Abstract
Peak bone mass achieved in adolescence is a determinant of bone mass in later life. In order to identify genetic variants affecting bone mineral density (BMD), we performed a genome-wide association study of BMD and related traits in 1518 children from the Avon Longitudinal Study of Parents and Children (ALSPAC). We compared results with a scan of 134 adults with high or low hip BMD. We identified associations with BMD in an area of chromosome 12 containing the Osterix (SP7) locus, a transcription factor responsible for regulating osteoblast differentiation (ALSPAC: P = 5.8 × 10−4; Australia: P = 3.7 × 10−4). This region has previously shown evidence of association with adult hip and lumbar spine BMD in an Icelandic population, as well as nominal association in a UK population. A meta-analysis of these existing studies revealed strong association between SNPs in the Osterix region and adult lumbar spine BMD (P = 9.9 × 10−11). In light of these findings, we genotyped a further 3692 individuals from ALSPAC who had whole body BMD and confirmed the association in children as well (P = 5.4 × 10−5). Moreover, all SNPs were related to height in ALSPAC children, but not weight or body mass index, and when height was included as a covariate in the regression equation, the association with total body BMD was attenuated. We conclude that genetic variants in the region of Osterix are associated with BMD in children and adults probably through primary effects on growth.
INTRODUCTION
Bone accrual during childhood and adolescence is a determinant of peak bone size and mass traits that are important determinants of bone structural strength in later life (1). Although several environmental factors are thought to contribute to the population variance in peak bone mass and bone size, genetic factors account for the majority of the variance in these traits as reported in twin and family studies (2). Indeed, familial resemblance of bone mineral density (BMD) is greater in adolescence and early adulthood than in later life (3,4). Furthermore, these familial influences upon BMD are expressed as early as pre-puberty (5).
Contrary to the case in older individuals in whom bone mass is also affected by age-related bone loss, bone mass in childhood largely reflects those processes involved in bone mass acquisition, and as such shows distinct patterns of genetic associations. For example, many of the genetic studies published in relation to childhood BMD have reported associations with markers in osteoblast-related genes, such as collagen type 1α2, osteocalcin, PTHR1, LRP5 and ESR1 (6–8).
Our understanding of the genetic influences on adult BMD was recently advanced by two genome-wide association studies (GWAS) in which BMD was analysed in relation to >300 000 markers identified from the human HapMap project (9,10), and a series of very large candidate gene studies from the GENOMOS consortium (11–14). The two genome-wide studies reported several associations with SNPs in genes related to the OPG/RANKL system, which is predominantly involved in regulating bone resorption. Interestingly, the genome-wide studies and one of the candidate gene studies found evidence of association with SNPs in LRP5 (9,11) and ESR1 (10), suggesting shared genetic influences on BMD in children and adults. In order to identify additional genetic determinants of BMD, we applied the genome-wide association approach to 1518 children from a large population-based cohort in which BMD was assessed by total DXA scans performed at 9.9 years of age (the ‘ALSPAC discovery set’) (15) as well as an Australian sample of adult individuals with extremes of higher or lower bone mineral densities (the ‘Australian Extremes’ set). We subsequently identified a region near a promising candidate gene that displayed suggestive evidence of association in both scans. A meta-analysis of two existing genome-wide studies (9,10) revealed strong combined evidence of association at this locus in adults. We subsequently replicated the association in a further 3692 children from the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort who had whole body measures of BMD and DNA available (the ‘ALSPAC replication set’).
RESULTS
Table 1 displays means and standard deviations of BMD, bone mineral content (BMC), bone area, area-adjusted BMC (aBMC) and other related measures for each of the three datasets used in this study. Values were comparable between the ALSPAC discovery and replication sets, although individuals in the replication set were slightly older, heavier and had higher BMC and bone area values. The correlation between the different bone phenotypes was high (i.e. rBMC-AREA=0.98, rBMD-BMC=0.89, rBMD-Area=0.77 in the ALSPAC cohort) indicating that we might expect SNPs to exhibit association with more than one bone-related phenotype. Adult individuals in the Australian cohort were taller and heavier than the ALSPAC children. While it does not make sense to compare the Australian and ALSPAC cohorts in terms of their bone phenotypes (since one involves whole body DXA and the other measurement at the femoral neck), we note that the distribution of BMD and BMC in the Australian sample was bimodal reflecting the selection procedure applied to this cohort.
Table 1.
Mean and standard deviations of Study Population Characteristics in the ALSPAC GWAS discovery sample, the ALSPAC replication set and the Australian Extremes Sample
| ALSPAC discovery set (total = 1518a: females = 778; males = 740) |
ALSPAC replication set (total = 4178a: females = 2062; males = 2116) |
Australian extremes (total = 134a: females = 134; males = 0) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| n | Mean | SD | n | Mean | SD | n | Mean | SD | |
| Age (years) | 1518 | 9.85 | 0.26 | 4178 | 9.94 | 0.34 | 134 | 67.01 | 7.67 |
| Height (cm) | 1507 | 139.61 | 6.18 | 4178 | 139.58 | 6.33 | 109 | 158.61 | 6.55 |
| Weight (kg) | 1516 | 34.20 | 6.77 | 4176 | 34.87 | 7.58 | 109 | 64.31 | 14.97 |
| BMI (kg/m2)b | 1506 | 17.44 | 2.58 | 4176 | 17.77 | 2.94 | 108 | 25.39 | 5.65 |
| BMD (g/cm2)b | 1418 | 0.78 | 0.05 | 3875 | 0.78 | 0.05 | – | – | – |
| BMC (g)b | 1418 | 884.87 | 172.02 | 3875 | 897.60 | 184.87 | – | – | – |
| Area (cm2)b | 1418 | 1132.95 | 156.99 | 3875 | 1143.04 | 165.01 | 118 | 4.89 | 0.50 |
aTotal number of individuals genotyped in each dataset. Phenotypic data were not available for all individuals genotyped. In these situations, n displays the total number of individuals that have phenotypic information available for the relevant trait.
bBone measurements refer to whole body less head values in the case of the ALSPAC datasets, and the femoral neck for the Australian extremes cohort. Individuals in the Australian cohort were specifically selected to have low or high BMD and thus average BMD and BMC values are not reported for this dataset.
Tables S1–S4 in the Supplementary Material display a full list of SNPs that exhibit nominal evidence of association (P < 10−4) with the bone phenotypes in the ALSPAC genome-wide association scan. While no SNP passed a stringent threshold required for genome-wide significance in the ALSPAC discovery set (0.05/315807=1.6 × 10−7), a number of SNPs in candidate genes displayed nominal associations with BMD, BMC, bone area and/or aBMC obtained from whole body DXA scans. These included the SNPs rs525735 (BMD: T = 3.16, P = 0.002; aBMC: T = 4.30, P = 1.9 × 10−5) and rs168568 (BMD: T = 3.48, P = 0.0005; BMC: T = 2.58, P = 0.01; aBMC: T = 3.07, P = 0.002) in the gene OSMR, the SNP rs1798802 (BMD: T = −3.18, P = 0.002; aBMC: T = −3.70, P = 0.0002) in the gene CTNNB1, and several SNPs residing in a linkage disequilibrium block near the gene Osterix (Fig. 1 and Table 2). In contrast, SNPs in or near RANKL (rs9594759, rs9594738), OPG (rs6993813, rs6469804), ZBTB40 (rs7524102, rs6696981), the MHC (rs3130340) and SNPs in or close to ESR1 (rs2504063, rs851982, rs9479055, rs4870044, rs1038304, rs6929137, rs1999805), which had previously displayed association with hip and spine BMD in a recent Icelandic genome-wide association study, did not show strong evidence of association in our study (P > 0.01 across all phenotypes). Similarly, we did not find any evidence of association for the SNP rs4355801 (near TNFRSF11B) which was recently identified in a European study of hip and spine BMD involving 8557 participants (9), although we did find some evidence of association between rs3736228 (in LRP5) and BMD in the same direction as reported by Richards et al. (9) (P = 0.017).
Figure 1.
A graphical representation of the region on chromosome 12 that displays nominal association in both the ALSPAC and Australian genome-wide association scans. Gene tracks, location of SNPs (blue lines), linkage disequilibrium between markers and –log10 P-values for ALSPAC (upper) and Australian (bottom) genome scans are shown. The strongest signals occur in and around the genes Osterix and AAAS, although linkage disequilibrium is strong and extends across a wider region that includes the genes ESPL1, PFDN5 and MYG1.
Table 2.
Association results for four SNPs in the region of Osterix in the initial ALSPAC genome-wide association scan, the ALSPAC replication dataset and the two ALSPAC datasets combined
| Phenotype | SNP | Minor Allele | MAFa | Initial GWASb |
Replication setb |
Combined setb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | BETAc | SEc | P-value | n | BETAc | SEc | P-value | n | BETAc | SEc | P-value | ||||
| BMD (g/cm2) | |||||||||||||||
| rs2016266 | G | 0.33 | 1418 | 0.133 | 0.040 | 0.00082 | 3689 | 0.067 | 0.025 | 0.0063 | 5275 | 0.085 | 0.020 | 0.000031 | |
| rs4759021 | A | 0.32 | 1416 | 0.134 | 0.039 | 0.00058 | 3692 | 0.054 | 0.025 | 0.031 | 5272 | 0.075 | 0.020 | 0.00026 | |
| rs6580942 | C | 0.33 | 1416 | 0.118 | 0.039 | 0.0025 | 3691 | 0.071 | 0.025 | 0.0045 | 5273 | 0.082 | 0.020 | 0.000054 | |
| rs10876432 | G | 0.29 | 1418 | 0.131 | 0.041 | 0.0013 | 3610 | 0.062 | 0.025 | 0.0142 | 5160 | 0.081 | 0.021 | 0.00012 | |
| BMC (g) | |||||||||||||||
| rs2016266 | G | 0.33 | 1418 | 0.130 | 0.040 | 0.0012 | 3689 | 0.066 | 0.025 | 0.0087 | 5275 | 0.082 | 0.020 | 0.000061 | |
| rs4759021 | A | 0.32 | 1416 | 0.124 | 0.039 | 0.0017 | 3692 | 0.045 | 0.025 | 0.077 | 5272 | 0.066 | 0.020 | 0.0013 | |
| rs6580942 | C | 0.33 | 1416 | 0.108 | 0.039 | 0.0063 | 3691 | 0.062 | 0.025 | 0.013 | 5273 | 0.072 | 0.020 | 0.00041 | |
| rs10876432 | G | 0.29 | 1418 | 0.124 | 0.041 | 0.0027 | 3610 | 0.070 | 0.026 | 0.0068 | 5160 | 0.087 | 0.021 | 0.000043 | |
| Bone area (cm2) | |||||||||||||||
| rs2016266 | G | 0.33 | 1418 | 0.126 | 0.041 | 0.0019 | 3689 | 0.065 | 0.025 | 0.0091 | 5275 | 0.081 | 0.020 | 0.000078 | |
| rs4759021 | A | 0.32 | 1416 | 0.116 | 0.040 | 0.0038 | 3692 | 0.040 | 0.025 | 0.116 | 5272 | 0.061 | 0.021 | 0.0029 | |
| rs6580942 | C | 0.33 | 1416 | 0.100 | 0.040 | 0.0125 | 3691 | 0.055 | 0.025 | 0.028 | 5273 | 0.065 | 0.021 | 0.0014 | |
| rs10876432 | G | 0.29 | 1418 | 0.119 | 0.042 | 0.0044 | 3610 | 0.071 | 0.026 | 0.0057 | 5160 | 0.088 | 0.021 | 0.00003 | |
| aBMC (g) | |||||||||||||||
| rs2016266 | G | 0.33 | 1418 | 0.022 | 0.040 | 0.5844 | 3689 | 0.008 | 0.025 | 0.749 | 5275 | 0.012 | 0.021 | 0.570 | |
| rs4759021 | A | 0.32 | 1416 | 0.041 | 0.040 | 0.300 | 3692 | 0.026 | 0.025 | 0.287 | 5272 | 0.027 | 0.021 | 0.187 | |
| rs6580942 | C | 0.33 | 1416 | 0.040 | 0.040 | 0.308 | 3691 | 0.037 | 0.025 | 0.136 | 5273 | 0.038 | 0.020 | 0.063 | |
| rs10876432 | G | 0.29 | 1418 | 0.028 | 0.042 | 0.499 | 3610 | 0.000 | 0.026 | 0.994 | 5160 | 0.004 | 0.021 | 0.853 | |
aMinor allele frequency in combined sample.
bDiscrepancy in sample size between the combined dataset and the sum of the initial and replication datasets is a result of missing genotype data in the genome-wide scan. The combined dataset represents single SNP genotyping carried out across the entire ALSPAC cohort at selected SNPs—i.e. not a mixture of genome-wide association data and single SNP genotypes.
cStandardized beta coefficients and standard errors. The sign of the beta coefficient indicates the direction of effect per addition of minor allele.
Given the small size of our genome-wide scan, and consequently the low power to detect quantitative trait loci, it is likely that many of the top ‘hits’ listed in Supplementary Material, Tables S1–S4 reflect type I error. Therefore, in order to identify genuine variants that exhibit nominal levels of association but do not meet the stringent levels required for genome-wide significance, we compared our complete set of genome-wide results to an Australian genome-wide association scan, which examined a sample of adult individuals with high (66 individuals) or low (68 individuals) hip BMD, and identified regions of nominal association in the same direction in both scans (i.e. P < 0.01). Several SNPs showed evidence of association in both the ALSPAC discovery set and the Australian extremes study (see Supplementary Material, Table S5), including SNPs in a large area of linkage disequilibrium (∼500 kb) on chromosome 12 which contained the strongest statistical evidence of association in both scans (P < .001 in both scans, Supplementary Material, Table S5, Fig. 1 and Table 2). While the SNP rs4759021 which displayed the strongest evidence of association with BMD in both scans (ALSPAC: standardized beta = .134, SE = 0.039, P = 0.0006; Australian scan: OR = 2.75, P = 0.001) lies in the gene AAAS, the extensive linkage disequilibrium across this region means that a putative functional variant might lie within any one of a number of different genes. Notably, the region included the likely candidate gene Osterix (SP7), which encodes a transcription factor responsible for regulating osteoblast differentiation (16). The same SNPs were also reported to have moderate associations in an Icelandic GWAS of hip (P = 3.5 × 10−2 to 3.3 × 10−3) and lumbar spine (P = 1.1 × 10−4 to 1.0 × 10−6) BMD (10), as well as nominal association in a European GWAS of femoral neck (P = 8.5 × 10−3 to 5.0 × 10−2) and lumbar spine (P = 3.0 × 10−2 to 2.2 × 10−1) BMD (9), but did not achieve genome-wide significance in either study. Two of these SNPs (rs10876432 and rs2016266) were subsequently followed up in the Icelandic study, and showed further evidence of association to spine BMD in an independent Icelandic sample (P = 4.0 × 10−7 and P = 2.6 × 10−5, respectively), but not in a Danish and Australian replication cohort (P > 0.05).
In order to quantify the amount of pre-existing evidence for an association between these SNPs and BMD, we combined results from the previously published Icelandic (10) and Richards et al. (9) studies using Fisher’s inverse chi-square test (Table 3). Using this method, we calculated an overall significance value for lumbar spine BMD of P = 9.9 × 10−11 at rs10876432, and an overall P = 2.8 × 10−10 at rs2016266. For BMD measured at the femoral neck, the P-values were 3.1 × 10−5 in the case of rs10876432 and 7.9 × 10−4 at rs2016266. There is thus strong evidence for an association between these SNPs and adult lumbar BMD, and suggestive evidence of association between these SNPs and adult femoral neck BMD. Given the strong pre-existing evidence for association at these SNPs, the presence of moderate association signals in the ALSPAC and Australian genome-wide scans, together with SP7’s obvious candidate gene status, we were interested in whether these signals might replicate in the remainder of the ALSPAC children. We chose not to follow up the other SNPs listed in Supplementary Material, Table S5 because of weaker evidence of association and (lack of) candidacy.
Table 3.
Meta-analysis of P-values from two previous genome-wide association studies
| Reference | Sample | rs10876432 |
rs2016266 |
|||
|---|---|---|---|---|---|---|
| Increaser allele | P-value | Increaser allele | P-value | |||
| Lumbar spine BMD | ||||||
| Styrkarsdottir et al. (10) | Icelandic GWAS discovery | G | 1.0 × 10−6 | G | 1.04 × 10−6 | |
| Styrkarsdottir et al. (10) | Icelandic replication | G | 4.0 × 10−7 | G | 2.6 × 10−5 | |
| Styrkarsdottir et al. (10) | Australian and Danish replication | G | 0.19 | G | 0.069 | |
| Richards et al. (9) | Twins UK | G | 0.22 | G | 0.029 | |
| Overall | G | 9.9 × 10−11 | G | 2.8 × 10−10 | ||
| Femoral neck BMD | ||||||
| Styrkarsdottir et al. (10) | Icelandic GWAS discovery | G | 0.0052 | G | 0.0033 | |
| Styrkarsdottir et al. (10) | Icelandic replication | G | 0.0053 | G | 0.27 | |
| Styrkarsdottir et al. 2008 (10) | Australian and Danish replication | G | 0.021 | G | 0.21 | |
| Richards et al. 2008 (9) | Twins UK | G | 0.052 | G | 0.0085 | |
| Overall | G | 3.1 × 10−5 | G | 0.00079 | ||
P-values are listed for each study and these are combined by Fisher’s method to produce an overall level of significance.
Table 2 presents the results of the ALSPAC genome-wide association study, the replication analysis in the full ALSPAC cohort and the two analyses combined for the four SNPs that exhibited the strongest evidence of association in the discovery sets. In each case, the association is with BMD, BMC and bone area, but not with aBMC. In addition, when subject’s height is included as a covariate in the regression equation, the association with BMD, BMC and bone area attenuates (Table 4). Since bone size is determined by skeletal growth, which in turn reflects a contribution of longitudinal and periosteal bone growth, we examined association of the SNPs with other growth-related phenotypes including height, sitting height, leg length, weight and BMI. Table 5 presents results of these analyses. All SNPs were associated with measures of height, but not with weight or BMI (although the SNP rs10876432 showed some evidence of an effect on weight in the same direction). Finally, in order to determine whether the association between the SNPs and the different bone measures was robust, we only included pre-pubertal children in the analyses (∼80% of children). Although there was some increase in P-values (which is to be expected given the smaller sample size), the estimated standardized regression coefficients were roughly similar to the total analysis (data not shown).
Table 4.
Association results for four SNPs in the region of Osterix in the combined analysis after adjusting for height (combined set)
| Combined set |
|||||
|---|---|---|---|---|---|
| Phenotype | SNP | n | BETAa | SEa | P-value |
| BMD (g/cm2) | |||||
| rs2016266 | 5226 | 0.033 | 0.017 | 0.045 | |
| rs4759021 | 5225 | 0.038 | 0.017 | 0.021 | |
| rs6580942 | 5223 | 0.044 | 0.017 | 0.009 | |
| rs10876432 | 5114 | 0.019 | 0.012 | 0.058 | |
| BMC (g) | |||||
| rs2016266 | 5226 | 0.013 | 0.013 | 0.310 | |
| rs4759021 | 5225 | 0.018 | 0.013 | 0.160 | |
| rs6580942 | 5223 | 0.020 | 0.013 | 0.109 | |
| rs10876432 | 5114 | 0.022 | 0.013 | 0.092 | |
| Bone area (cm2) | |||||
| rs2016266 | 5226 | 0.008 | 0.011 | 0.486 | |
| rs4759021 | 5225 | 0.010 | 0.011 | 0.360 | |
| rs6580942 | 5223 | 0.010 | 0.011 | 0.372 | |
| rs10876432 | 5114 | 0.019 | 0.012 | 0.097 | |
aStandardized beta coefficients and standard errors. The sign of the beta coefficient indicates the direction of effect per addition of minor allele.
Table 5.
Association of SNPs in and around Osterix with growth-related phenotypes at age 9 (combined set)
| rs2016266 |
rs4759021 |
rs6580942 |
rs10876432 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | BETAa | SEa | P-value | n | BETAa | SEa | P-value | n | BETAa | SEa | P-value | n | BETAa | SEa | P-value | |
| Height | 5624 | 0.082 | 0.020 | 0.000037 | 5621 | 0.058 | 0.020 | 0.0035 | 5614 | 0.065 | 0.020 | 0.0011 | 5503 | 0.077 | 0.021 | 0.00022 |
| Sitting height | 5669 | 0.069 | 0.020 | 0.00046 | 5664 | 0.053 | 0.020 | 0.0072 | 5660 | 0.062 | 0.020 | 0.0018 | 5545 | 0.064 | 0.021 | 0.0018 |
| Leg length | 5669 | 0.077 | 0.020 | 0.00010 | 5664 | 0.051 | 0.020 | 0.010 | 5660 | 0.058 | 0.020 | 0.0036 | 5545 | 0.070 | 0.021 | 0.00067 |
| Weight | 5670 | 0.034 | 0.020 | 0.087 | 5665 | 0.016 | 0.020 | 0.408 | 5661 | 0.017 | 0.020 | 0.383 | 5546 | 0.041 | 0.020 | 0.043 |
| BMI | 5620 | −0.005 | 0.020 | 0.806 | 5617 | −0.013 | 0.020 | 0.525 | 5610 | −0.016 | 0.020 | 0.422 | 5499 | 0.0095 | 0.021 | 0.643 |
aStandardized beta coefficients and standard errors. The sign of the beta coefficient indicates the direction of effect per addition of minor allele.
DISCUSSION
To our knowledge, this report describes the first genome-wide association study of BMD in a population of children. Using this approach, we were able to identify several common variants in an area of linkage disequilibrium on chromosome 12q13 that were associated with total body BMD as measured at 9.9 years. Given the high correlation between markers across this region, we cannot unequivocally localize the association signal to any one gene. For example, one of the SNPs rs4759021 lies in the AAAS gene, which has been implicated in ‘Triple A Syndrome’, in which osteopenia occasionally occurs (17,18). We also note that our association signal is very strong around the candidate gene Osterix. Based upon studies of osterix null mice, Osterix is thought to act as an osteoblast-specific transcription factor down-stream to Runx-2, serving as a ‘master gene’ in determining osteoblast differentiation (19). In spite of the strong rationale for a role of genetic variants in Osterix influencing bone mass acquisition, we are not aware of any previous genetic studies in humans highlighting this association.
Although our discovery set was underpowered for genome-wide association analysis, we observed a similar association in our replication cohort. Our findings are also consistent with a large genome-wide association study of Icelandic individuals that documented strong evidence of association between markers in the same region and adult hip and lumbar spine BMD (10), and by an association between the SNPs and hip BMD in a small Australian sample of adults with extremely high and low BMD. In fact, while this manuscript was under review, a follow-up study from the same Icelandic group found significant evidence of association between the marker rs10876432 near the Osterix gene with lumbar spine BMD (20).
Whereas the variants identified in this study were related to total body BMC, BMD and bone area, which are all size-dependent, no association was observed with aBMC (a measure which has been adjusted for size) suggesting that these markers predominantly influence bone size. Increments in bone size occur through a combination of longitudinal bone growth (involving endochondral bone formation) and radial expansion (involving periosteal apposition), which are closely matched to ensure that bone shape is maintained throughout the growth. Since the SNPs were related to height and the associations with BMC, BMD and bone area were attenuated adjusting for height, associations between variants in the region and measures of bone mass may reflect a primary influence on longitudinal growth. Alternatively, due to the close relationship between longitudinal and periosteal bone growth, variants in the Osterix region might influence bone development in a fundamental way that impacts equally on longitudinal and periosteal bone growth.
Although an association between markers in the Osterix region and BMD was observed in both child and adult populations, in a previous genome-wide analysis performed in adults, no association was observed between these markers and adult height (21). This contrasts with the case for HMGA2 variants, for which equivalent associations were reported in children (based on analyses in ALSPAC) and adults (22). Any tendency for the variants to affect height in childhood but not in adults might reflect a role in the rate of skeletal maturation without influencing final height achieved. That delayed skeletal maturation adversely affects peak bone mass achieved without affecting final stature, as may be the case for the variants identified in this study, is consistent with previous reports that pubertal age is inversely related to peak bone mass in boys and girls (23,24).
In summary, we identified associations between four polymorphisms in the region of the Osterix gene and total body BMD as assessed in the ALSPAC cohort at age 9 and with hip BMD in an adult population. Although this association in children was explained at least in part by an effect on bone size, these markers do not appear to be related to height in adults, suggesting they may affect rate of growth but not the final height achieved. Based on our findings, further studies are justified to examine the role of osteoblast transcription factors such as Osterix in regulating the rate of skeletal maturation and to what extent the latter impacts on peak bone mass and bone size. In particular, we are keen to extend our studies to examine associations with more accurate measures of bone dimensions, for example using peripheral quantitative computer tomography (pQCT), which is currently being undertaken in the ALSPAC cohort.
MATERIALS AND METHODS
ALSPAC participants
The Avon Longitudinal Study of Parents and their Children (ALSPAC) is a population-based birth cohort study consisting initially of over 13 000 women and their children recruited in the county of Avon, UK in the early 1990s (15). Both mothers and children have been extensively followed from the 8th gestational week onwards using a combination of self-reported questionnaires, medical records and physical examinations. Biological samples including DNA have been collected for 10121 of the children from this cohort. Ethical approval was obtained from the ALSPAC Law and Ethics committee and relevant local ethics committees, and written informed consent provided by all parents.
Whole body BMD of 5333 children with DNA was measured by Dual energy X-ray absorptiometry (DXA) when they reached 9 years of age (Lunar Prodigy). Total body less head values were expressed as BMD, BMC and bone area. Conventional BMD measures, obtained by dividing BMC by bone area, represent an ‘areal’ density that is only partially corrected for skeletal size. To provide a more accurate estimate of bone mass independent of size, we derived a further parameter from BMC, termed aBMC, by linear regression of BMC on bone area. Adjusting BMC for bone area by linear regression analysis in this way generates a variable which, unlike areal BMD, is fully independent of skeletal size (25). Scans exhibiting evidence of artefacts were removed from further analyses. All bone measures were approximately normally distributed and therefore not transformed. Based upon comparisons performed on 122 children who had two BMD scans performed on a single day, the coefficient of variation for total body BMD was 0.8. Children’s standing height, sitting height and leg length was also measured using a Harpenden Stadiometer. Weight was quantified using a Tanita Body Fat Analyser. Body mass index (BMI) was calculated as weight in kilograms divided by height in metres squared. Both weight and BMI were log10 transformed prior to analysis in order to improve the shape of the distributions. Children’s pubertal development was assessed by Tanner score. Children were defined as pre-pubertal (i.e. Tanner Stage I) and pubertal otherwise (i.e. Tanner Stage II and above).
Australian participants
One hundred and thirty-five unrelated subjects were recruited from four Australian population studies, including the Dubbo Osteoporosis Epidemiology Study (26), Geelong Osteoporosis Study (27), Tasmanian Older Adult Cohort (28), and the Calcium Intake Fracture Outcome Study (29), as well as from the Oxford Family Osteoporosis Study (30), a study of British families recruited via probands with low BMD (z-score <−2). Subjects selected from these cohorts were unrelated white Caucasian women >55 years of age but >80 years of age, and <5 years post-menopausal. Screening had been performed for secondary causes of osteoporosis, and subjects had not been taking osteoporosis agents prior to bone densitometry measures. In addition, subjects were selected by bone densitometry criteria (measured at the total hip), having either low BMD (z-scores >−4 but <−1.5, n = 69), or high BMD (z-scores >+1.5 but <+4, n = 66). All subjects provided written, informed consent, and each study was approved by its Ethics review committee.
Genotyping
One thousand five hundred and forty-three ALSPAC individuals and 135 Australian unrelated subjects were initially genotyped using the Illumina HumanHap317K SNP chip. This chip contains 317 504 SNPs and provides ∼75% genomic coverage of the Utah CEPH (CEU) HapMap samples for common SNPs at r2 > 0.8 (31). Markers with minor allele frequency <1%, SNPs with >5% missing genotypes and, any marker that failed an exact test of Hardy–Weinberg equilibrium (P < 10−7) were excluded from further analyses. After data cleaning, 315 807 SNPs were left in the ALSPAC genome-wide association analysis, and 312 726 SNPs in the Australian analysis. Follow-up genotyping of four SNPs (rs2016266, rs4759021, rs6580942, rs10876432) that showed the best evidence of association across both datasets was carried out in the entire ALSPAC cohort for whom DNA was available (i.e. 10 121 individuals) by K-Biosciences (http://www.kbioscience.co.uk), who employ a novel form of competitive allele specific PCR (KASPar) and Taqman™ system for genotyping.
Statistical analysis
Genome-wide identity by state sharing was calculated for each pair of individuals in both cohorts to identify cryptic relatedness. In order to identify individuals who might have ancestries other than Western European, we merged data from both cohorts with the 60 western European (CEU) founder, 60 Nigerian (YRI) founder and 90 Japanese (JPT) and Han Chinese (CHB) individuals from the International HapMap Project (32). Genome-wide IBS distances for each pair of individuals were calculated on markers shared between the HapMap and the Illumina 317K SNP chip, and then the multidimensional scaling option in R was used to generate a two-dimensional plot based upon individuals’ scores on the first two principal coordinates from this analysis. Samples that did not cluster with the CEU individuals were excluded from subsequent analyses. In addition, we plotted the proportion of missing data for each individual against their genome-wide heterozygosity. Any individual, who did not cluster with others (i.e. with >5% missing data, a genome-wide heterozygosity >36.4% or <34.3%) was removed from further analyses. Finally, we removed a single-male individual in the ALSPAC cohort who scored heterozygous at many loci on the X chromosome. After data cleaning, we were left with 1518 individuals in the ALSPAC cohort (1418 of whom had bone density measurements), and 66 individuals with high and 68 individuals with low bone density in the Australian study.
Association analyses were performed in the ALSPAC cohort for BMD, BMC, bone area and aBMC using least squares linear regression assuming an additive model for genotypes and including sex as a covariate. Tests for association in the Australian cohort were performed using the Cochrane Armitage trend test. Analyses were performed using STATA and PLINK (33). QQ plots as well as small values of λ (from 1.005 to 1.02), the genomic-control inflation factor (34), indicated only a small degree of population substructure in both samples. We therefore report uncorrected results for all our analyses.
Meta-analysis
We combined results from the previously published Richards et al. (9) and Icelandic (10) genome-wide association studies in the Osterix region using Fisher’s inverse chi-square test (35) in order to estimate overall significance:
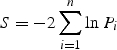 |
where n is the number of independent studies being combined and Pi is the P-value from the ith test. The statistic is distributed as a chi-square statistic with 2N degrees of freedom under the null hypothesis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a Medical Research Council New Investigator Award (MRC G0800582 to D.M.E). This work was supported by the Wellcome Trust. The Australian Extreme study was funded by the National Health and Medical Research Council (Australia). Twins UK is funded in part by the Wellcome Trust; NIHR (T.D.S.), NIHR Biomedical Research Centre (grant to Guys’ and St Thomas’ Hospitals and King’s College London); the Chronic Disease Research Foundation; and the Canadian Institutes of Health Research (J.B.R.). Funding to Pay the Open Access Charge was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council, the Wellcome Trust and the University of Bristol provide core support for ALSPAC. We thank the Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute for generating the ALSPAC GWA data. We thank twins UK for generously sharing the results of their genome-wide association study. This publication is the work of the authors and they will serve as guarantors for the contents of this paper.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rosen C.J. Pathogenesis of osteoporosis. Baillieres Best Pract. Res. Clin. Endocrinol. Metab. 2000;14:181–193. doi: 10.1053/beem.2000.0068. [DOI] [PubMed] [Google Scholar]
- 2.McGuigan F.E., Murray L., Gallagher A., Davey-Smith G., Neville C.E., Van’t Hof R., Boreham C., Ralston S.H. Genetic and environmental determinants of peak bone mass in young men and women. J. Bone Miner. Res. 2002;17:1273–1279. doi: 10.1359/jbmr.2002.17.7.1273. [DOI] [PubMed] [Google Scholar]
- 3.Gueguen R., Jouanny P., Guillemin F., Kuntz C., Pourel J., Siest G. Segregation analysis and variance components analysis of bone mineral density in healthy families. J. Bone Miner. Res. 1995;10:2017–2022. doi: 10.1002/jbmr.5650101223. [DOI] [PubMed] [Google Scholar]
- 4.Jones G., Nguyen T.V. Associations between maternal peak bone mass and bone mass in prepubertal male and female children. J. Bone Miner. Res. 2000;15:1998–2004. doi: 10.1359/jbmr.2000.15.10.1998. [DOI] [PubMed] [Google Scholar]
- 5.Ferrari S., Rizzoli R., Slosman D., Bonjour J.P. Familial resemblance for bone mineral mass is expressed before puberty. J. Clin. Endocrinol. Metab. 1998;83:358–361. doi: 10.1210/jcem.83.2.4583. [DOI] [PubMed] [Google Scholar]
- 6.Koay M.A., Tobias J.H., Leary S.D., Steer C.D., Vilarino-Guell C., Brown M.A. The effect of LRP5 polymorphisms on bone mineral density is apparent in childhood. Calcif. Tissue Int. 2007;81:1–9. doi: 10.1007/s00223-007-9024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobias J.H., Cook D.G., Chambers T.J., Dalzell N. A comparison of bone mineral density between Caucasian, Asian and Afro-Caribbean women. Clin. Sci. (Lond.) 1994;87:587–591. doi: 10.1042/cs0870587. [DOI] [PubMed] [Google Scholar]
- 8.Vilarino-Guell C., Miles L.J., Duncan E.L., Ralston S.H., Compston J.E., Cooper C., Langdahl B.L., Maclelland A., Pols H.A., Reid D.M., et al. PTHR1 polymorphisms influence BMD variation through effects on the growing skeleton. Calcif. Tissue Int. 2007;81:270–278. doi: 10.1007/s00223-007-9072-7. [DOI] [PubMed] [Google Scholar]
- 9.Richards J.B., Rivadeneira F., Inouye M., Pastinen T.M., Soranzo N., Wilson S.G., Andrew T., Falchi M., Gwilliam R., Ahmadi K.R., et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Styrkarsdottir U., Cazier J.B., Kong A., Rolfsson O., Larsen H., Bjarnadottir E., Johannsdottir V.D., Sigurdardottir M.S., Bagger Y., Christiansen C., et al. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 11.Van Meurs J.B., Trikalinos T.A., Ralston S.H., Balcells S., Brandi M.L., Brixen K., Kiel D.P., Langdahl B.L., Lips P., Ljunggren O., et al. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–1290. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langdahl B.L., Uitterlinden A.G., Ralston S.H., Trikalinos T.A., Balcells S., Brandi M.L., Scollen S., Lips P., Lorenc R., Obermayer-Pietsch B., et al. Large-scale analysis of association between polymorphisms in the transforming growth factor beta 1 gene (TGFB1) and osteoporosis: the GENOMOS study. Bone. 2008;42:969–981. doi: 10.1016/j.bone.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Uitterlinden A.G., Ralston S.H., Brandi M.L., Carey A.H., Grinberg D., Langdahl B.L., Lips P., Lorenc R., Obermayer-Pietsch B., Reeve J. The association between common vitamin D receptor gene variants and osteoporosis: a participant-level meta-analysis. Ann. Intern. Med. 2006;145:255–264. doi: 10.7326/0003-4819-145-4-200608150-00005. [DOI] [PubMed] [Google Scholar]
- 14.Ioannidis J.P., Ralston S.H., Bennett S.T., Brandi M.L., Grinberg D., Karassa F.B., Langdahl B., van Meurs J.B., Mosekilde L., Scollen S. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA. 2004;292:2105–2114. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 15.Golding J., Pembrey M., Jones R. ALSPAC—the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr. Perinat. Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y., Jheon A., Nourkeyhani H., Kobayashi H., Ganss B. Molecular cloning, structure, expression, and chromosomal localization of the human Osterix (SP7) gene. Gene. 2004;341:101–110. doi: 10.1016/j.gene.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Barat P., Goizet C., Tullio-Pelet A., Puel O., Labessan C., Barthelemy A. Phenotypic heterogeneity in AAAS gene mutation. Acta. Paediatr. 2004;93:1257–1260. doi: 10.1080/08035250410027706. [DOI] [PubMed] [Google Scholar]
- 18.Dusek T., Korsic M., Koehler K., Perkovic Z., Huebner A., Korsic M. A novel AAAS gene mutation (p.R194X) in a patient with Triple A Syndrome. Hormone Res. 2006;65:171–176. doi: 10.1159/000092003. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 20.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Snorradottir S., Center J.R. New sequence variants associated with bone mineral density. Nat. Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 21.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weedon M.N., Lettre M.N., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chevalley T., Bonjour J.P., Ferrari S., Rizzoli R. Influence of age at menarche on forearm bone microstructure in healthy young women. J. Clin. Endocrinol. Metab. 2008;93:2594–2601. doi: 10.1210/jc.2007-2644. [DOI] [PubMed] [Google Scholar]
- 24.Kindblom J.M., Lorentzon M., Norjavaara E., Hellqvist A., Nilsson S., Mellstrom D., Ohlsson C. Pubertal timing predicts previous fractures and BMD in young adult men: the GOOD study. J. Bone Miner. Res. 2006;21:790–795. doi: 10.1359/jbmr.020602. [DOI] [PubMed] [Google Scholar]
- 25.Tobias J.H., Steer C.D., Vilarino-Guell C., Brown M.A. Estrogen receptor alpha regulates area-adjusted bone mineral content in late pubertal girls. J. Clin. Endocrinol. Metab. 2007;92:641–647. doi: 10.1210/jc.2006-1555. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen T., Sambrook P., Kelly P., Jones G., Lord S., Freund J., Eisman J. Prediction of osteoporotic fractures by postural instability and bone density. BMJ. 1993;307:1111–1115. doi: 10.1136/bmj.307.6912.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders K.M., Pasco J.A., Ugoni A.M., Nicholson G.C., Seeman E., Martin T.J., Skoric B., Panahi S., Kotowizc M.A. The exclusion of high trauma fractures may underestimate the prevalence of bone fragility fractures in the community: the Geelong Osteoporosis Study. J. Bone Miner. Res. 1998;13:1337–1342. doi: 10.1359/jbmr.1998.13.8.1337. [DOI] [PubMed] [Google Scholar]
- 28.Zhai G., Blizzard L., Srikanth V., Ding C., Cooley H., Cicuttini F., Jones G. Correlates of knee pain in older adults: Tasmanian Older Adult Cohort Study. Arthritis Rheum. 2006;55:264–271. doi: 10.1002/art.21835. [DOI] [PubMed] [Google Scholar]
- 29.Prince R.L., Devine A., Dhaliwal S.S., Dick I.M. Effects of calcium supplementation on clinical fracture and bone structure: results of a 5-year, double-blind, placebo-controlled trial in elderly women. Arch. Intern. Med. 2006;166:869–875. doi: 10.1001/archinte.166.8.869. [DOI] [PubMed] [Google Scholar]
- 30.Duncan E.L., Brown M.A., Sinsheimer J., Bell J., Carr A.J., Wordsworth B.P., Wass J.A. Suggestive linkage of the parathyroid receptor type 1 to osteoporosis. J. Bone Miner. Res. 1999;14:1993–1999. doi: 10.1359/jbmr.1999.14.12.1993. [DOI] [PubMed] [Google Scholar]
- 31.Barrett J.C., Cardon L.R. Evaluating coverage of genome-wide association studies. Nat. Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 32.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 35.Fisher R.A. Statistical Methods for Research Workers. London: Oliver and Boyd; 1932. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



