Abstract
Introduction
Tissue microarrays (TMA) enable rapid analysis of biomarkers in large-scale studies involving archival tumor specimens, however their utility in heterogeneous tumors such as ovarian cancer is limited.
Methods
In this study, immunohistochemical (IHC) analysis was performed on TMAs comprised of epithelial ovarian cancer (EOC) to estimate the prevalence of loss of expression of three mismatch repair (MMR) proteins. TMAs were initially created using cores sampled from the center of donor tissue blocks from 59 EOC cases. Full sections were subsequently created and levels of expression were compared between tissues sampled from the central portion versus the periphery. Follow-up analyses were performed by obtaining cores from the periphery of up to 5 additional donor blocks per case. A linear mixed model for each protein was used to investigate differences between results from the initial and follow-up blocks.
Results
In the original TMAs created using centrally sampled cores, loss of MMR expression from was noted in 17 (29%) of the 59 cases. By comparison, analyses from peripherally sampled cores revealed loss of expression in only 6 of these 17 cases. For each protein, significant differences (p<0.05) were detected between results from the initial donor block and the majority of the follow-up blocks.
Conclusions
Our investigations, based on EOC, suggest that sampling variability in protein expression may result when TMAs are used. Thus, at least for EOC, it is important to preferentially sample from the periphery of tumor blocks where exposure to tissue fixatives is optimal.
Keywords: Tissue microarray (TMA), ovarian cancer, mismatch repair, biomarkers, sampling strategy, immunohistochemistry (IHC)
INTRODUCTION
The development of tissue microarrays (TMA) for high-throughput molecular profiling of tumor specimens (1) has led to a rapid, relatively inexpensive and efficient technique to analyze biomarkers in archived tumor specimens from large population studies. In contrast, conventional methods for the molecular analysis of tumor specimens, which typically require full section slides to be created from an original donor block (2), are labor intensive and time consuming. TMAs are constructed by obtaining small core biopsies from morphologically representative areas of paraffin-embedded tumor tissues and subsequently assembling the cores on a recipient paraffin block (3). A primary advantage of TMAs is that by their design, they allow for simultaneously analysis of specimens from a large number of cases on one slide, thus enabling all specimens to be exposed to uniform experimental conditions. Additionally, since cores rather than full sections are used, there is minimal destruction to the original donor material (1, 3). These notable benefits notwithstanding, a major challenge when using TMAs is to achieve accurate representation of the parent tumor tissues from which they were derived (4-6). Indeed, construction of TMAs must be completed under the guidance of a skilled pathologist in order to ensure accurate estimates of the prevalence and/or level of expression of the biomarker under investigation. Although investigators comparing molecular expression data between TMA cores and full sections have reported high rates of concordance (4, 7-12), factors that affect accurate representation include tissue heterogeneity, which varies according to tumor type, as well as tissue fixation. Epithelial ovarian cancers (EOC) are a particularly heterogeneous type of tumor (13-16), in part due to their large size. Furthermore, as with other tumor types, there is biological variability within and between individual EOC tumor blocks (17).
Another source of variability in biomarker studies is the use of immunohistochemical analysis (IHC), which provides a means to measure protein expression in various tumors. While IHC is widely used in surgical pathology as a diagnostic, prognostic, and therapeutic tool, there is variable consistency and poor reproducibility of results (2, 18-21). Published studies comparing IHC results from core- vs. full-section-derived methods when using EOC specimens are lacking.
Ovarian cancers, diagnosed in over 22,000 women per year in the United States, cause more deaths than any other gynecologic malignancy(22). While understanding of the molecular pathways underlying ovarian tumorigenesis is currently lacking, there is evidence that a proportion of cases are due to defects in the mismatch repair (MMR) genes, primarily MLH1, MSH2, and MSH6 (23-29). Mutations in these genes lead to Hereditary Non Polyposis Colorectal Cancer Syndrome (HNPCC) (30, 31), an inherited cancer predisposing condition. IHC analysis has been a useful strategy for investigating MMR defects in colorectal cancer (32, 33), although there is limited information as to its utility in other HNPCC-associated cancers, such as ovarian cancer (34-37). Thus, evaluation of the role of MMR deficiency in the pathogenesis of EOC is of considerable interest (36-39). To date, investigation of MMR protein expression in EOC has yielded frequency estimates of loss of expression between 2% and 10% (34-37). To improve on the precision of these estimates using TMAs, we performed a validation study.
MATERIALS AND METHODS
Participants and Tissue Samples
EOC tumor blocks used in the current investigation were obtained from participants in the Tampa Bay Ovarian Cancer Study (TBOCS), a population-based study of incident epithelial ovarian cancer in a heavily populated 2-county region of west central Florida. The study was approved by the institutional review board of the University of South Florida. Further details about study design, population, and data collection methods have been published previously (40). Briefly, cases included 232 women aged 18 to 80 with histologically confirmed invasive or borderline epithelial ovarian cancer diagnosed between December 13, 2000 and September 30, 2003, on whom paraffin-embedded EOC tumor blocks were obtained on 85% of the study sample.
Construction of TMAs
A TMA was constructed from paraffin-embedded tumor specimens from 59 of the original 232 subjects. Archived, formalin-fixed, paraffin-embedded EOC tissue blocks that had been stored at room temperature were used. Hematoxylin-and-eosin-stained full sections were reviewed to select representative areas of tumor in the center of an initial donor block from which cores were acquired for the microarray. The TMA blocks were constructed with a precision instrument (Chemicon model ATA-100, Chemicon Int'l, Temecula, CA, USA) as previously described (3). For each case, three replicate 1 mm cores were sampled from the center of the donor tissue block and placed side-by-side on a separate recipient block. Normal control tissue (fallopian tissue) was included in the block. A heated glass slide was used to even the surface of the recipient block.
The distribution of histologic subtypes of EOC was representative of the general population. Sample tracking was based on coordinate positions for each tissue core in the TMA recipient block; 4μm sections were transferred onto separate TMA slides for IHC staining of each of the three MMR proteins under investigation (hMLH1, hMSH2, and hMSH6).
IHC Staining for MMR Proteins
Deparaffinized, formalin-fixed paraffin-embedded tissues were microwaved in 1X EDTA (Chemicon Int'l, Temecula, CA) (hMSH2) or Borg Decloaker (BioCare Medical, Concord, CA) (hMLH1 and hMSH6), cooled at room temperature for 20 minutes, rinsed with deionized water and placed in TBS/Tween for 5 minutes. Immunostaining was carried out on the Dako Autostainer using the Vector Elite Mouse IgG - HRP detection kit (Vector Laboratories, Burlingame, CA) following avidin/biotin blocking (Vector Laboratories). Slides were incubated in mouse monoclonal hMLH1 (clone G168-15, BioCare Medical, Concord, CA) at 1:40 or hMSH6 (clone BC/44, BioCare, Concord, CA) at 1:70 overnight at 4°C or hMSH2 (Clone FE11, Zymed/Invitrogen, Carlsbad, CA) at 1:200 for 30 minutes at room temperature. For overnight incubations, slides were removed from the autostainer, placed in a humid chamber in the refrigerator, and returned to the autostainer the following day. 3,3'-Diaminobenzidine (Dako, Carpinteria, CA) was the chromogen. Slides were counterstained with modified Mayer's hematoxylin, dehydrated through ascending grades of ethanol, cleared with xylene and mounted with resinous mounting medium.
Loss of MMR expression was defined as absence of detectable nuclear staining of tumor cells in the presence of retained nuclear staining in lymphocytes and/or in non-neoplastic epithelial or stromal cells, which served as internal positive controls. Two pathologists with expertise in ovarian pathology (SN, NV) independently reviewed all stainings. Stainings were classified based on nuclear staining intensity and distribution using a semi-quantitative ordinal scoring system in which a combined expression score of 0 represents total absence of expression and a combined expression score of 9 represents total presence of expression. After taking into account the expected level of immunoreactivity, specimen size, the amount of target antigen in the specimen, and clinical appropriateness (18), the study pathologist defined cases with reduced or absent staining as having a mean core expression score of ≤4.
Creation of Regular Tumor Sections and Subsequent Follow-up Analysis
For the 17 cases showing reduced or absent staining, full tumor sections were created from the respective donor paraffin block (from which the cores were derived) and were subsequently stained in order to evaluate a larger tumor area. Follow-up analyses for these cases was performed by creating a new TMA comprised of representative cores obtained from the periphery of up to 5 additional donor tissue blocks (triplicate cores per block) per case. The number of blocks available per case (ranging from 1 to 20) determined the number of additional donor tissue blocks sampled in the follow-up analysis. Additionally, five of the 42 cases in which staining was present, full sections were created from the original donor block, and stained for IHC expression of MMR proteins.
Statistical analysis
Descriptive statistics including graphical illustrations were generated for each protein, and summary statistics and distributions of expression scores were examined by case and by block. The distribution of protein values by block was analyzed using the Anderson-Darling statistic to gauge the need for data transformations. General linear mixed effects model (GLMM) were used to investigate differences between IHC expression scores from the initial donor block and follow-up blocks for each protein, with the main interest variable being the block (41, 42), while adjusting for potential confounding factors. In the GLMM, the variable of case nested within the core was included as a random intercept. The correlation among the multiple observations within the same core (also nested within each block) was accounted for, assuming the compound symmetry correlation structure in the model. We applied the small-sample inference method for the fixed effects to the GLMM, proposed by Kenward and Roger (43) to adjust for the small number of cases. Several covariates were included in the initial model: tumor age (year), tumor size, number of cores, and number of blocks. Specific contrasts comparing the expression score for the initial block with the score for each subsequent block were tested for each protein model, using the F test statistic. The final model included the fixed effect of the block and the random effect of intercept. All tests were two-sided and claimed statistically significant at the level of 5%. No adjustment for multiple comparisons was applied, as the nature of this investigation was hypothesis-generating. SAS software was used for all statistical analyses (SAS Institute, Version 9.1, SAS Institute Inc., Cary, NC, USA, 2007).
RESULTS
Initial IHC analysis of the 59 cases for expression of 3 MMR proteins (ie: MLH1, MSH2 and MSH6) revealed loss of expression of at least one MMR protein (characterized by a mean expression score < 4) for 17 of the 59 (29%) cases. The tumor samples used to create the TMAs for these cases were sampled from the central portion of the respective original donor block. Characteristics of the tumors under study are shown in Table 1. Of the 17 cases in which there was initial loss of expression, the mean age of these tumor blocks was 59.5 months (range: 45-74; standard deviation (SD): 2.6). The mean number of blocks sampled upon follow-up was 3.6 (range: 1-5; SD: 0.4). The mean number of cores sampled upon follow-up was 10.8 (range 3.15; SD: 1.1).
Table 1.
Histopathologic characteristics of epithelial ovarian cancer cases
| Total (n=59) | Initial Loss of Expression (n=17) | No Initial Loss of Expression (n=42) | |
|---|---|---|---|
| Histologic Subtype | #cases (%) | #cases (%) | #cases (%) |
| Serous | 26 (44.0) | 5 (29) | 21 (50) |
| Endometrioid | 11 (18.6) | 5 (29) | 6 (14) |
| Mucinous | 8 (13.6) | 3 (18) | 5 (12) |
| Mixed Cell, NOS | 8 (13.6) | 3 (18) | 5 (12) |
| Adenocarcinoma, NOS | 2 (3.4) | 1 (6) | 1 (2) |
| Clear Cell | 1 (1.7) | 0 | 1 (2) |
| Other |
3 (5.1) |
0 |
3 (7) |
| Stage | |||
| I | 16 (27.1) | 5 (29) | 11 (26) |
| II | 5 (8.5) | 2 (12) | 3 (7) |
| III | 33 (55.9) | 10 (59) | 23 (55) |
| IV |
5 (8.5) |
0 |
5 (12) |
| Grade | |||
| I | 17 (28.8) | 3 (18) | 14 (33) |
| II | 14 (23.7) | 3 (18) | 11 (26) |
| III | 26 (44.1) | 9 (52) | 17 (40) |
| NA | 2 (3.4) | 2 (12) | 0 |
To evaluate the potential that the location of the core to create the TMA influenced the staining results, we compared staining from the same block, but on cores taken from the periphery of the paraffin-embedded tumor tissue, as compared to those taken from the center. Results of staining of full sections showed that 11 of the 17 tumor sections had lack of expression in the central portion, but positive expression in the periphery. Tumor sections for the other 6 cases revealed lack of expression in both the center and periphery. Additional analyses were performed on a subset of the 42 cases that showed positive expression on the initial TMA, all of which were sampled centrally. Full sections were created on 5 of the 42 cases for IHC analysis, and results showed consistent protein expression in both the center and periphery. Additionally, a subset of 19 randomly selected cases not used in the TMA were stained with Vimentin, which is an antigen particularly sensitive to inappropriate fixation (44). Of these cases, 16 showed positive Vimentin staining, demonstrating adequate fixation. In 3 cases, the Vimentin stain was negative, suggesting inadequate fixation.
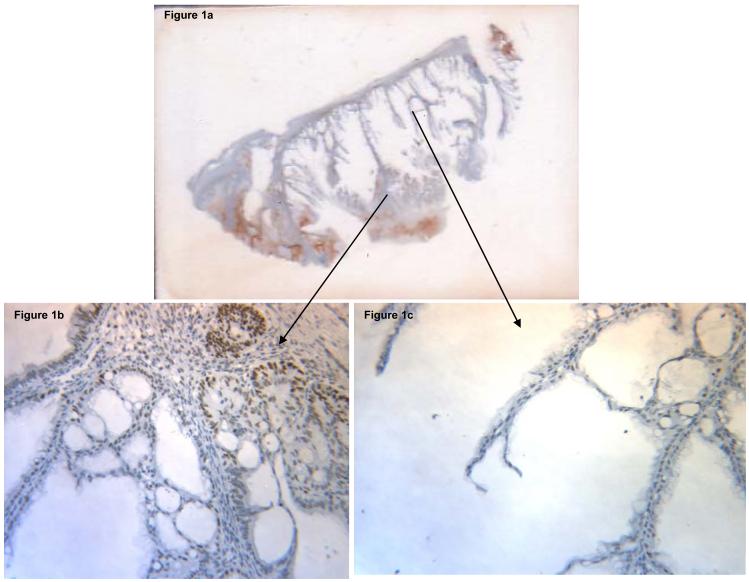
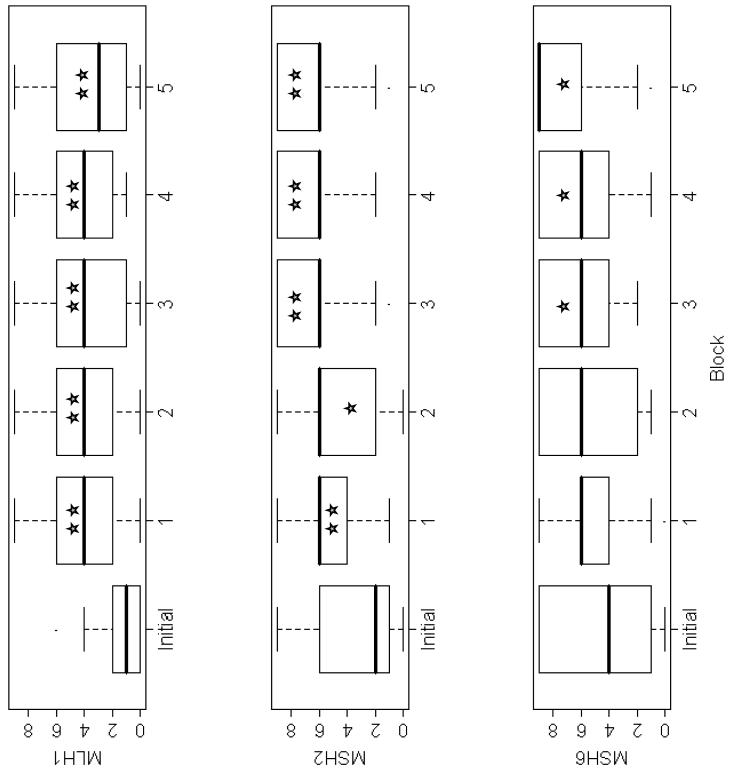
In order to clarify the initial IHC results, follow-up analyses were performed through construction of a new TMA comprised of peripheral cores created on the 17 cases. The results of IHC expression for the three MMR proteins showed loss of expression in the same 6 cases (ie: 10% of the total sample) that had lacked both central and peripheral protein expression on full sections. Sample images comparing MLH1 expression from cores sampled centrally versus peripherally from a mucinous adenocarcinoma are illustrated in Figure 1 and these observations were consistently seen in those cases with initial loss of MMR protein expression when sampled centrally, which upon preferential peripheral sampling, indicated presence of expression. These results were not attributed to an edge effect, as the criteria used to distinguish an edge effect from true positive staining is that edge effect generally involves all components of the tissue affected, thus would not be specifically localized to the nuclei, in contrast to findings illustrated in Figure 1. Statistical analyses were performed to compare results of the initial and follow-up IHC analyses. For each of the three MMR proteins, the overall mean expression score was lower for the initially centrally-sampled block as compared to the overall mean expression score for each of the peripherally-sampled follow-up blocks. Specifically, the difference in results between the initial donor block and follow-up blocks for each protein were statistically significant for MLH1 (all p-values ≤ 0.001) and MSH2 (all p-values ≤ 0.001) in all cases and in three of 5 comparisons for MSH6 (at p=0.001 level). Thus, there were lower overall mean expression scores in the initially centrally-sampled block as compared to the overall mean expression score for each of the peripherally-sampled follow-up blocks. Initially, each linear mixed model was fitted to account for potential confounding factors such as tumor size, tumor block age, and the number of blocks sampled. None of these variables altered the overall mean expression score across blocks in the models. Box plots were generated to visually portray the difference in expression scores when comparing central versus peripheral sampling from the initial versus follow-up blocks, respectively (Figure 2). The observed differences could not be accounted for by tumor size, tumor block age, the number of blocks sampled, or the number of cores punched per block, for any of the three proteins examined.
Figure 1a.
Full section slide of a mucinous adenocarcinoma stained for hMLH1, demonstrating lack of protein expression centrally and presence of expression peripherally (magnification: 100X); Figure 1b: Centrally sampled core demonstrating lack of hMLH1 expression (magnification: 200X); Figure 1c: Peripherally-sampled core demonstrating positive hMLH1 expression (magnification: 200X).
Figure 2.
Comparison of initial and follow-up results from MMR protein expression analysis: The box plots of the results of the staining of each of the 3 proteins (ie: MLH1, MSH2, and MSH6) is demonstrated. The `initial' block refers to the block initially analyzed. Blocks 1-5 refer to the blocks used during the follow-up analyses. Specifically, the numbers of follow-up blocks were: 1 (17 cases), 2 (14 cases), 3 (13 cases), 4 (10 cases), and 7 (5 cases). For comparisons of `initial' block with each follow-up block, statistically significant findings are indicated as follows: ★indicates P<0.05; ★★indicates P<0.001.
DISCUSSION
The results of the current study illustrate the potential impact that tissue sampling strategy can have on biomarker studies when TMAs are used as the primary platform for protein expression analysis. Specifically, our results indicate that peripheral, rather than central, sampling of tumor blocks has the potential to yield more accurate results about the presence of MMR protein expression in a given EOC tumor. The impetus for this study stemmed from findings obtained during the conduct of a larger population-based study investigating MMR protein expression in EOC. In that previous investigation, TMA platforms were used to determine MMR expression levels and observed loss of expression estimates were much higher than the expected 2-10% reported in the literature (34-37). Motivated by this discrepancy, we sought to explore possible explanations for this difference. Further investigations subsequently implicated that tissue core sampling strategy within the tumor specimens during the creation of TMAs was the likely source of the difference.
In the current study, improper tissue fixation is the most likely etiology of the discrepant finding between central and peripheral sampling. Previous investigations have shown that formaldehyde-based fixation of tumor tissue (2, 45, 46) is a factor when interpreting IHC results. Cross-linking of proteins is one of the most critical molecular changes induced by formalin, and the ideal fixation time for a 5μm thick tissue block is 12-24 hours.(46) Underfixation occurs when formalin diffuses slowly into tissues, resulting in strong staining near the periphery of a tumor block and less staining in the center (45), where fixation is poor, due to inadequate preservation. Tissue fixation is one of the least controlled phases of the IHC staining process because fixation conditions (i.e. time to fixation, total fixation time, rate of fixative penetration in tissues of different types and thicknesses) can vary by specimen and institution. (47, 48) Tissue underfixation as the cause of discrepant TMA results has previously been reported in prostate cancer(45), although there are no such reports in EOC. Our findings highlight the need to consider the effect of improper fixation on biomarker expression in TMA-based studies. Previous investigations have shown that tumors should be fixed within 30 minutes of surgical removal of the tissue, as delayed fixation causes increased proteolytic degradation. Depending on the antigen, this may lead to irreversible weak or absent staining.(46) The specimens should remain in the fixative for at least 12 hours to avoid underfixation, yet no longer than 24 hours to minimize overfixation. (46)
Another source of variation of IHC results is the presence of tissue heterogeneity which affects the extent to which TMA-derived cores are representative of the parent tumor from which they are derived. For example, colorectal cancers are often large and highly heterogeneous with marked stromal areas between glandular structures. This results in an increased chance that a core biopsy may miss tumor-cell-rich regions. In contrast, in the case of thyroid malignancies where tumors consist of more homogeneous regions packed with cancer-rich cells, there is less concern that a sampled core may misrepresent the tumor as a whole(8). This concern regarding tissue heterogeneity is not unique to cores, however, since conventional tumor sections also represent a small fraction of the volume of most tumors (4). Validation studies have compared molecular expression data derived from cores and conventional full sections, and have shown high concordance between the methods (4, 7-12, 49), although the concordance varies according to tumor type and the respective degree of heterogeneity. The degree of heterogeneity is influenced by intratumor levels of ischemia and/or proximity to vessels or stroma, resulting in effects on delivery of chemokines, growth factors, or other modulators(50).
Heterogeneity is frequently seen in epithelial ovarian cancers, which are typically large in size(51-53). Furthermore, as with other tumor types, there is biological variability within and between individual EOC tumor blocks (17). However, to our knowledge, no studies to date have compared IHC results from core- vs. full-section-derived methods when using EOC specimens. In the current study, an internal validation study was completed prior to using TMAs in this investigation, which involved the comparison of full-section versus TMA-derived IHC results in a small subset of EOC cases, and findings showed concordance of 95%, 90%, and 75% for MLH1, MSH2, and MSH6, respectively (data not shown). Although reassuring, large validation studies are warranted when investigating biomarkers in heterogeneous tumors like EOC, as the use of TMAs may be limited due to inadequate sampling of representative tumor tissue due to tumor heterogeneity. Overall, the higher the degree of tumor heterogeneity, the less likely TMAs may be to adequately represent the tumor of origin.
Although staining artifacts at the edge of tissue sections, referred to as leading edge effects, are a known phenomenon in IHC (8, 46), such artifacts are unlikely to be responsible for the discrepant biomarker expression scores when sampling from the periphery versus the center of the individual tumor blocks. This is because the cores with lack of expression were distributed throughout the tumor blocks, derived from various sections of the original tumor.
Another factor to consider when constructing TMAs for biomarker expression studies are both the chacteristics of the specific biomarker, as well as the expression pattern. For example, investigations of proteins involved in angiogenesis are particularly sensitive to ischemia during sample collection, which could greatly affect protein pattern (54). In contrast, vimentin, an ubiquitous antigen, is usually used as a reporter molecule, to assess fixation and processing of tissue. It has been shown that vimentin is particularly sensitive to overfixation, and may be destroyed by this process(44). Therefore, the vimentin monoclonal antibody, V9, is used to assess the degree of formalin fixation. The MMR antibodies used in the current investigations are known to be relatively stable, based on their reliability in accurately identifying colorectal tumors with loss of MMR protein expression, in which germline mutations have subsequently been identified(32, 38, 55-57). Ultimately, when contemplating the use of a particular biomarker, systematic analyses should be performed to determine the level of expression and the degree of uniformity required to accurately describe the outcome of interest (6, 50)
There were several strengths in the current investigation, including the prior quality work completed to validate correlation of IHC protein expression results between TMAs and full sections. In addition, we applied rigorous statistical methods to evaluate differences between central and peripheral punches. Despite these strengths, there remain a few limitations, including the relatively small sample size, sources of variability inherent in the multi-step process of IHC analysis of archived tumor tissue, as well as tumor heterogeneity. Additionally, although it would have been preferable to normalize the staining intensity in cancer cells to adjacent normal cells, this was not feasible given the study resources, thus more research is needed to evaluate whether or not this enhances the ability to utilize sub optimally fixed tumor specimens for IHC analysis.
Findings from this study suggest that preferentially sampling from the periphery of archival tumor blocks when constructing TMAs in preparation for IHC may be important because exposure to tissue fixatives is optimal in this location. This may reduce the likelihood of tissue fixation as a contributor to the lack of protein expression, and improve the reliability of the staining interpretation and overall validity of study results. Alternatively, another option is to stain full section slides from tumor blocks selected for TMA construction with Vimentin, and only proceed accordingly. Ultimately, the introduction of the TMA enhances the ability to conduct biomarker research on a large number of tumor specimens in a cost-effective and efficient manner under uniform experimental conditions; however, data from the current investigation underscore the notion that there are several issues that should be considered when contemplating clinical research using TMAs for protein expression analysis of archival tumor tissue. Furthermore, it is critical to perform additional research to develop uniform processing procedures interpretation, including the development of optimal tissue fixation guidelines (i.e.: thickness of sections, time to fixation, type of fixative used), with subsequent implementation. These standards have recently been implemented for processing of breast cancer samples (50, 58), and evaluating the implementation of these standards for all tumor processing should be considered. .
Acknowledgments
Financial Support: This work was supported by grants #R01CA111914 (TP) and #K07CA108987 (TP) from the National Cancer Institute.
REFERENCES
- 1.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 2.Cregger M, Berger AJ, Rimm DL. Immunohistochemistry and quantitative analysis of protein expression. Arch Pathol Lab Med. 2006;130:1026–30. doi: 10.5858/2006-130-1026-IAQAOP. [DOI] [PubMed] [Google Scholar]
- 3.Fedor HL, De Marzo AM. Practical methods for tissue microarray construction. Methods Mol Med. 2005;103:89–101. doi: 10.1385/1-59259-780-7:089. [DOI] [PubMed] [Google Scholar]
- 4.Bentzen SM, Buffa FM, Wilson GD. Multiple biomarker tissue microarrays: bioinformatics and practical approaches. Cancer Metastasis Rev. 2008 doi: 10.1007/s10555-008-9145-8. [DOI] [PubMed] [Google Scholar]
- 5.Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10:657–62. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang H, Zhang W, Fuller GN. Tissue microarrays: applications in neuropathology research, diagnosis, and education. Brain Pathol. 2002;12:95–107. doi: 10.1111/j.1750-3639.2002.tb00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriks Y, Franken P, Dierssen JW, et al. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469–77. doi: 10.1016/S0002-9440(10)63841-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoos A, Cordon-Cardo C. Tissue microarray profiling of cancer specimens and cell lines: opportunities and limitations. Lab Invest. 2001;81:1331–8. doi: 10.1038/labinvest.3780347. [DOI] [PubMed] [Google Scholar]
- 9.Jourdan F, Sebbagh N, Comperat E, et al. Tissue microarray technology: validation in colorectal carcinoma and analysis of p53, hMLH1, and hMSH2 immunohistochemical expression. Virchows Arch. 2003;443:115–21. doi: 10.1007/s00428-003-0833-z. [DOI] [PubMed] [Google Scholar]
- 10.Griffin MC, Robinson RA, Trask DK. Validation of tissue microarrays using p53 immunohistochemical studies of squamous cell carcinoma of the larynx. Mod Pathol. 2003;16:1181–8. doi: 10.1097/01.MP.0000097284.40421.D6. [DOI] [PubMed] [Google Scholar]
- 11.Rubin MA, Dunn R, Strawderman M, Pienta KJ. Tissue microarray sampling strategy for prostate cancer biomarker analysis. Am J Surg Pathol. 2002;26:312–9. doi: 10.1097/00000478-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Zu Y, Steinberg SM, Campo E, et al. Validation of tissue microarray immunohistochemistry staining and interpretation in diffuse large B-cell lymphoma. Leuk Lymphoma. 2005;46:693–701. doi: 10.1080/10428190500051844. [DOI] [PubMed] [Google Scholar]
- 13.Chiaffarino F, Parazzini F, Bosetti C, et al. Risk factors for ovarian cancer histotypes. Eur J Cancer. 2007;43:1208–13. doi: 10.1016/j.ejca.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 14.Eltabbakh GH, Natarajan N, Piver MS, Mettlin CJ. Epidemiologic differences between women with borderline ovarian tumors and women with epithelial ovarian cancer. Gynecol Oncol. 1999;74:103–7. doi: 10.1006/gyno.1999.5459. [DOI] [PubMed] [Google Scholar]
- 15.Kurian AW, Balise RR, McGuire V, Whittemore AS. Histologic types of epithelial ovarian cancer: have they different risk factors? Gynecol Oncol. 2005;96:520–30. doi: 10.1016/j.ygyno.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Risch HA. Estrogen replacement therapy and risk of epithelial ovarian cancer. Gynecol Oncol. 1996;63:254–7. doi: 10.1006/gyno.1996.0315. [DOI] [PubMed] [Google Scholar]
- 17.Scott M, McCluggage WG. Current concepts in ovarian epithelial tumorigenesis: correlation between morphological and molecular data. Histol Histopathol. 2006;21:81–92. doi: 10.14670/HH-21.81. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein NS, Bosler D. An approach to interpreting immunohistochemical stains of adenocarcinoma in small needle core biopsy specimens: the impact of limited specimen size. Am J Clin Pathol. 2007;127:273–81. doi: 10.1309/ATHVF5R4CQUKB7LX. [DOI] [PubMed] [Google Scholar]
- 19.O'Leary TJ. Standardization in immunohistochemistry. Appl Immunohistochem Mol Morphol. 2001;9:3–8. [PubMed] [Google Scholar]
- 20.Seidal T, Balaton AJ, Battifora H. Interpretation and quantification of immunostains. Am J Surg Pathol. 2001;25:1204–7. doi: 10.1097/00000478-200109000-00013. [DOI] [PubMed] [Google Scholar]
- 21.Wick MR, Mills SE. Consensual interpretive guidelines for diagnostic immunohistochemistry. Am J Surg Pathol. 2001;25:1208–10. doi: 10.1097/00000478-200109000-00014. [DOI] [PubMed] [Google Scholar]
- 22.American Cancer Society I Cancer Facts & Figures 2007. 2007 [Internet] [cited 2007 2007]; Available from: http://www.cancer.org/docroots/STT/stt_0.asp.
- 23.Watson P, Lynch HT. Cancer risk in mismatch repair gene mutation carriers. Fam Cancer. 2001;1:57–60. doi: 10.1023/a:1011590617833. [DOI] [PubMed] [Google Scholar]
- 24.Vasen HF, Wijnen JT, Menko FH, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–7. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 25.Aarnio M, Mecklin JP, Aaltonen LA, Nystrom-Lahti M, Jarvinen HJ. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer. 1995;64:430–3. doi: 10.1002/ijc.2910640613. [DOI] [PubMed] [Google Scholar]
- 26.Watson P, Lynch HT. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer. 1993;71:677–85. doi: 10.1002/1097-0142(19930201)71:3<677::aid-cncr2820710305>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Brown GJ, St John DJ, Macrae FA, Aittomaki K. Cancer risk in young women at risk of hereditary nonpolyposis colorectal cancer: implications for gynecologic surveillance. Gynecol Oncol. 2001;80:346–9. doi: 10.1006/gyno.2000.6065. [DOI] [PubMed] [Google Scholar]
- 28.Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81:214–8. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 29.Cederquist K, Emanuelsson M, Wiklund F, Golovleva I, Palmqvist R, Gronberg H. Two Swedish founder MSH6 mutations, one nonsense and one missense, conferring high cumulative risk of Lynch syndrome. Clin Genet. 2005;68:533–41. doi: 10.1111/j.1399-0004.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 30.Arzimanoglou, Lallas T, Osborne M, Barber H, Gilbert F. Microsatellite instability differences between familial and sporadic ovarian cancers. Carcinogenesis. 1996;17:1799–804. doi: 10.1093/carcin/17.9.1799. [DOI] [PubMed] [Google Scholar]
- 31.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst. 1995;87:1114–25. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 32.Marcus VA, Madlensky L, Gryfe R, et al. Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol. 1999;23:1248–55. doi: 10.1097/00000478-199910000-00010. [DOI] [PubMed] [Google Scholar]
- 33.Rigau V, Sebbagh N, Olschwang S, et al. Microsatellite instability in colorectal carcinoma. The comparison of immunohistochemistry and molecular biology suggests a role for hMSH6 [correction of hMLH6] immunostaining. Arch Pathol Lab Med. 2003;127:694–700. doi: 10.5858/2003-127-694-MIICC. [DOI] [PubMed] [Google Scholar]
- 34.Domanska K, Malander S, Masback A, Nilbert M. Ovarian cancer at young age: the contribution of mismatch-repair defects in a population-based series of epithelial ovarian cancer before age 40. Int J Gynecol Cancer. 2007;17:789–93. doi: 10.1111/j.1525-1438.2007.00875.x. [DOI] [PubMed] [Google Scholar]
- 35.Geisler JP, Goodheart MJ, Sood AK, Holmes RJ, Hatterman-Zogg MA, Buller RE. Mismatch repair gene expression defects contribute to microsatellite instability in ovarian carcinoma. Cancer. 2003;98:2199–206. doi: 10.1002/cncr.11770. [DOI] [PubMed] [Google Scholar]
- 36.Malander S, Rambech E, Kristoffersson U, et al. The contribution of the hereditary nonpolyposis colorectal cancer syndrome to the development of ovarian cancer. Gynecol Oncol. 2006;101:238–43. doi: 10.1016/j.ygyno.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 37.Rosen DG, Cai KQ, Luthra R, Liu J. Immunohistochemical staining of hMLH1 and hMSH2 reflects microsatellite instability status in ovarian carcinoma. Mod Pathol. 2006;19:1414–20. doi: 10.1038/modpathol.3800672. [DOI] [PubMed] [Google Scholar]
- 38.Chiaravalli AM, Furlan D, Facco C, et al. Immunohistochemical pattern of hMSH2/hMLH1 in familial and sporadic colorectal, gastric, endometrial and ovarian carcinomas with instability in microsatellite sequences. Virchows Arch. 2001;438:39–48. doi: 10.1007/s004280000325. [DOI] [PubMed] [Google Scholar]
- 39.Singer G, Kallinowski T, Hartmann A, et al. Different types of microsatellite instability in ovarian carcinoma. Int J Cancer. 2004;112:643–6. doi: 10.1002/ijc.20455. [DOI] [PubMed] [Google Scholar]
- 40.Pal T, Permuth-Wey J, Betts JA, et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–16. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 41.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. Journal of the American Statistical Association. 1993;88:9–25. [Google Scholar]
- 42.Wolfinger ROCM. Generalized linear mixed models: A pseudo-likelihood approach. Journal of Statistical Computation and Simulation. 1993;4:233–43. [Google Scholar]
- 43.Kenward MGRJ. Small Sample Inference for Fixed Effects from Restricted Maximum Likelihood. Biometrics. 1997;53:983–97. [PubMed] [Google Scholar]
- 44.Battifora H. Assessment of antigen damage in immunohistochemistry. The vimentin internal control. Am J Clin Pathol. 1991;96:669–71. doi: 10.1093/ajcp/96.5.669. [DOI] [PubMed] [Google Scholar]
- 45.De Marzo AM, Fedor HH, Gage WR, Rubin MA. Inadequate formalin fixation decreases reliability of p27 immunohistochemical staining: probing optimal fixation time using high-density tissue microarrays. Hum Pathol. 2002;33:756–60. doi: 10.1053/hupa.2002.126187. [DOI] [PubMed] [Google Scholar]
- 46.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am J Surg Pathol. 2000;24:1016–9. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 47.Leong AS. Quantitation in immunohistology: fact or fiction? A discussion of variables that influence results. Appl Immunohistochem Mol Morphol. 2004;12:1–7. doi: 10.1097/00129039-200403000-00001. [DOI] [PubMed] [Google Scholar]
- 48.Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–24. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 49.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest. 2000;80:1943–9. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 50.Moeder CB, Giltnane JM, Harigopal M, et al. Quantitative justification of the change from 10% to 30% for human epidermal growth factor receptor 2 scoring in the American Society of Clinical Oncology/College of American Pathologists guidelines: tumor heterogeneity in breast cancer and its implications for tissue microarray based assessment of outcome. J Clin Oncol. 2007;25:5418–25. doi: 10.1200/JCO.2007.12.8033. [DOI] [PubMed] [Google Scholar]
- 51.Ashworth A, Balkwill F, Bast RC, et al. Opportunities and challenges in ovarian cancer research, a perspective from the 11th Ovarian cancer action/HHMT Forum, Lake Como, March 2007. Gynecol Oncol. 2008;108:652–7. doi: 10.1016/j.ygyno.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 52.Naora H. The heterogeneity of epithelial ovarian cancers: reconciling old and new paradigms. Expert Rev Mol Med. 2007;9:1–12. doi: 10.1017/S1462399407000324. [DOI] [PubMed] [Google Scholar]
- 53.Khalique L, Ayhan A, Weale ME, Jacobs IJ, Ramus SJ, Gayther SA. Genetic intra-tumour heterogeneity in epithelial ovarian cancer and its implications for molecular diagnosis of tumours. J Pathol. 2007;211:286–95. doi: 10.1002/path.2112. [DOI] [PubMed] [Google Scholar]
- 54.Atkin GK, Daley FM, Bourne S, Glynne-Jones R, Northover JM, Wilson GD. The impact of surgically induced ischaemia on protein levels in patients undergoing rectal cancer surgery. Br J Cancer. 2006;95:928–33. doi: 10.1038/sj.bjc.6603362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonis PA, Trikalinos TA, Chung M, et al. Evid Rep Technol Assess (Full Rep) 2007. Hereditary nonpolyposis colorectal cancer: diagnostic strategies and their implications; pp. 1–180. [PMC free article] [PubMed] [Google Scholar]
- 56.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–56. [PubMed] [Google Scholar]
- 57.Thibodeau SN, French AJ, Roche PC, et al. Altered expression of hMSH2 and hMLH1 in tumors with microsatellite instability and genetic alterations in mismatch repair genes. Cancer Res. 1996;56:4836–40. [PubMed] [Google Scholar]
- 58.Ross JS, Symmans WF, Pusztai L, Hortobagyi GN. Standardizing slide-based assays in breast cancer: hormone receptors, HER2, and sentinel lymph nodes. Clin Cancer Res. 2007;13:2831–5. doi: 10.1158/1078-0432.CCR-06-2522. [DOI] [PubMed] [Google Scholar]