Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a progressive muscle disorder that has been associated with a contraction of 3.3-kb repeats on chromosome 4q35. FSHD is characterized by a wide clinical inter- and intrafamilial variability, ranging from wheelchair-bound patients to asymptomatic carriers. Our study is unique in comparing the gene expression profiles from related affected, asymptomatic carrier, and control individuals. Our results suggest that the expression of genes on chromosome 4q is altered in affected and asymptomatic individuals. Remarkably, the changes seen in asymptomatic samples are largely in products of genes encoding several chemokines, whereas the changes seen in affected samples are largely in genes governing the synthesis of GPI-linked proteins and histone acetylation. Besides this, the affected patient and related asymptomatic carrier share the 4qA161 haplotype. Thus, these polymorphisms by themselves do not explain the pathogenicity of the contracted allele. Interestingly, our results also suggest that the miRNAs might mediate the regulatory network in FSHD. Together, our results support the previous evidence that FSHD may be caused by transcriptional dysregulation of multiple genes, in cis and in trans, and suggest some factors potentially important for FSHD pathogenesis. The study of the gene expression profiles from asymptomatic carriers and related affected patients is a unique approach to try to enhance our understanding of the missing link between the contraction in D4Z4 repeats and muscle disease, while minimizing the effects of differences resulting from genetic background.
Keywords: expression profiling, microarray, skeletal muscle
Facioscapulohumeral muscular dystrophy (FSHD) is an autosomal dominant disorder, the locus of which has been mapped to the subtelomeric portion of chromosome 4, at 4q35 (1). This region is characterized by a series of 3.3-kb repeats termed D4Z4. The D4Z4 array may vary from 11 to more than 100 units in the general population, whereas most FSHD patients have a partial deletion of an integral number of these repeats, and exhibit 10 or fewer units (2). Similarly sized D4Z4 regions are observed in affected relative members within families.
FSHD is typically characterized initially by facial muscle weakness. During progression, weakness and atrophy of shoulder girdle muscles is observed in almost all cases. A gradual spread to abdominal and foot-extensor muscles, followed by clinical involvement of upper arm and pelvic girdle muscles, is seen in most patients. Asymmetry of muscle involvement is common. Among the extramuscular features that may be associated with FSHD are retinal abnormalities and high-tone hearing loss (3). Depression, muscle pain, and fatigue are also often observed among FSHD patients (4–6).
Although it is not possible to predict the course of the disease, there tends to be an inverse relationship between the residual repeat size, the age at onset, and the severity of the disease. Patients with 1 to 3 repeat units are usually very severely affected, whereas patients with 4 to 10 repeats tend to have a milder course (7).
FSHD is also characterized by interfamilial and intrafamilial variability, with severity ranging from asymptomatic carriers to loss of ambulation (8). Some families also show clinical anticipation, although all affected members carry the same deleted fragment (9, 10). Males are on average more often and more severely affected than females, with ≈20% of individuals related to FSHD patients remaining asymptomatic (11, 12). Furthermore, these nonpenetrant cases seem to be more common in particular families (13).
Several observations have suggested that FSHD is caused by a complex and uncommon mechanism. Despite the 98% of homology between the D4Z4 repeats at 4q35 and 10q26, and the equal frequency of translocations observed between these 2 regions, FSHD is uniquely associated with contractions on chromosome 4. Contraction of a translocated 4-type allele on chromosome 10 does not cause FSHD (14–16). As monosomy of 4qter is not associated with FSHD, it is believed that the presence of a small number of D4Z4 repeats is crucial to the process leading to the disease (17). In addition, studies of 4 different polymorphic markers showed that FSHD is restricted to one specific haplotype at 4q35ter, 4qA161 (18). In addition, the proximal unit of D4Z4 is significantly hypomethylated in affected and asymptomatic carriers, while in type-2 FSHD, a form of the disease that is not linked to contractions of D4Z4 repeats at 4q35, both alleles are significantly hypomethylated (19).
Some studies have proposed that FSHD is caused by the transcription of a putative gene encoded by the D4Z4 repeats, termed double homeobox 4 (DUX4) (20, 21). The over-expression of DUX4 is generally toxic to cells, leading to apoptosis and activation of PITX1 (paired-like homeodomain transcription factor 1). Changes in DUX4 or PITX1, both homeodomain proteins, could explain several of the key features in FSHD, including the left–right asymmetry, atypical inflammatory responses, and defects in myoblasts reported in FSHD patients (20, 21). Another model suggests that over-expression of critical genes upstream of the D4Z4 repeats at 4q35 in FSHD patients causes a loss of position-effect variegation (22, 23). The findings that a repressor complex composed of YY1, HMGB2, and nucleolin bind to D4Z4 and that a nuclear matrix attachment site (S/MAR), located immediately upstream of D4Z4, dissociates from the nuclear matrix in FSHD patients, are consistent with a model for dysregulation of genes in cis as a primary event in FSHD (24, 25). Data consistent with such dysregulation have not been supported by several studies using gene array or quantitative RT-PCR, however.
Despite the substantial effort to elucidate the molecular mechanism underlying FSHD, the exact mechanism is still unclear. It is also significant that neither of the proposed mechanisms can explain the clinical variability characteristic of FSHD, clinical anticipation, or why some individuals with 10 or fewer D4Z4 repeats remain asymptomatic. We postulated that these asymptomatic individuals might provide important clues about the pathogenesis of FSHD and the molecular mechanisms to suppress it. To test this, we compared the gene-expression profiles from affected individuals, asymptomatic carriers, and normal controls through microarray analysis, looking for genes that might be implicated in suppression or enhancement of the disease expressivity. We focused our studies on samples from related individuals to minimize variations caused by genetic background. The contribution of microRNAs to FSHD was also evaluated.
Results
Expression Profiles in Affected Patients and Asymptomatic Carriers.
The gene-expression profiles from 3 related members (affected, asymptomatic carrier, and control) from 5 different families were compared through microarrays. Using the criteria described in Materials and Methods for microarray data analysis to identify the differentially expressed genes among affected individuals, asymptomatic carriers, and normal controls, 180 loci-annotated probes were found to be significantly dysregulated. When we compared the expression levels between the affected and asymptomatic, 147 probes were significantly dysregulated, of which 13 were up-regulated in affected patients compared to asymptomatic relatives [supporting information (SI) Tables S1 and S2]. In comparisons of affected individuals and healthy controls, only 56 probes were differentially expressed, of which 20 were up-regulated in affected relative to controls (Table S3 and Table S4). Comparisons of asymptomatic carriers and healthy controls identified 12 probes with a significant fold-change, all of which were up-regulated. Surprisingly, 5 of these probes represent genes from chromosome 4q (Table 1). The only gene similarly and significantly dysregulated in affected and asymptomatic carriers relative to control is IGHA1 (see Table 1), encoding an Ig heavy chain, suggesting that its up-regulation might be related to the presence of the FSHD repeat contraction in these individuals.
Table 1.
Expression fold-changes of significantly dysregulated genes in asymptomatic vs. control
| Probe set | Gene | Loci | Fold-change (asy/cont) | Fold-change (asy/aff) | Fold-change (aff/cont) |
|---|---|---|---|---|---|
| 217022_s_at | IGHA1 | 14q32.33 | 13.85 | 0.69 | 19.98 |
| 203915_at | CXCL9 | 4q21 | 11.44 | 1.85 | 6.19 |
| 211122_s_at | CXCL11 | 4q21.2 | 10.66 | 8.52 | 1.25 |
| 210163_at | CXCL11 | 4q21.2 | 8.91 | 12.13 | 0.73 |
| 205890_s_at | UBD | 6p21.3 | 8.60 | 8.13 | 1.06 |
| 1557690_x_at | NPAS2 | 2q11.2 | 4.62 | 4.99 | 0.92 |
| 1558340_at | DIXDC1 | 11q23.1 | 4.00 | 5.16 | 0.78 |
| 1565935_at | LOC91431 | 4q25 | 3.98 | 4.65 | 0.86 |
| 216968_at | MASP2 | 1p36.3-p36.2 | 3.98 | 4.60 | 0.87 |
| 228230_at | PRIC285 | 20q13.33 | 3.95 | 4.32 | 0.91 |
| 206835_at | STATH | 4q11-q13 | 3.92 | 3.11 | 1.26 |
| 243874_at | LPP | 3q28 | 3.49 | 4.87 | 0.72 |
Underlined fold-changes represent those deemed significant.
The categories of biological processes that our results identified as altered in FSHD were categorized in the DAVID program as described in Materials and Methods. The biological processes most affected in FSHD patients, compared to controls, are involved in histone acetyltransferase and synthesis of GPI-anchors (Table 2). In asymptomatic carriers, however, there is a clear prevalence of processes related to chemokines (see Table 2), and in affected versus asymptomatic samples, most of the categories are related to regulation of transcription (see Table 2).
Table 2.
Gene ontology (GO) categories enriched in the set of differentially expressed genes (P < 0.05) ranked by the fold-enrichment
| GO category | Fold enrichment |
|---|---|
| Affected vs. Control | |
| Histone acetyltransferase binding | 80.6 |
| GPI anchor metabolic process | 32.9 |
| GPI-anchor biosynthesis | 32.3 |
| Phosphoinositide metabolic process | 16.7 |
| Lipoprotein metabolic process | 13.4 |
| Glycerophospholipid metabolic process | 12.2 |
| Glycan structures– biosynthesis 2 | 11.1 |
| Membrane lipid biosynthetic process | 9.5 |
| Nucleolus | 7.0 |
| Membrane lipid metabolic process | 6.0 |
| Cell adhesion | 5.3 |
| Intracellular non-membrane-bound organelle | 2.1 |
| Nonmembrane-bound organelle | 2.1 |
| Asymptomatic vs. Control | |
| CXC chemokine | 190.8 |
| Small chemokine, C-X-C | 153.4 |
| Small chemokine, interleukin-8-like | 65.2 |
| Chemokine activity | 58.8 |
| Chemokine receptor binding | 57.6 |
| Inflammatory response | 51.1 |
| SCY | 46.8 |
| G-protein-coupled receptor binding | 38.4 |
| Locomotory behavior | 20.2 |
| Inflammatory response | 12.8 |
| Behavior | 12.0 |
| Response to wounding | 9.0 |
| Immune response | 8.4 |
| Immune system process | 6.3 |
| Secreted | 4.9 |
| Direct protein sequencing | 3.7 |
| Affected vs. Asymptomatic | |
| Ligase activity, forming phosphoric ester bonds | 48.1 |
| Double-stranded RNA binding | 18.2 |
| Domain: helix-loop-helix motif | 7.9 |
| Basic helix-loop-helix dimerisation region (bHLH) | 7.4 |
| Helix-loop-helix DNA-binding | 6.9 |
| HLH | 6.8 |
| DNA-binding region: basic motif | 6.3 |
| Immune response | 6.0 |
| Membrane lipid biosynthetic process | 5.2 |
| Compositionally biased region: pro-rich | 2.8 |
| Immune response | 2.8 |
| RNA-binding | 2.5 |
| Immune system process | 2.4 |
| RNA binding | 2.3 |
| Defense response | 2.2 |
Expression of 4q35 Genes.
To check if the genes previously reported to be dysregulated from the region in cis to 4q35 are specifically dysregulated in our affected-patient population, we compared their expression among our 3 groups of samples. There was no hybridization signal for 2 genes: DUX4 and FRG2 (Table S5). There were no significant alterations in transcriptional level among affected, asymptomatic, and controls for FRG1, PDLIM3, and ANT1 (see Table S5). However, as reported elsewhere (21), the probes on U133 Plus 2.0 chip for the DUX4 and FRG1 genes do not target the chromosome 4q35 genes specifically. The only significant differences in this set of genes were for ANKRD37 and F11 (see Table S5), which were significantly dysregulated only in affected patients, compared to asymptomatic carriers. Their products could be involved in the pathogenesis of FSHD.
Validation of Microarray Results.
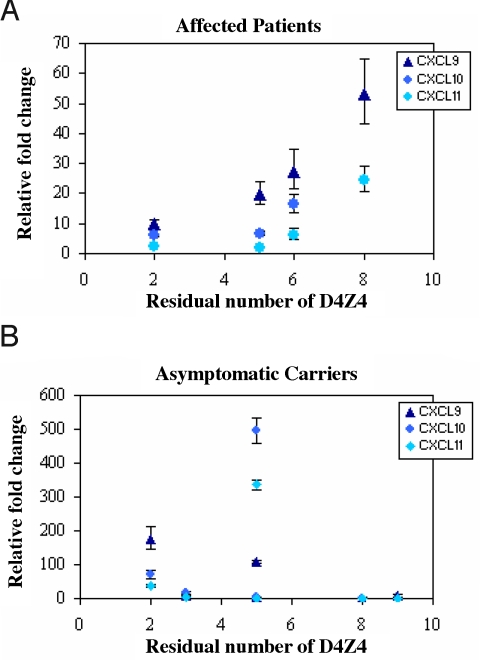
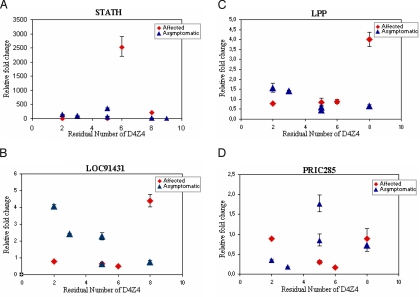
The most interesting dysregulated genes were in asymptomatic muscle biopsies relative to control (see Table 1) and were from chromosome 4q. It was necessary to validate their expression using all possible samples to determine if these genes would be candidates to suppress the FSHD phenotype in asymptomatic carriers. (Please note that RNA samples were not available from all of the original samples used in generating the microarrays for this purpose). The 4 dysregulated genes from chromosome 4q (CXCL9, CXCL11, LOC91431, and STATH) were selected along with 2 other genes, the expression of which appeared more homogeneous among the samples (PRIC285 and LPP), and also CXCL10, which is clustered between CXCL9 and CXCL11 on 4q21, for validation by quantitative RT-PCR. Comparisons of the expression levels of these genes in paired samples from the same family that showed CXCL9, CXCL10, CXCL11, STATH, and LOC91431 are significantly up-regulated in asymptomatic carriers compared to affected individuals (Table 3). However, when other samples from unrelated affected and asymptomatic individuals with different numbers of residual D4Z4 are included, there is no difference in the relative expression of those genes by RT-PCR in affected and asymptomatic groups (Table S6). Interestingly, a graph of these results, considering the fold-changes for affected and asymptomatic as a function of the residual number of D4Z4 repeats (Fig. 1), revealed a correlation between the expression levels of the chemokines and the residual number of D4Z4 in affected patients, but not in asymptomatic carriers. In contrast, negative correlations were found for LOC91431 in asymptomatic carriers (Fig. 2). Given the small sample size available for the validation through RT-PCR, some of the calculated correlations are not significant (P > 0.05).
Table 3.
Expression fold-change in affected and asymptomatic carriers relative to healthy controls
| Gene | Fold-change/affected | Fold-change/asymptomatic |
|---|---|---|
| CXCL9 | 14.1 (11.2–17.8) | 137.8 (113.5–167.4) |
| CXCL10 | 6.4 (5.5–7.4) | 186.9 (154.7–225.8) |
| CXCL11 | 2.1 (1.7–2.5) | 113.8 (102.7–126.2) |
| STATH | 2.4 (2.0–2.8) | 225.4 (206.2–246.2) |
| LOC91431 | 0.7 (0.6–0.8) | 3.0 (2.8–3.3) |
| PRIC285 | 0.5 (0.4–0.6) | 0.8 (0.7–0.9) |
| LPP | 0.8 (0.7–1.0) | 1.0 (0.8–1.1) |
Only the samples analyzed on microarrays were considered.
Fig. 1.
Validation of the expression of the chemokines through RT-PCR. The expression levels of CXCL9, CXCL10, and CXCL11 were calculated relative to the mean value from normal controls in the function of residual number of D4Z4. (A) In affected patients, the rank correlations are 1.000* (CXCL9), 1.000* (CXCL10), and 0.800*** (CXCL11); while the line correlations are 0.933** (CXCL9), 0.730*** (CXCL10), and 0.805*** (CXCL11). (B) In asymptomatic carriers, the rank correlations are –0,464*** (CXCL9), –0.564*** (CXCL10), and –0.527*** (CXCL11); while the line correlations are –0.595*** (CXCL9), –0,004*** (CXCL10), and –0.191*** (CXCL11). *, P < 0.0001; **, P = 0.07; ***, P > 0.1.
Fig. 2.
Relative expression of the other genes validated through RT-PCR in function of the number of D4Z4 repeats. The calculated correlation coefficients for the expression level and the number of repeats for STATH (A) in affected patients are 0.8000*** (rank) and 0.2645***(line) and in asymptomatic carriers are –0.7537** (rank) and –0.4417***(line); for LOC91431 (B) in affected patients are 0.2000*** (rank) and 0.6908*** (line) and in asymptomatic carriers are –0.8208** (rank) and –0.8280** (line); for LPP (C) in affected patients are 1.000* (rank) and 0.7503*** (line) and in asymptomatic carriers are –0.6669*** (rank) and –0.7650*** (line); for PRIC285 (D) in affected patients are 0.2000*** (rank) and –0.1361*** (line) and in asymptomatic carriers are 0.5643*** (rank) and 0.4008*** (line). *, P < 0.0001; **, P = 0.08; ***, P > 0.1.
Characterization of Polymorphisms from 4q35.
As it was previously reported that only contraction of D4Z4 repeats in 4qA161 haplotype was found to cause FSHD, while contractions in other common 4q haplotypes are nonpathogenic (18), we tested if the affected and related asymptomatic individuals, sharing the same FSHD contraction, have different haplotypes that could be associated with the expression of the disease. We examined several polymorphisms on 4qter: the subtelomeric variations distal to D4Z4 (A and B), which can be distinguished by the presence of a beta satellite DNA on A-type alleles, the G/C SNP within the most proximal D4Z4 unit, and a simple sequence-length polymorphism (SSLP) located 3.5-kb proximal to D4Z4. The polymorphisms, when compared in affected and related asymptomatic carriers, can determine their correlation with the pathogenicity of the contracted allele. All tested individuals (from families 1 to 5, Table S7) shared the 4qA allele and the same SNP in D4Z4 (G). In addition, the 4qA161 haplotype was carried in all families tested, with the exception of family 4, in which the affected individual has the 4qA161/4qA166 haplotypes, while the asymptomatic carrier has the 4qA159/4qA166 haplotypes (data not shown).
microRNAs in FSHD.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that are increasingly recognized for their ability to regulate gene expression posttranscriptionally. Among other targets, miRNAs in skeletal muscle regulate the expression of transcription factors and signaling mediators important for muscle biology, which in turn can influence proliferation and differentiation during myogenesis (26, 27).
The expression profiles of miRNAs in 10 different groups of muscle disorders, including FSHD, were recently compared (28) and 62 miRNAs that were differentially expressed in FSHD muscle—all up-regulated—were identified. The predicted targets for these differentially expressed miRNAs were compared to the gene products identified here as down-regulated in FSHD patients, compared to healthy controls. MAMI (meta mir:target inference) was used to predict the targets for the dysregulated miRNAs. Several different miRNAs target the same gene (Table S8), such as ATG16L1, EPAS1, and PCDH9, which are down-regulated in biopsy samples from affected patients. These same genes, ATG16L1, EPAS1, and PCDH9, are also down-regulated in affected individuals relative to asymptomatic carriers (see Table S2), which suggests that their down-regulation by miRNAs may be linked to the pathophysiology of FSHD. Perhaps significantly, most of these predicted targets take part in histone acetylation and the synthesis of GPI anchors, both of which are dysregulated in affected patients vs. healthy controls (see Table 2).
Discussion
Although FSHD was among the first of the muscular dystrophies to have its locus mapped, the molecular mechanism leading to the disease is still unclear. The unspecific histological alterations in muscle and the high clinical variability observed among patients do not give any clues about a possible mechanism underlying FSHD. One possible way to understand the pathology would be to examine the differences in the expression of genes in patients with FSHD and in their close relations, who are asymptomatic carriers. The present study is unique in making such a comparison, which minimizes the effects deriving from differences in genetic background, and likely gives more consistent and reliable results with microarray analysis (29, 30). In addition, all muscle biopsies were collected by the same physician and were processed in the same way, keeping variations from technical sources to a minimum. The results support the evidence that the expression of some genes in FSHD, and particular those at chromosome 4q, is abnormal in FSHD patients.
The finding that the genes from the chemokine cluster on 4q21, that are located more than 100 Mbp from D4Z4 repeats, have a similar expression pattern (CXCL9 > CXCL10 > CXCL11) in all tested affected patients through RT-PCR suggests that these 3 chemokines may be under control of the same regulators in FSHD patients, in contrast to asymptomatic carriers. The IFN-γ-induced chemokines CXCL9, CXCL10, and CXCL11 are ligands for CXCR3 receptor, and are thought to play a key role in directing activated T cells and other cell types (such as natural killer cells and macrophages) to sites of inflammation. The up-regulation of these chemokines have not been described in either FSHD or other forms of muscular dystrophy (31–35), suggesting that their expression is unlikely to be linked to inflammatory cell infiltrates. Moreover, the muscle histology from asymptomatic carriers looks similar to the normal controls, not showing a major inflammatory component, while in affected patient muscle it is possible to observe general dystrophic features (Fig. S1). The fact that there was no difference in the expression levels of these chemokines as well as the other genes validated—STATH, LOC91431, LPP, and PRIC285—between affected patients and asymptomatic carriers when additional samples were included, lessens the likelihood that these genes could be suppressing the FSHD phenotype in asymptomatic carriers. On the other hand, our microarray and RT-PCR data suggest that the contraction of repeats in affected patients and asymptomatic carriers might disturb the gene expression in cis. This observation is supported by the finding that the contracted allele is significantly hypomethylated in both affected individuals and asymptomatic carriers (19). These studies did not look at the differential expression of FRG1 or DUX4 in FSHD patient biopsies. There was evidence from the microarray data that indicated that from chromosome 4q35, ANKRD37 and F11 genes are differentially expressed when compared with affected patients and asymptomatic individuals, suggesting that these 2 groups may have some specific differences on 4q35. In the 4 families tested, it appeared that the affected patients and related asymptomatic carriers share the same 4qA161 haplotype. Thus, these polymorphisms by themselves cannot explain the pathogenicity linked to the contraction of D4Z4 repeats, and other factors, perhaps acting in cis, are likely to be important in the disease mechanism. Further studies concerning the differences between affected patients and asymptomatic carriers are of utmost importance and will certainly help in our knowledge about factors necessary to trigger FSHD.
The data presented here indicated that the biological process that may be specifically impaired in FSHD patients involves the synthesis of GPI anchors. The structure of the GPI anchor is composed of short chains of sugars, specifically mannose and glucosamine, which are assembled in the endoplasmic reticulum and linked to the inositide residues of phosphatidylinositol. After synthesis, the entire glycolipid is transferred to C-terminal regions of proteins posttranslationally, thereby anchoring these proteins to the outer leaflet of the cell membrane, where they tend to associate with lipid rafts (36). GPI anchors also have roles in membrane diffusion, intracellular protein sorting, and signaling (37). A major subclass of rafts is caveolae, which are invaginations of the cell membrane characterized by the abundance of caveolin. Caveolae contain clusters of GPI-anchored proteins, the most well-studied of which is the folate receptor. However, evidence also exists for the presence of other GPI-anchored proteins, such as alkaline phosphatase, Thy-1, and prion PrP(C) in caveolae (38). No caveolae abnormalities in muscle plasma membranes from FSHD patients were found in one study (39). However, alterations in the sarcolemmal reorganization in muscle tissue from FSHD patients have been reported (40). The steps in GPI assembly, and the enzymes that carry them out, are highly conserved. The genes found to be down-regulated in biopsies from FSHD patients presented here, and which take part in this biological process, are involved in the catalytic steps of the GPI biosynthetic pathway. The importance of GPI anchoring in mammals is underscored by the fact that abrogation of GPI biosynthesis results in embryonic lethality (41). Defects in O-glycan biosynthesis are associated with congenital muscular dystrophies; further studies concerning alterations in the GPI anchor and other glycan structures from caveolae and other proteins associated with the sarcolemma in FSHD should be pursued.
The changes in gene expression observed here may also be linked to changes in histone acetylation, which occur only in affected patients and not in asymptomatic carriers. Histone modifications are likely to control the structure and/or function of the chromatin fiber. The pattern of histone modification may correlate with transcriptional repression/activation and also global chromatin dynamics (42). Although previous studies (43, 44) on histone H4 acetylation levels in the proximal region of D4Z4 did not find any difference between affected individuals and healthy controls, the specific state of the chromatin at the D4Z4 repeats of chromosome 4q35 has not yet been studied in affected patients because of the lack of specific primers, but such studies could yield interesting findings.
Our study suggests that changes in the expression of miRNAs may also be pathogenic in FSHD. Our results are consistent with another study that profiled the expression of miRNAs in FSHD, and suggest that the down-regulation of mRNA levels encoding ATG16L1, EPAS1, and PCDH9 in affected individuals, but not in asymptomatic carriers or healthy controls, may be mediated at least in part by increases in the levels of miRNA that target these gene products.
In summary, there appears to be a profound difference in the transcriptional regulation between affected patients and asymptomatic carriers. The contraction of D4Z4 repeats seems to affect the transcription of genes at 4q, in both groups, but they suggest that factors in other regions of the genome are also associated with the pathogenesis of FSHD. Nevertheless, perturbations in histone acetylation may affect the chromatin structure specifically in the affected patients, leading to the pathogenic changes associated with the disease. Similarly, differences in the synthesis of GPI-anchored proteins may also be linked to muscle pathology in affected individuals. In addition, our results indicate that the miRNAs play a role in the regulatory network in FSHD, increasing the complexity of the mechanism underlying the disease. The study of expression profiles from asymptomatic carriers is a unique approach that has revealed these new and interesting findings, and in the future should contribute to a better understanding of FSHD.
Materials and Methods
Muscle Biopsies.
Muscle biopsies were taken from related members (affected, asymptomatic carrier, and normal control) belonging to 5 unrelated families (see families 1 to 5 in Table S7). Asymptomatic carriers were considered those older than 30 years old, without any FSHD clinical signal, but who share the deleted fragment in common with their clinically affected relative. Additional biopsies were also collected from single members of additional families (families 6 to 10 in Table S7). The asymptomatic carriers corresponding to number 16, 17, 19, and 20 samples also have at least one related affected who shares the same FSHD allele. The presence of mosaicism had been previously excluded through pulsed field-gel electrophoresis. For the microarray studies, muscle biopsies were taken from the biceps in 3 families and from the deltoid in the remaining 2 families, because the clinically affected patients had a severe atrophy. The same muscle was taken from all individuals within each family to minimize variability. One portion of each biopsy was frozen for molecular studies and another for histological analysis (see Fig. S1). All of the biopsies were taken at the Human Genome Research Center (University of São Paulo, Brazil) after informed consent approved by the institutional review board.
RNA Isolation.
The frozen muscle tissue was ground in dry ice in a mortar, and the total RNA was isolated from muscle tissue using the TRIReagent method (Molecular Research Center, Inc.) following the manufacturer's protocol. For details, see the SI Text.
Microarray Assay.
mRNA microarray was performed with the Human Genome U133 Plus 2.0 chip (Affymetrix), following the procedures for target labeling according to “One-Cycle Eukaryotic Target Labeling” protocol as described in the SI Text.
Microarray Data Analysis.
A probe-by-probe differential fold analysis was performed between any 2 sample populations—control vs. asymptomatic, control vs. affected, asymptomatic vs. affected—using a geometric logarithmic fold method. For details see the SI Text.
Gene Ontology Enrichment Analysis.
We performed a gene ontology (GO) enrichment analysis of the list of probes of interest using DAVID 2008 (http://david.abcc.ncifcrf.gov) data freeze June 2008. These lists were restricted to probes that had an Entrez gene assignment and duplicated/redundant Entrez genes were removed. The background gene set was 19,501 unique Entrez genes assayed on the Affymetrix Human Genome GeneChip Human Genome U133 Plus 2.0 microarray. We identified ontologic categories that had a Fisher exact P-value (EASE score) <0.05 (45). This analysis was performed on the significantly dysregulated genes.
RT-PCR Validation of Microarray Data.
The validation of microarray results was done through quantitative RT-PCR, using TaqMan Gene Expression Assays (Applied Biosystems) as described in the SI Text.
Characterization of 4q35 Alleles.
After isolation of DNA from peripheral blood, the DNA was digested with HindIII and separated by electrophoresis on agarose gels, followed by transfer to Hybond membrane (GE Healthcare) by Southern blotting. The membrane was hybridized with probes that specifically distinguish these alleles, as described in ref. 10. The D4Z4 SNP and the SSLP of all individuals studied in the present work were defined as in ref. 18.
Supplementary Material
Acknowledgments.
We thank all families, whose collaboration was essential for this study, and also Constância Urbani. We also thank Dr. Mariz Vainzof, Lydia Yamamoto, and Dr. Ivo Pavanello for helping with the muscle biopsies; Richard J.L.F. Lemmers for helping with SSLP genotyping; all members from M.Z.'s laboratory, and Genri Kawahara, Juan C. Casar, Hart Lidov, and Anu Balasubramanian for helpful suggestions. This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo-Centro de Pesquisa, Inovação e Difusão (P.A. and M.Z.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (M.Z.), Facioscapulohumeral Society Research Fellowship Grant FSHS-FS-005 (to P.A. and M.Z.), Instituto Nacional de Células-Tronco em Doenças Genéticas (M.Z.), and Wellstone Center Grant 1U54HD060848–01 (to L.M.K. and M.Z.). L.M.K. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE15090).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901573106/DCSupplemental.
References
- 1.Wijmenga C, et al. Location of facioscapulohumeral muscular dystrophy gene on chromosome 4. Lancet. 1990;336:651–653. doi: 10.1016/0140-6736(90)92148-b. [DOI] [PubMed] [Google Scholar]
- 2.van Deutekom JCT, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Molec Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 3.Padberg GW, et al. On the significance of retinal vascular disease and hearing loss in facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S73–S80. [PubMed] [Google Scholar]
- 4.Bungener C, Jouvent R, Delaporte C. Psychopathological and emotional deficits in myotonic dystrophy. J Neurol Neurosurg Psychiatry. 1998;65:353–356. doi: 10.1136/jnnp.65.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalkman JS, Schillings ML, Zwarts MJ, van Engelen BGM, Bleijenberg G. Psychiatric disorders appear equally in patients with myotonic dystrophy, facioscapulohumeral dystrophy, and hereditary motor and sensory neuropathy type I. Acta Neurol Scand. 2007;115:265–270. doi: 10.1111/j.1600-0404.2006.00737.x. [DOI] [PubMed] [Google Scholar]
- 6.Pandya S, King WM, Tawil R. Facioscapulohumeral dystrophy. Phys Ther. 2008;88(1):105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 7.Lunt PW, et al. Phenotypic-genotypic correlation will assist genetic counseling in 4q35- facioscapulohumeral muscular dystrophy. Muscle Nerve. 1995;2:S103–S109. [PubMed] [Google Scholar]
- 8.Padberg GW, et al. Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve. 1995;2:S81–S84. [PubMed] [Google Scholar]
- 9.Zatz M, et al. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am J Hum Genet. 1995;56(1):99–105. [PMC free article] [PubMed] [Google Scholar]
- 10.Tonini MMO, et al. Homozygosity for autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) does not result in a more severe phenotype. J Med Genet. 2004;41(2):e17–e22. doi: 10.1136/jmg.2003.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padberg GW, Lunt PW, Koch M, Fardeau M. Diagnostic criteria for facioscapulohumeral muscular dystrophy. Neuromuscul Disord. 1991;1:231–234. doi: 10.1016/0960-8966(91)90094-9. [DOI] [PubMed] [Google Scholar]
- 12.Zatz M, et al. The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severly and more frequently than females. Am J Med Genet. 1998;77(2):155–161. [PubMed] [Google Scholar]
- 13.Tonini MMO, et al. Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD) Neuromuscul Disord. 2004;14(1):33–38. doi: 10.1016/j.nmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Deidda G, et al. Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur J Hum Genet. 1995;3(3):155–167. doi: 10.1159/000472291. [DOI] [PubMed] [Google Scholar]
- 15.van Deutekom JCT, et al. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counseling and etiology of FSHD1. Hum Mol Genet. 1996;5:1997–2003. doi: 10.1093/hmg/5.12.1997. [DOI] [PubMed] [Google Scholar]
- 16.Lemmers RJ, et al. Inter- and intrachromosomal sub-telomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis. Hum Mol Genet. 1998;7:1207–1214. doi: 10.1093/hmg/7.8.1207. [DOI] [PubMed] [Google Scholar]
- 17.Tupler R, et al. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet. 1996;33:366–370. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmers RJ, et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Overveld PGM, et al. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 20.Kowaljow V, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Dixit M, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hewitt JE, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Molec Genet. 1994;3:1287–1295. doi: 10.1093/hmg/3.8.1287. [DOI] [PubMed] [Google Scholar]
- 23.Winokur ST, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 24.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 25.Petrov A, et al. Chromatin loop domain organization within the 4q35 locus in facioscapulohumeral dystrophy patients versus normal human myoblasts. Proc Natl Acad Sci USA. 2006;103:6982–6987. doi: 10.1073/pnas.0511235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callis TE, Deng Z, Chen JF, Wang DZ. Muscling through the microRNA world. Exp Biol Med. 2008;233(2):131–138. doi: 10.3181/0709-MR-237. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg I, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A, et al. Assessing natural variations in gene expression in humans by comparing with monozygotic twins using microarrays. Physiol Genomics. 2005;21(1):117–123. doi: 10.1152/physiolgenomics.00228.2003. [DOI] [PubMed] [Google Scholar]
- 31.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 32.Celegato B, et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 33.Winokur ST, et al. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum Mol Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 34.Campanaro S, et al. Gene expression profiling in dysferlinopathies using a dedicated muscle microarray. Hum Mol Genet. 2002;11:3283–3298. doi: 10.1093/hmg/11.26.3283. [DOI] [PubMed] [Google Scholar]
- 35.Bakay M, Zhao P, Chen J, Hoffman EP. A Web-accessible complete transcriptome of normal human and DMD muscle. Neuromuscul Disord. 2002;12(S1):S125–S141. doi: 10.1016/s0960-8966(02)00093-7. [DOI] [PubMed] [Google Scholar]
- 36.Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 38.Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 39.Bonilla E, Fischbeck K, Schotland DL. Freeze-fracture studies of muscle caveolae in human muscular dystrophy. Am J Pathol. 1981;104(2):167–173. [PMC free article] [PubMed] [Google Scholar]
- 40.Reed P, et al. Sarcolemmal reorganization in facioscapulohumeral muscular dystrophy. Ann Neurol. 2006;59:289–297. doi: 10.1002/ana.20750. [DOI] [PubMed] [Google Scholar]
- 41.Nozaki M, et al. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest. 1999;79(3):193–199. [PubMed] [Google Scholar]
- 42.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Jiang G, et al. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Molec Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 44.Yang F, Shao C, Vedanarayanan V, Ehrlich M. Cytogenetic and immuno-FISH analysis of the 4q subtelomeric region, which is associated with facioscapulohumeral muscular dystrophy. Chromosoma. 2004;112:350–359. doi: 10.1007/s00412-004-0280-x. [DOI] [PubMed] [Google Scholar]
- 45.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(9):R60–R60.11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.