as yet been replicated. We find a non-significant trend for support for this locus, as it was deleted in 0.82% in cases and in 0.50% in controls in the current study (OR = 1.7, n.s.). This finding is strengthened, as one of the cases had a sibling affected with schizophrenia, who was found (with Agilent arrays) to also have inherited this deletion from the unaffected mother. The region was filtered out in the ISC study (4), as CNVs in this region (deletions and duplications) were found in >1% of the sample. We have now re-analysed the unfiltered ISC data. As there were a number of different-sized CNVs in this region, we limited the analysis to the region flanked by segmental duplications, which is also the region covered by genes, i.e. between 20.35 and 20.65 Mb (Fig. 1), and selected only those deletions that intersected any part of this region. We identify a more than 2-fold excess of deletions in this locus in cases over controls in the ISC study: 26 versus 11, which is nominally significant on its own (P = 0.03, OR = 2). In the combined Cardiff and ISC studies, the rate of deletions affecting this locus is 0.78% versus 0.4%, in cases and controls, respectively (P = 0.026).
Figure 1.
Positions of the deletions in 15q11.2, between 20.35 and 20.6 Mb (the interval containing genes), that were identified in the current (Cardiff) and ISC studies (4). Red, cases; brown, controls. The positions of genes and segmental duplications in the region are also shown. Produced with the UCSC Genome Browser (http://www.genome.ucsc.edu).
We found no deletions in cases in either of the other two implicated loci (1q21.1 or 15q13.3). However, we confirm the very low rate of deletions in both loci in our control population (Tables 2 and 3). Analysis of each of the three CNVs identified previously (4,5) across all these three data sets (including the present one) revealed highly significant (P < 10−7) evidence for association for all three loci (Table 3). Thus, all three CNVs can be regarded as susceptibility factors for schizophrenia.
Table 3.
Results for the three candidate loci identified in the two previous large schizophrenia studies (4,5) and the current one
| 1q21.1: 144.9–146.3 |
15q11.2: 20.3–20.8 |
15q13.3: 28.7–30.3 |
||||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| Stefansson et al. (5) | 11 of 4718 | 8 of 41199 | 26 of 4718 | 79 of 41194 | 7 of 4213 | 8 of 39800 |
| ISC (4) | 10 of 3391 | 1 of 3181 | 26 of 3391 | 11 of 3181 | 9 of 3391 | 0 of 3181 |
| Cardiff study (current) | 0 of 471 | 2 of 2792 | 4 of 471 | 14 of 2792 | 0 of 471 | 0 of 2792 |
| Totala | 17 of 7918 | 11 of 46502 | 49 of 7918 | 103 of 46497 | 15 of 7413 | 8 of 45103 |
| OR | 9.1 (4.2–19.4) | 2.8 (2.0–3.9) | 11.4 (4.8–27.0) | |||
| P-value (Fisher's exact test) | 2.52 × 10−8 | 4.46 × 10−8 | 2.81 × 10−8 | |||
| Totala (Icelandic controls excluded) | 17 of 7918 | 3 of 14060 | 49 of 7918 | 45 of 14055 | 15 of 7413 | 1 of 12661 |
| OR | 10.0 (2.9–34.2) | 1.94 (1.3–2.9) | 25.7 (3.4–194.4) | |||
| P-value (Fisher's exact test; Icelandic controls excluded) | 9.6 × 10−6 | 0.0017 | 3.34 × 10−6 | |||
aThe total excludes overlaps from the sample from Aberdeen (Scotland) that was analysed in both ISC (4) and Stefansson et al. (5) studies. P-values are two-tailed, based on Fisher's exact test and are presented separately for the whole sample, and (bottom row) after excluding the 32 442 Icelandic controls, which could potentially bias the sample because of their disproportionally greater number.
DISCUSSION
In this study, we confirm earlier observations of an increased rate of large CNVs in cases of schizophrenia. We find more than a 4-fold increase of deletions that are more than 1 Mb in size. Deletions, as a general class, have previously been reported to be in excess in schizophrenia (7,8), but so far the strongest evidence comes from the ISC study (4) which reported that deletions larger than 500 kb were 1.67 times more common in patients. Although this was not the most enriched category in our study, we nevertheless find an excess at the same threshold of >500 kb, with a 2.18-fold increase in cases. Unlike the larger ISC study (4), we do not find an overall increase for all CNVs combined or for duplications in the 100–200 kb range. This may reflect the reduced power of our study, because of a smaller sample size, or a reduced sensitivity to detect smaller CNVs with the Affymetrix 500K platform, compared with the Affymetrix 5.0 or 6.0 that were used in the ISC (4).
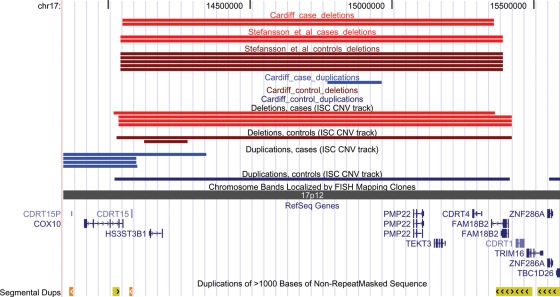
Two of the large deletions are in the 22q11.2 region that causes VCFS/DiGeorge Syndrome when deleted and has been confirmed in multiple studies as a genuine susceptibility factor for psychosis and autism (2–5,16). Another large deletion that was found in two schizophrenia cases but not in controls is on 17p12. The region affected by the 17p12 deletion, and the genes covered by it, are shown in Figure 2. To confirm the provisional association with the 17p12 deletion, we examined the data from the two recent large CNV studies of schizophrenia (4,5), ensuring that samples that had been included in both studies were only used once in the analysis (the sample from Aberdeen was genotyped in both studies). The deletion was observed in 4 of 3391 cases and 1 of 3181 controls in the ISC study (4) and in 2 of 1438 cases and 5 of 33246 controls in the study by Stefansson et al. (5). The combined evidence for association in these two independent studies is significant (Fisher's exact test, P = 0.001), as it is in all three studies combined (P = 5 × 10−5). Across all three studies, the 17p12 deletion is approximately 10 times more common in schizophrenic patients than in controls (0.15% versus 0.015%).
Figure 2.
Positions of CNVs on 17p12 between 14.05 and 15.36 Mb that were identified in the current (Cardiff), ISC (4) and Stefansson et al. (5) studies. Deletions in cases are in red. The positions of genes and segmental duplications in the region are also shown. Produced with the UCSC Genome Browser (http://www.genome.ucsc.edu).
17p12 deletions are found in cases of hereditary neuropathy with liability to pressure palsies (HNPP, MIM 162500) (18), and duplications of 17q12 cause Charcot–Marie–Tooth disease Type 1A (CMT1A, MIM 118220) (19). The neurological phenotypes are known to be caused by deletion and duplication of peripheral myelin protein 22 (PMP22) (18,19). This gene might also influence the psychiatric phenotype, because there is strong evidence for abnormal myelination in schizophrenia (20). Reductions in PMP22 mRNA have been observed in post mortem brains of patients affected with schizophrenia (21) and major depression (22). There is at least one other report on a patient with schizophrenia with this deletion without neurological symptoms (23). In contrast, we are not aware of any reports of excess psychiatric morbidity in HNPP. However, such an association might have been obscured by the incomplete penetrance for both phenotypes. The prevalence of HNPP is reported in 1:6000 people, about the same as in the control populations analysed in the current studies. This association of schizophrenia with 17p12 deletions will need to be examined in other samples in order to confirm or refute it. Ethical constraints do not currently allow us to re-contact the schizophrenic patients with the deletion to undertake detailed neurological testing (no neurological disorder in any person of this family was recorded at the time of the recruitment).
We cannot speculate about the role of any of the remaining large deletions, as they were found only once in our patients (Table 2). At least one other deletion is intriguing however, as it affects another good candidate gene, neurexophilin (NXPH2), which interacts with neurexins, of which NRXN1 is a member (see Table S1, Supplementary Material). Deletions in NRXN1 have been shown to be associated with both autism (11,24,25) and schizophrenia (6,7,26), especially if they disrupt exons (26). In the current study, we found one deletion in NRXN1 in a case and three in controls, all disrupting exons: a 2-fold increased rate in cases (0.2% versus 0.1%), which is not significant (P = 0.5).
Large duplications of >1 Mb were also significantly more common in our cases (Table 1). Two of them appear very good candidates: duplications at 16p13.1, and of the PWS/AS critical region, as both CNVs have been implicated in the aetiology of autism. We reasoned previously (6) that schizophrenia has certain features that suggest a partially overlapping aetiology with autism. These include a tendency to show delayed development (27), cognitive deficits present even before the onset of illness (28), mean premorbid IQ scores approximately one-half of a standard deviation below that of healthy controls (29), language and communication problems and a higher rate of minor physical anomalies (30). An overlapping aetiology has already been shown for patients with 22q11.2 deletion syndrome, who have increased rates of autism and schizophrenia (31). An overlapping aetiology between schizophrenia and autism is also emerging for carriers of deletions in NRXN1 as pointed out above and for the deletion at 1q21.2 (4,5,32).
Duplications of 16p13.1, at 14.9–16.4 Mb, were reported by Ullmann et al. (12) in four autistic patients from three families, out of 182 autistic individuals, and the deletion of the same interval was present in three mentally retarded patients. This region was not reported in the other recent studies on autism. Another study identified the duplication in five cases out of 1027 patients who had mental retardation or multiple congenital anomalies (0.49%) but also in five out of 1682 normal controls (0.29%) (17). That study found the same reciprocal deletion in five cases (0.49%) but in no controls. The presence of the duplication in several normal controls in this, and in our study, makes its pathogenic role less certain than that of the deletion, however all these studies find the duplication at very similar rates, and at approximately 2-fold increased rate in patients with autism, mental retardation or schizophrenia. This finding receives support in the ISC study (4) where there are 13 cases with a duplication and 3 with a deletion (a total of 0.47%) and 7 duplications and 1 deletion in controls (a total of 0.24%), a very similar, 2-fold increased rate in cases (Fisher's exact test, P = 0.1). The region contains an excellent candidate gene, NDE1, which interacts with DISC1, a leading candidate gene for schizophrenia (33).
The second intriguing duplication, which involves the PWS/AS critical region, has been implicated in autism and developmental delay in multiple publications. It is regarded as the most prevalent cytogenetic abnormality found in autism (10,13).
Finally, we also provide new data and a re-analysis of previous data that convincingly implicates as a schizophrenia risk factor a CNV at 15q11.2 which was previously implicated by Stefansson et al. (5).
In summary, our work confirms the association with large CNVs with schizophrenia and adds several specific novel loci to the growing list of CNVs that are emerging as susceptibility factors for schizophrenia. These associations should be replicated in future studies, but the evidence that a number of CNVs increase susceptibility to this disorder is getting stronger.
MATERIALS AND METHODS
Subjects
We selected 520 unrelated patients suffering from schizophrenia or schizoaffective disorder recruited in the UK. Of those, 471 survived filtering and were used for analysis of CNVs (447 schizophrenia and 24 schizoaffective of which 315 were males and 156 were females). Diagnoses were made according to DSM-IV criteria, following assessment by a psychiatrist using the Schedules for Clinical Assessment in Neuropsychiatry (34) and hospital notes. All patients received information and signed a consent form for participation in genetic studies. Ethics committee approval was obtained from all regions where patients were recruited. Controls were the same 2998 subjects used in the WTCCC study (14): 1498 volunteers from the National Blood Service and 1500 individuals born in the UK during 1 week in 1958, described in more detail in the Supplementary Material to that paper. Of those, 2792 survived filtering and were used for the analysis of CNVs (1384 males and 1408 females).
Microarray analysis
The DNA samples were hybridized on Affymetrix GeneChip® Human Mapping 250K NspI and StyI arrays at the Affymetrix Service Laboratory, San Francisco, USA. All samples were typed during the same pipeline as the samples used in the WTCCC study (14), which provided the data on the controls. For establishment of copy number state, we used the Genotyping Console v2.1 software (http://www.affymetrix.com). We used the recommended parameters of the software to exclude samples not passing the initial quality control (QC) threshold: QC call rate<0.93. For reference sets, we used arrays produced with the same batches of hybridizations, as we noticed that using the same reference set for all samples would produce poor-quality results.
To minimize the false-positive rate for calling CNVs, we employed very stringent selection criteria. We identified a CNV on one array only where it was >100 kb and contained ≥10 probes (the default of the software is five probes). The quality of arrays for copy number analysis was measured with the IQR. Arrays above the default IQR > 0.40 were excluded. Samples with an IQR > 0.40 on either array produced, with almost no exception, >20 CNVs (or even >100 CNVs when IQR was >0.50), indicating that samples with >20 CNVs were very likely false-positive findings. Therefore, we also excluded any sample that had >20 CNVs on either array, even if it had an IQR < 0.40. The filtering process led to the exclusion of 49 cases and 206 controls. We required every CNV to be identified independently by both arrays, with >100 kb of its length overlapping between the two arrays. Matching of the two arrays according to these criteria was achieved with the use of an in-house written software (available upon request). We also removed CNVs that were outliers on low SNP density, using a cut-off of <3 SNPs/100 kb as they usually overlapped with large segmental duplications and were clear artefacts.
Validation
Validation was performed with Agilent Human Genome CGH 4×44K microarrays using the manufacturer's protocols (http://www.chem.agilent.com). This array has over 43 000 probes covering the genome at 43 kb overall median probe spatial resolution. We validated the presence of CNVs in cases for all 13 deletions >1 Mb listed in Table 2 (as they provided the most significant results in the study), as well as all CNVs in cases that are individually discussed in the paper (e.g. 15q11.2, 17p12, 16p13.1). Each CNV was confirmed, reflecting the very stringent inclusion criteria employed. Additional family members, where available, were also tested for these loci.
Association analysis
Association analysis was performed using PLINK v1.03 (15), available from http://pngu.mgh.harvard.edu/purcell/plink/. We filtered out CNVs with a frequency >1% in the sample, while also removing any CNV that had >50% of its length spanning a region with more than 33 CNVs in the total sample (1%), in a manner similar to that employed in the ISC study (4). To estimate the significance of any difference in the rate of CNVs between cases and controls (Table 1), we used PLINK to produce a permutation P-value. To estimate the significance of the increased rate of very rare specific CNVs, we used Fisher's exact test.
All coordinates in this paper are according to the March 2006 human reference sequence (NCBI Build 36, hg18).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None.
FUNDING
This research was supported by grants from the MRC, the Wellcome Trust and by NIMH (USA) CONTE: 2 P50 MH066392-05A1. This study makes use of data generated by the Wellcome Trust Case Control Consortium (full list of contributors is presented in the Supplementary Material). Funding for that project was provided by the Wellcome Trust under award 076113.
Supplementary Material
REFERENCES
- 1.Gottesman I.I. Schizophrenia Genesis: The Origins of Madness. New York: Henry Holt & Company, Inc.; 1991. [Google Scholar]
- 2.Murphy K.C., Jones L.A., Owen M.J. High rates of schizophrenia in adults with velocardio-facial syndrome. Arch. Gen. Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov D., Kirov G., Norton N., Williams H.J., Williams N.M., Nikolov I., Tzwetkova R., Stambolova S.M., Murphy K.C., Toncheva D., et al. A molecular genetic study of the vcfs region in patients with early-onset psychosis. Br. J. Psychiatry. 2003;183:409–413. doi: 10.1192/bjp.183.5.409. [DOI] [PubMed] [Google Scholar]
- 4.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirov G., Gumus D., Chen W., Norton N., Georgieva L., Sari M., O'Donovan M.C., Erdogan F., Owen M.J., Ropers H.H., Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 7.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A., et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 8.Xu B., Roos J.L., Levy S., van Rensburg E.J., Gogos J.A., Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 9.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2008;320:539–543. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullmann R., Turner G., Kirchhoff M., Chen W., Tonge B., Rosenberg C., Field M., Vianna-Morgante A.M., Christie L., Krepischi-Santos A.C., et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum. Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 13.Freitag C.M. The genetics of autistic disorders and its clinical relevance: a review of the literature. Mol. Psychiatry. 2007;12:2–22. doi: 10.1038/sj.mp.4001896. [DOI] [PubMed] [Google Scholar]
- 14.Wellcome Trust Case Control Consortium. Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassett A.S., Marshall C.R., Lionel A.C., Chow E.W., Scherer S.W. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum. Mol. Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannes F.D., Sharp A.J., Mefford H.C., de Ravel T., Ruivenkamp C.A., Breuning M.H., Fryns J.P., Devriendt K., Van Buggenhout G., Vogels A., et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J. Med. Genet. 2008 doi: 10.1136/jmg.2007.055202. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 19.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A., et al. DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell. 1991;66:219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 20.Davis K.L., Stewart D.G., Friedman J.I., Buchsbaum M., Harvey P.D., Hof P.R., Buxbaum J., Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch. Gen. Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- 21.Dracheva S., Davis K.L., Chin B., Woo D.A., Schmeidler J.H., Haroutunian V. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol. Dis. 2006;21:531–540. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Aston C., Jiang L., Sokolov B.P. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol. Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 23.Kunugi H., Ozeki Y., Mizuguchi T., Hirabayashi N., Ogawa M., Ohmura N., Moriuchi M., Harada N., Matsumoto N. A case of schizophrenia with chromosomal microdeletion of 17p11.2 containing a myelin-related gene PMP22. Open Psychiatry J. 2008;2:1–4. [Google Scholar]
- 24.Kim H.G., Kishikawa S., Higgins A.W., Seong I.S., Donovan D.J., Shen Y., Lally E., Weiss L.A., Najm J., Kutsche K., et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahir F.R., Baross A., Delaney A.D., Eydoux P., Fernandes N.D., Pugh T., Marra M.A., Friedman J.M. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J. Med. Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 26.Rujescu D., Ingason A., Cichon S., Pietiläinen O.P., Barnes M.R., Toulopoulou T., Picchioni M., Vassos E., Ettinger U., Bramon E., et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum. Mol. Genet. 2008 doi: 10.1093/hmg/ddn351. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray R.M., Jones P.B., Susser E., van Os J., Cannon M. The Epidemiology of Schizophrenia. Cambridge: Cambridge University Press; 2002. p. 454. [Google Scholar]
- 28.Reichenberg A., Weiser M., Rapp M.A., Rabinowitz J., Caspi A., Schmeidler J., Knobler H.Y., Lubin G., Nahon D., Harvey P.D., Davidson M. Premorbid intra-individual variability in intellectual performance and risk for schizophrenia: a population-based study. Schizophr. Res. 2006;85:49–57. doi: 10.1016/j.schres.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 29.Woodberry K.A., Giuliano A.J., Seidman L.J. Premorbid IQ in schizophrenia: a meta-analytic review. Am. J. Psychiatry. 2008;165:579–587. doi: 10.1176/appi.ajp.2008.07081242. [DOI] [PubMed] [Google Scholar]
- 30.Schiffman J., Ekstrom M., LaBrie J., Schulsinger F., Sorensen H., Mednick S. Minor physical anomalies and schizophrenia spectrum disorders: a prospective investigation. Am. J. Psychiatry. 2002;159:238–243. doi: 10.1176/appi.ajp.159.2.238. [DOI] [PubMed] [Google Scholar]
- 31.Ousley O., Rockers K., Dell M.L., Coleman K., Cubells J.F. A review of neurocognitive and behavioral profiles associated with 22q11 deletion syndrome: implications for clinical evaluation and treatment. Curr. Psychiatry Rep. 2007;9:148–158. doi: 10.1007/s11920-007-0085-8. [DOI] [PubMed] [Google Scholar]
- 32.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D., et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burdick K.E., Kamiya A., Hodgkinson C.A., Lencz T., DeRosse P., Ishizuka K., Elashvili S., Arai H., Goldman D., Sawa A., Malhotra A.K. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Hum. Mol. Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wing J.K., Babor T., Brugha T., Burke J., Cooper J.E., Giel R., Jablenski A., Regier D., Sartorius N. SCAN: Schedules for Clinical Assessment in Neuropsychiatry. Arch. Gen. Psychiatry. 1990;47:589–593. doi: 10.1001/archpsyc.1990.01810180089012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.