Abstract
Here we use published 16S rRNA gene sequences to compare the bacterial assemblages associated with humans, other mammals, other metazoa, and free-living microbial communities spanning a range of environmental conditions. The composition of the vertebrate gut microbiota is influenced by diet, host morphology and phylogeny, and in this respect the human gut bacterial community is typical for an omnivorous primate. However, a wider view reveals that the vertebrate gut microbiota is highly differentiated from free-living communities not associated with animal body habitats. The recently initiated international Human Microbiome Project should strive to include a broad representation of humans, as well as other mammals and environmental samples: comparative analyses of microbiotas and their microbiomes are a powerful way to explore the evolutionary history of the biosphere.
Introduction
Diverse microbes, and a diversity of microbial communities, are a feature of modern life on Earth, and likely necessary for life as we know it to have evolved1 Microbes formed spatially organized communities as early as 3.25 billion years ago when some left their mark in the fossil record2. Today, microbial life is found in diverse communities all over the biosphere. The high level of novelty necessary for microbes to develop a diversity of cell lineages and inhabit a vast diversity of habitats likely required whole communities to have exchanged innovations1. Comparative studies of microbial communities are starting to reveal which environmental features, such as biogeography, salinity, or redox-potential, have important effects on the organization of microbial diversity 3-6. These types of analyses are now being extended to the microbial communities that populate a globally ubiquitous but ephemeral habitat: the body surfaces of animals, including those of humans.
Multicellular eukaryotes have existed for at least a quarter of Earth's history, or 1.2 billion years7. Thus, an already long history of interaction between multicellular life-forms and microbial communities preceded, and likely shaped, the evolution of the vertebrates. The legacy of ancient associations between hosts and their epibiotic microbial communities is evident in the present day effects that the gut microbiota exerts on host biology, ranging from the structure and functions of the gut and the innate and adaptive immune systems, to host energy metabolism8-11. Host responses to microbial colonization are evolutionarily conserved among diverse vertebrates including zebrafish, mice and humans12. The scripts that dictate our interactions with our microbial partners thus provide some of the foundations of our Homo sapiens genome.
If microbial communities are and have always been so intricately associated with their vertebrate hosts, then how specialized are body-associated microbial lineages to vertebrates, and how distinct are they from those that populate the non-living environments of the biosphere? In this review we place our human gut microbiota in the context of many other diverse microbiotas, from our close relatives the primates, to more distantly related mammals, to other metazoans, and finally to `free-living' microbial communities. This evolutionary ecology perspective helps put the recently initiated international Human Microbiome Project13 in the context of the biosphere within which we humans and our microbes evolved.
Diet and the evolution of modern humans
Food is central to the evolution of Homo sapiens. During the first half of the evolution of our lineage, Australopithecus species split from prehistoric apes, persisting from ~4.4 Mya until about 2.5 Mya14. This early split has been associated with a dietary shift to seed and soft fruits, based on comparisons of australopithecine and prehistoric ape tooth morphologies14. The second half of our evolutionary story saw Homo erectus appear, about 2 Mya, and persist for ~1My15. H. erectus was a large-bodied, bipedal hominid, similar in size and form to modern humans, but with a brain size and life-span intermediate between that of modern H. sapiens and chimpanzees15. The appearance and persistence of the early Homo species occurred as Earth's climate became cooler, drier and more variable, and as C4-plant dominated grasslands expanded.
Climate-driven changes in habitat are thought to have favored significant alterations in the foraging behavior and diet of early Homo species16,17. In one view, termed the “grandmother hypothesis”, foraging and sharing of so-called “underground storage units” (plant roots, bulbs and tubers) by older females gained importance and transformed Homo biology, ecology and societies15. The grandmother hypothesis was derived in part from observations of the Hadza people: in this small population of traditional foragers inhabiting arid savannah woodlands of the Eastern Rift in Tanzania, older women's foraging for tubers is essential for the nourishment of children. Grandmother support of grandchildren allows more pregnancies in daughters and thus greater reproductive fitness15. This kind of behavior in our ancestors would have favored post-menopausal longevity, a distinguishing trait of humans15. An increased role of roots, bulbs and tubers in the diet of early Homo species is consistent with the stable isotope composition of early hominin tooth enamel and bone apatite, which are similar to those of the African mole rat, a present-day consumer of such foods18. A second major shift in diet occurred during the Pleistocene when Homo sapiens began eating more meat: this may have been critical for the development of a larger brain [the “Expensive Tissue Hypothesis” 19]. By the Middle Pleistocene, stone-tool manufacturing and big-game hunting were widespread20. A third major change in diet came with adoption of agricultural practices and the domestication of animals, particularly dairy animals, and involved a subset of the population, albeit one that eventually largely replaced the hunter-gatherer societies.
Changes in the human diet over time have left their mark on our genomes, and comparisons between the human genome and the genomes of our close relatives have uncovered genetic and structural differences involved in the development of human-lineage specific traits21. The importance of starch in human ecological history is reflected in the population genetics of the gene for salivary amylase (AMY1), the enzyme responsible for starch hydrolysis. The ancestral amylase gene was duplicated in the human genome after the split of humans from the chimpanzee lineage22. Human AMY1 shows extensive variation in copy number, with roughly three times more copies than in the chimpanzee genome. Furthermore, higher copy number correlates positively with salivary amylase protein levels in humans23. Remarkably, individuals from human populations with high-starch diets (e.g., agricultural societies) have, on average, more AMY1 copies than those with traditionally low-starch diets (e.g., rainforest and circumarctic hunter-gatherers) 23. The more recent addition of milk to the human diet led to the prevalence of lactose tolerance in modern humans. Lactose tolerance in current human populations coincides geographically with milk protein gene diversity in cattle, and with the locations of Neolithic cattle farming sites24. Human reliance on domesticated milk-producing animals has only occurred in the last 10,000 years or so: fixation of this trait, which is dominant in families, is very recent in our evolution16. Comparisons between different human lactase haplotypes and with the chimpanzee gene sequence enabled the ancestral haplotype to be determined, and provided evidence for positive selection in northern Europeans associated with lactase persistence 25.
The gut microbiota in the evolution of humans from primate ancestors
Initial studies indicate that different aspects of the gut microbiota can distinguish human populations according to their histories and lifestyles, including diet. The stomach-associated bacterium Helicobacter pylori exemplifies coevolution between microbes and humans: patterns of Homo sapiens migration out of Africa and across the globe can be traced from the strain diversity of this species26. Furthermore, urinary levels of gut microbial-host co-metabolites such as hippurate, phenylacetylglutamine and methylamines differentiate present day East Asian and Western populations27,28.
Contrasting the human gut microbiota and human gut microbiome (the microbiota's full complement of genes; see box for a glossary of terms) with those of other primates and mammals could, in principle, reveal if there is a `core' set of gut microbial genes and lineages that are shared by most if not all humans. Such comparisons could shed light on how attributes specific to modern human biology and nutrition, such as the availability of foods that are more diverse, abundant, and often heavily processed, affect our microbial partners. Evidence gathered to date from a limited number of people indicates that composition of the gut microbiota, as measured by 16S rRNA gene sequences, varies substantially between individuals29,30,31. We have placed inter-human variability in the broader context of inter-mammal variability by performing a 16S rRNA gene sequence-based comparative analysis of the human gut microbiota with the fecal samples from 17 non-human primate species and 42 other mammal species representing 11 taxonomic orders32. Despite large variability between human fecal bacterial communities from healthy men and women from three continents, spanning 27 to 94 years of age, the human-associated communities were more similar to one another than to those associated with members of other mammalian species.
This finding emerged from two complementary and related types of analysis. In the first, `UniFrac'33 distances between samples were compared: these distances represent the fraction of branch length shared by any two samples' communities in a phylogenetic tree built from16S rRNA sequence data from all samples. UniFrac distances between samples were significantly less when the samples belonged to the same species (conspecifics) than to different species. Similarly, UniFrac distances between human samples were significantly smaller than distances between humans and other animals32.
The second approach considered a single level of phylogenetic resolution in the data (i.e., the level of bacterial genera, or operational taxonomic units (OTUs) that share ≥96% identity in their 16S rRNA gene sequences). The OTU-based analysis employed a bipartite network in which mammal hosts were designated as one node type, and bacterial genera were designated as a second node type in the network: a given host was connected to a given bacterial genus node (via an `edge') if the genus was detected in the host. The results showed that human samples shared more genus-level OTUs with other humans than with other species of mammal and that conspecific hosts in general were more highly connected (shared more genera) to one another than to hosts of different species32.
Besides humans, the primates in this study included four other hominids (great apes, represented by the Bonobo, Chimpanzee, Orangutan and Gorilla), allowing comparisons between the human fecal microbiotas and those of closer primate relatives. Clustering of samples by UniFrac indicated that the human fecal microbiotas were more similar to those of other primates than to non-primates, but not to other hominids specifically. Instead, diet appeared to be of principal importance in clustering among primates. Human samples clustered with those of other omnivores (e.g., Ring-tailed Lemur, Black Lemur, Mongoose Lemur, Bonobo, Spider Monkey), but the other hominids, which tend have a diet that is more dominated by plant materials, clustered in an intermediate position between the omnivorous primates and non-primate herbivores such as the Artiodactyla (sheep and their relatives32).
Of the hominids surveyed, the Bonobo's diet includes the most fruit, and its microbiota clustered most closely with the humans. Thus, based on comparative measurements of the gut microbiotas of humans and primates alone, the human species might be viewed as unspecialized frugivores, one whose flexible diet includes seeds and meat depending on availability34. However, our close primate relatives are best described as “raw-foodists”, whereas our modern lifestyle employs technology to make preferred foods more available (with agriculture and elaborate transportation systems) but also more easily digested (by refining, fermentation, cooking). Yet despite the varied and sometimes very synthetic nature of our modern diets, the composition of our gut microbiotas does not depart dramatically in composition from the microbiotas of other omnivorous primates. What differences do exist between human and other primate microbiotas could result from diet, our modern lifestyle, or other facets of our biology. An understanding of how these factors, and their interplay, shape the human gut microbiota will require wider surveys of humans, with different lifestyles and cultures, in addition to a wider sampling of primates in the wild and captivity.
Primate microbiotas in the context of mammalian evolution
Ancestral mammals had teeth best suited for eating insects and meat, or fruit35. The incorporation of a wider set of plant parts into the diet came later in mammalian evolution. In the Jurassic, the massive radiation of mammals into herbivorous niches was likely spurred by competitive release, once the largely herbivorous dinosaurs became extinct36. Mammals of different lineages have independently evolved an herbivorous lifestyle many times: it is estimated that 80% of extant mammalian species are herbivorous.
Gut microbes were necessary for mammals to move into herbivorous niches. To liberate sugars from complex plant polysaccharides, the same solution was arrived at by different mammal lineages over evolutionary time: i.e, fermentation by microbes that possess the glycoside hydrolases and polysaccharide lyases needed for breakdown of complex polysaccharides that their hosts lack37,38. Digesta retention times had to be prolonged: this was accomplished by enlargement of parts of the gut in order to retain microbes (other solutions include re-passaging digesta by coprophagy). In what is a classic example of convergent evolution, mammals from various lineages have used two broad strategies: enlargement of the gut upstream of the stomach [foregut ferementers such as sheep, pig, cow, and some primates (e.g., Colobine monkeys)], and downstream of the stomach38 (hindgut fermenters such as the horse, rhinoceros, gorilla). However, it remains unclear whether the same microbes perform the same fermentative roles in all herbivorous mammals, or if the gut communities of herbivorous mammals have evolved this capacity independently, just as disparate mammal lineages have expanded the gut independently.
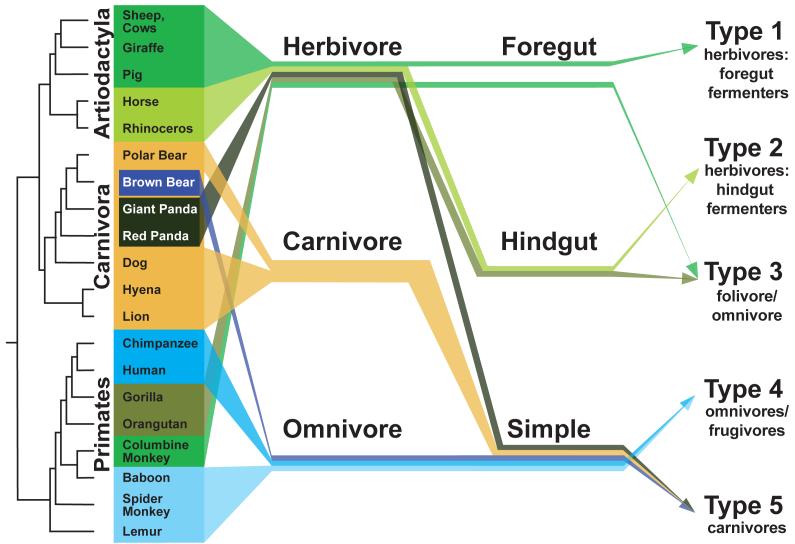
Our recent comparison of mammalian gut microbiotas32 addresses this question in part by including fecal microbial communities of related and unrelated mammals that have similar physiologies and diets. Overall, diet is a powerful predictor of fecal microbiota composition (Fig. 1). When mammal hosts are classified as herbivores, omnivores and carnivores, their microbiotas cluster into groups that corresponded to these categories. However, gut physiology is also a strong predictor of the composition of the fecal microbiotas. For example, herbivore-associated fecal microbiotas split into two groups according to whether they were hindgut or foregut fermenters. Remarkably, these similarities occur irrespective of the taxonomic order of the hosts (a cluster of foregut-fermenters included the Red Kangaroo and sheep, and a cluster of hindgut fermenters included elephants and horses). It remains to be seen if the fecal microbiotas are different between foregut and hindgut digesters because of downstream metabolic processing of the digesta once it passes out of the fermentation pouch, or because the communities within the area of the gut where the fermentation occurs differ.
Figure 1. Factors shaping the mammalian gut microbiota.
Schematic representation of the impact of host phylogeny, gut morphology and diet on fecal bacterial community composition, 32. A host phylogeny (far left) shows the evolutionary relationships of a representative set of mammals whose names are colored according to diet. The gut morphologies of the mammals are indicated with arrows, as well as their diet categories and the type of fecal bacterial community recovered (Types 1-532). Related animals typically have the same type of gut, diet and gut bacterial community (e.g., the Artiodactyla, green arrows). Other branches of the mammalian tree have `outlier' species, such as the Columbine monkeys in the Primates, whose gut physiology (foregut enlargement) and diet (herbivory) are unusual for the lineage. Note that although sampled animals with simple gut morphologies can have a variety of diets, they only hd community types 3,4 and 5; carnivores only had type 5 fecal bacterial communities, even though type 5 communities were found in animals with a variety of diets.
In some mammal lineages, gut morphology trumps host phylogeny and diet in determining fecal microbial community composition. These three aspects of an animal are inter-related, and often confounded, so it is helpful to take a look at `outliers', or animals whose diets are unusual for their gut morphology (Fig. 1). The Giant and Red Pandas are herbivores with the simple gut of other members of the order Carnivora (most of which, as their name implies, eat a meat-rich diet). These carnivorous mammals share a set of fecal bacterial lineages that set them apart from the omnivores and herbivores32. Remarkably, despite their herbivorous diet, the panda fecal microbiotas were more similar to those of the other Carnivora than to unrelated herbivores. This finding complements the findings of Hackstein and colleagues39,40 who, based on an analysis of methane emission from 253 species of animals, proposed that the common ancestor of some phylogenetic groups of animals have lost the ability to harbor gut methanogens. They noted that the giant and red pandas, like their carnivore relatives, do not produce CH4. Red and giant pandas have a rapid passage rate of their digesta, and use plant cell contents rather than cell walls for their nutrition41.
Leaf-eating primates (e.g., Columbine Monkeys) are another `outlier' group, but one that presents a more complex scenario. These animals' gut microbiotas are in an intermediate position between the omnivorous primates (including humans) and other herbivores. It appears that incorporation of fiber-rich plant material into their diet is accompanied by the acquisition of fermentative microbial lineages that are typically dominant in true herbivores: however, they also retain lineages found in other omnivorous primates.
Mammal microbiotas in the context of the microbial biosphere
Are the physical and chemical niche attributes of the gut (structural configuration, flow rate, temperature, pH, nature of substrates) the principal factors governing microbial community composition of the mammal gut? Are all mammals, when born germ-free, best viewed as empty vessels with no host-mediated control over the inhabitants of the gut? To address these questions, we have placed the mammalian gut microbiotas into the greater context of `free-living' microbial communities not associated with the body surfaces of multicellular eukaryotes and those associated with non-vertebrates, non-mammal vertebrates, and other human habitats, using published 16S rRNA surveys. This dataset combines the gut mammalian dataset described above32 with the `environmental dataset' of Lozupone and Knight (2007), composed of 202 samples from 111 published studies of diverse free-living communities, including soils, seawater, hotsprings, sediments and lakes. The analysis of the free-living communities showed that microbial consortia of the biosphere fell clearly into two main groups: those adapted to saline conditions, and those adapted to non-saline conditions5. Another finding was that habitats in different locales harbored similar communities. Thus, despite the potential of horizontal gene transfer to confer any function to any lineage, it appears that the same phylogenetic lineages are performing the same functions under similar conditions in different places. The patterns observed for the gut microbiotas are generally similar: mammals with similar diets harbor similar microbiotas.
To allow for comparisons between the mammal gut and the guts of other vertebrate and non-vertebrates, and human associated habitats, we augmented this combined dataset substantially with data from other published studies (Table S1). To the gut mammalian dataset, which consisted entirely of fecal samples from healthy individuals, we added gut samples retrieved from mucosal tissue and rumen fluids, and from adult and infant humans with a range of physiologic and pathophysiologic phenotypes (obesity, antibiotic-resistant diarrhea, colonic diverticulosis). We augmented the `environmental dataset' with 34 samples from large sequencing efforts of free-living communities that were published after our initial analysis. In addition, we added samples from other human body habitats (including the mouth, skin, ear, vulva, and vagina), from the guts of non-mammal vertebrates, such as poultry and zebrafish, and from the gut or whole body of diverse metazoa, including termites, beetles, lice, earthworms, fruit flies, mosquitoes, bees, gypsy moth larvae, corals and sponges. The final dataset consisted of 99,801 16S rRNA sequences from 464 samples and 181 studies (see Table S1).
We anticipated that if physical and chemical attributes of the gut habitat are principal shapers of microbial community composition, those free-living communities most similar to the mammalian gut communities might be, for instance, those associated with anoxic environments with high levels of complex polysaccharides (for example, anoxic soils or bogs). We also expected that gut communities would be far less different from one another than communities from the other environments, given that the temperature, pH and other physical-chemical parameters are much more constrained.
The patterns that emerge from the combined dataset are very different from what we anticipated. Bacterial communities that occupy the majority of vertebrate guts are markedly different from non-animal (free-living) bacterial communities. Principal coordinate analysis (PCoA) based on UniFrac distances clearly separates bacterial communities obtained from the vertebrate gut from other types of communities (Fig. 2A). What is remarkable is that the separation along PC1 (the principal coordinate that accounts for the largest amount of variance between samples) of vertebrate gut-associated communities from free-living communities is more than twice as great as the separation between saline and non-saline communities, which was evident over PC3 (Fig. 2B). Samples that had intermediate values along this PC1 are from the human mouth, vaginal epithelium, and vulva, plus the guts of carnivorous vertebrates (Fig. 2A) and herbivorous/omnivorous bears, indicating that microbes that perform fermentation-based degradation of plant material in the intestinal tract contribute greatly to the extreme divergence of vertebrate gut communities. Moreover, one sample from an anoxic rice paddy soil76 clusters in an intermediate position between the free-living and vertebrate gut samples along PC1, supporting the notion that the anaerobic, polysaccharide-rich nature of the gut environment could be related to the dichotomy.
Figure 2. Variance in bacterial community composition between samples from vertebrate-gut associated and other `free-living' communities.
Principal Coordinates Analysis (PCoA) of unweighted UniFrac values33,80 for 464 environmental samples representing 99,801 16S rRNA sequences from free-living and animal-associated environments. Each point represents a sample, colored by habitats. The uppermost panel describes the color codes. The color designations in the middle section refer to all of the panels (A-F). The color designations to the right and left only refer to the panels in their respective columns. All sequences were aligned with NAST81 and added to the Greengenes core set tree82 using parsimony insertion in Arb83. Near identical sequences were removed from each environmental sample using Divergent Set84. Samples with less than 15 divergent sequences in the full dataset (panels A and B), and with less than four divergent sequences within a particular division (panels C-F) were removed from the analysis. (A,B) UniFrac PCoA clustering results for the bacteria, Principal Coordinate (PC) 1 versus PC2 (panel A) and PC1 versus PC3 (panel B). (C-F) PCoA of UniFrac values from selected bacterial phyla including the Bacteroidetes (panels C, D), the Firmicutes (panel E), and the Proteobacteria (panel F). The phylogenetic tree for each phylum was extracted from the same Arb parsimony insertion tree used in panel A. The percent of the variation described by the plotted PC axes are indicated.
Another striking pattern to emerge from this analysis is the distinction between vertebrate and invertebrate- associated communities. Almost all of the invertebrate gut communities cluster with the free-living communities, with the exception of termites and most of the samples from beetle larvae. The beetle samples that cluster between the noncarnivorous vertebrates and the free-living communities were all from the specialized anaerobic hindgut region of beetles with differentiated guts42,43, whereas a beetle sample from the whole gut of larvae of Anoplophora glabripennis44 clustered with the free-living communities. Similarly, the specialized gut structures of certain beetle taxa have been associated with methane production, and the presence of methanogens in terrestrial arthropods in general was found to be associated with taxon-specific traits45. These findings suggest a strong host phylogenetic effect on the structure of the microbiota of arthropods, as in mammals. This clustering is also consistent with the theme that one key factor that shapes gut differentiation is the provision of an anaerobic environment with abundant complex carbohydrates from plant materials.
Other nonvertebrate-associated communities, such as those from adult and larval bees, gypsy moth larvae, whole fruit flies, and the earthworm gut, clustered with the free-living communities. Exceptions included the casts of earthworms, which clustered more closely with soil. This section of the PC plot (Fig. 2A top left), shows an aggregation of bacterial communities associated with diverse complex organisms, such as bacteria that tightly associate with plant roots, the human skin, outer ear, and vulva, plus a subset of the coral and sponge samples. The free-living communities that also cluster in the top left section of the plot in Fig. 2A were almost exclusively from studies that used a culturing step prior to PCR-amplification of 16S rRNA genes. This association suggests that these animal-associated communities are composed of r-selected organisms (i.e. fast growing or `weedy') that can quickly utilize readily available nutrients. Indeed a high copy number of 16S rRNA genes, which correlates with fast growth rates45, is typical of microbes that are common in the vertebrate gut46. These observations suggest that r-selection may have been important for the earliest associations of bacteria with animal guts.
Finally, the split between saline and non-saline environmental communities extends to non-vertebrates that inhabit saline and non-saline habitats. The third principal component (PC3) in this analysis clearly separated the saline from the non-saline free-living communities - a split that had been previously described5. Mirroring this pattern, in the expanded dataset, the marine sponges and corals harbor bacterial communities most similar to free-living communities associated with saline environments, and terrestrial non-vertebrate hosts (e.g., earthworms, bees, the gypsy moth, fruit flies, chewing lice, and beetles) harbor communities more similar to the free-living communities from non-saline habitats along this PC axis (Fig. 2B).
A deep dichotomy replicated in multiple bacterial lineages
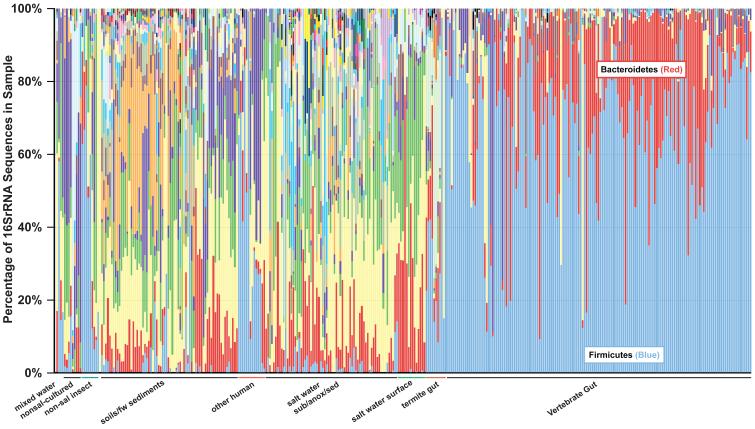
The distribution of phyla takes on very different patterns in the gut than in other types of habitats (Fig. 3). Across vertebrate gut samples, including the human gut, the Firmicutes and Bacteroidetes are the numerically dominant phyla. Although other phyla can make up a large proportion of the sequences recovered in some hosts (e.g., Actinobacteria in sheep), overall the Firmicutes and Bacteroidetes are the most ubiquitous and common (see Fig. 3). Moreover, although the Firmicutes and Bacteroidetes comprise a substantive fraction of a majority of communities in other types of habitats, other phyla tend to be more highly represented in non-gut samples. One important question is thus whether the UniFrac clustering patterns are due to differential representation of different phyla in different communities, or whether the patterns that relate groups of environments are replicated within each bacterial phylum.
Figure 3. Relative abundance of phyla in samples.
Bar graph showing the proportion of sequences from each sample that could be classified at the phylum level. The color codes for the dominant Firmicutes as well as the Bacteroidetes phyla are shown. For a complete description of the color codes see Supplementary Figure 1. `Other human' refers to body habitats other than the gut: i.e, mouth, ear, skin, vagina and vulva (see Table S1 for details).
The dichotomy between vertebrate gut and free-living communities observed at the whole-community level was indeed evident within the constituent phyla. We performed phylum-specific, UniFrac-based PCoA analyses for the three phyla most highly represented across all 462 gut- and non-gut associated microbial community samples: the Bacteroidetes, Firmicutes and Proteobacteria. Within the Bacteroidetes division alone, PC1 again separates the vertebrate and termite-associated gut microbiotas from free-living bacterial communities, and PC3 separates the saline and non-saline free-living communities (Fig. 2C,D). Remarkably, the variation between samples is greater for the vertebrate-gut associated Bacteroidetes than for the free-living Bacteroidetes. The dichotomy between vertebrate gut and free-living communities is also evident when the analysis is applied to the Proteobacteria and Firmicutes individually (Fig. 2E,F).
The phylum-specific analysis also helps to explain why samples from the guts of carnivores tend to cluster closer to free-living communities in the full analysis. Although this similarity may be attributed in part to the deficit of Bacteroidetes (Fig. 2C: most of the carnivores drop out of the analysis), even the Firmicutes within the carnivores are more similar to those in free-living communities than those that reside in the guts of herbivores and omnivores (Fig. 2E).
Genera that cross the divide
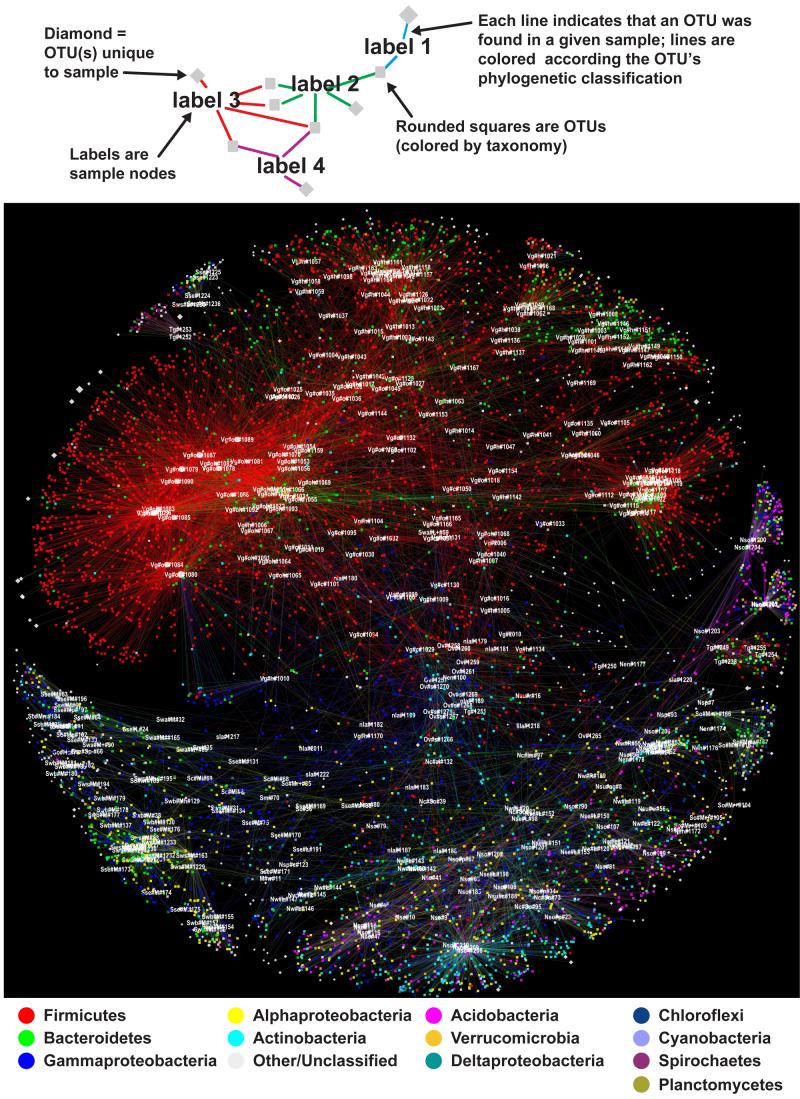
Another way to visualize the vertebrate gut/environmental dichotomy is with a network diagram that displays, in addition to the clustering of hosts with similar microbiotas, the bacterial genera that they share. In this representation of the data, the vertebrate gut samples are far more connected to one another than to the environmental samples (Fig. 4A,B). As in the UniFrac-based analysis, the non-gut human samples also occupy an intermediate position between the free-living and the gut communities. Fig. 5 shows the phylogenetic classification of the operational taxonomic units (OTUs) that are shared between samples: among humans, these are overwhelmingly Firmicutes, with some Bacteroidetes. In contrast, the free-living communities share OTUs from a wider range of phyla. Samples obtained from the guts of obese humans cluster away from the samples from healthy subjects and these are linked principally by Firmicutes: this observation is consistent with the finding that samples from obese individuals have a higher representation of the Firmicutes than do lean subjects31.
Figure 4. Network analysis of bacterial communities from animal-associated and free-living communities.
A simplified cartoon illustration of a host-gut microbe network is shown. In this Figure network diagrams are color-coded by habitat category. Host node colors in (A) highlight the human sample types, while the free-living are the same color. In (B), all vertebrate gut are colored the same, while free-living samples are colored differently from one another. Group abbreviations have the following meaning: VertGut, Vertebrate Gut; HumGut, Human Gut; TermGut, Termite Gut; NonsalInvert, Invertebrate from non-saline environments including diverse insects and earthworms; SalInvert, corals and sponges; HumSkin, Human Skin; HumMouth, Human Mouth; Plant, tightly adherent to plant roots; HumVagina, Human vagina; HumEar, Human Ear.
Figure 5.
Network analysis of bacterial communities from animal-associated and free-living communities where host node colors are all white, and genus-level OTUnodes are colored according to their phylogenetic classification at the phylum level.
Bacterial genera that connect the vertebrate-gut associated microbiotas to the free-living communities by inhabiting both can be viewed as cosmopolitan. As these analyses mainly capture the dominant members of a microbiota, these genera are presumed to grow and subsist in that environment (autochthonous members), and not simply be passing through (allochthonous members). Among these cosmopolitan groups were the Pseudomonadaceae family of the Gammaproteobacteria lineage, which contained OTUs detected both in the vertebrate gut, and free-living in saline and non-saline habitats. The Enterobacteriales (Gammaproteobacteria) were detected in the vertebrate gut, termite gut and in other invertebrates, but also in a surface soil sample and anoxic saline water. The Staphylococcaceae (Bacilli, Firmicutes), were common in the vertebrate gut samples but were also detected in soil and cultures derived from freshwater and saline habitats. Finally, members of Fusobacterium were detected in saltwater sediments in addition to the vertebrate gut. The cosmopolitan distribution of these organisms may have made them particularly important for introduction of novel functions during evolution of the gut microbiota, as they can bring new useful genes from the global microbiome into the gut microbiome via horizonal gene transfer. [A caveat: some of OTUs that are very common in humans and that occur at very low abundance in free-living communities may be contaminants of environmental samples introduced during handling47.]
In summary, the gut/non-gut dichotomy in community composition is evident across the bacterial tree, within phyla, and manifests as distinct sets of genera. This leads to the question of what types of selective pressures act on these many diverse lineages of gut microbes, driving them to `differentiate' into gut and non-gut groups?
Selection forces that have driven the vertebrate/free-living dichotomy
The ecological and evolutionary forces thought to be important in shaping the diversity of the gut have been discussed elsewhere48-50. However, the principal factors governing community composition are beginning to be tested experimentally. For example, the neutral model for community assembly has recently been applied to several microbial habitat types, including the human gut51,52. This simple conceptual model of community assembly, which has become central in the field of macro-ecology, states that composition at a local scale is shaped by random immigration, birth and death events, and assumes ecological equivalence between members53,54. Although the theory performed well for communities of tree holes, human lungs and lakes51,52, it failed to predict the community composition of fecal samples. Sloan and co-workers 48 suggested that genetic or nutritional differences between individuals were important in shaping the communities. This finding implies that individual hosts are too different from one another to be considered replicate habitats.
This type of analysis needs to be repeated with more datasets. If the conclusions are reliably reproducible, we can conclude that gut communities are governed by a different set of ecological rules than free-living communities. Niche differentiation and competition may play much larger roles in determining microbial community composition in the gut than in free-living communities. But the question remains, why is the gut so different? What could account for the large dichotomy between vertebrate-associated and other types of communities? We propose three possible, but not mutually exclusive, processes that might shape microbial community composition in the gut: the adaptive immune system; selection pressure on the host itself; and the nutrient/redox potential dichotomy.
The adaptive immune system
One feature that differentiates the vertebrate gut from other habitats is the adaptive immune system. Margaret McFall-Ngai has postulated55 that the evolution of the adaptive immune system in vertebrates allows for a level of complexity in their associated microbiota that invertebrates cannot manage with an innate immune system alone. The adaptive immune system mediates tolerance to the gut microbiota though IgA production in a way that suggests the need for perpetual modification of surface microbial epitopes56. Furthermore, the adaptive immune system is known to shape community composition in the gut: for example, IgA-deficiency leads to altered gut communities in mice57,58. Although their effect on gut microbes has not been determined, different major histocompatibility complex (MHC) haplotypes have been associated with distinct scents, produced in part by metabolites generated from their microbial communities, in otherwise genetically identical mice59. Constant selection pressure from the host, combined with high growth rates inherent to the gut, high cell densities, and a nutrient-rich milieu, might have created an environment of accelerated rates of evolution compared to free-living habitats, most of which are colder and far more oligotrophic.
The selective power of the immune system has so far only been tested against microbial communities that already have a legacy of selection by the immune system (e.g., mouse communities in mice). Controlled experiments in gnotobiotic mice with and without elements of the immune system, challenged with microbial communities from totally different habitats that are not pre-adapted to the gut, might show just how selective the immune system can be.
Selection pressure on the host
A second, fundamental difference between a living host and a natural environment is that the host itself is under selective pressure. What, if any, effect this might have on the evolutionary trajectory of the host's (gut) microbiota warrants further exploration both in theory and empirically. If the collective properties of the microbiota lead to reduced host fitness (for instance, poor resistance to the invasion of enteropathogens), the host will leave fewer offspring. Furthermore, a shorter-lived (unhealthy) host would disseminate its microbes to a lesser degree than a longer-lived, healthy host of the same species. Both scenarios lead to less dissemination of the host's microbiota, such that members of the microbiota will be less likely to be part of the meta-community that is available to other hosts for colonization49.
Access to beneficial microbes has been suggested as one of the added benefits of social behavior in animals60. Parental behavior in particular should facilitate host-to-host transmission of microbiotas, and indeed this has been observed in captive and wild animals: strong kinships effects on the cecal microbiota have been observed in laboratory mice61; similarly, the early environment, which is dominated by the parent's microbiota under natural conditions, strongly influences cloacal microbiota of chicks in two species of songbird62. Related humans share microbiotas to a greater extent than unrelated humans who co-habit63-66. The gut bacterial consortium that produces equol (4',7'-isoflavandiol, metabolized by gut bacteria from the isoflavone, daidzein) shows a pattern of inheritance in human populations consistent with autosomal dominant transmission66. Parental- and/or kin-to- offspring transmission of microbiotas could promote the co-evolution of whole communities with their host species by stabilizing the associations between particular host and microbial lineages.
A possible outcome of selection pressure on a host with vertically transmitted communities is co-diversification between the communities and the host species. Co-diversification is a form of co-evolution exemplified in the congruent phylogenetic trees of the endosymbiont Buchnera and their aphid host species67: it occurs when co-evolution of the microbes and hosts is so tightly orchestrated that their mutual evolution is reflected in overlapping phylogenies. To show that whole communities and hosts co-diversified, it is not the phylogeny of one species of bacteria that must match the host phylogeny, but rather the patterns of similarity between the bacterial community and the host. We have shown32 a subtle but significant effect of co-diversification in this way: the patterns of gut community similarity match the mammal phylogeny more often than what would be expected if no co-diversification has occurred.
A unique biochemical environment?
We propose a third feature that may be important in driving the community differences between in gut and non-gut environments: the vertebrate gut may be highly unusual as a microbial habitat by combining high abundance, diversity and flux of polysaccharides in an anoxic environment with a constant controlled temperature. Experiments that compare microbial selection in controlled anaerobic, oxidized-nutrient rich environments within an animal (e.g., gnotobiotic hosts) and in engineered systems such as bioreactors treating organic-rich wastes should help to further elucidate the contributions of host and non-host-associated factors in shaping the vertebrate gut microbiota.
Global change without and within: a call to sample diversity now
Our analyses indicate that gut-associated microbiotas are profoundly different from other free-living microbiotas from across the biosphere. Most of this biodiversity has yet to be explored, and high-throughput techniques from metagenomics to metabolomics promise an integrated view of the microbe-plus-host `supraorganism'68,69. But time is limited: the microbial communities we wish to study may be disappearing faster than the rate at which we develop the necessary hardware and software tools.
We live in a period of rapid loss of biodiversity. Plants and animals are heading to extinction at alarming rates: it is too early to know with certainty if the microbial diversity of the biosphere is also in decline70. However, a recent review of studies that investigated the sensitivity of terrestrial microbial community composition to forcers of global change (nitrogen, phosphorus, potassium, and organic carbon amendments, temperature change) showed that in the majority of cases, microbial community composition was indeed sensitive to disturbance71. Thus, loss of microbial diversity is a real possibility in the near future, at least in some biomes. Some have called for the preservation of microbial DNA from a range of environments thought to be at risk72, and have begun to preserve microbial life threatened by anthropogenic disturbance73.
Microorganisms that are undeniably threatened with extinction are those in symbiotic associations with critically endangered hosts74. Presently one quarter of mammal species alone face extinction75: combined with other vertebrate species under threat, the potential loss of microbial diversity this represents is staggering. In addition, the ability to maintain and successfully breed animals threatened with extinction in zoos or protected nature reserves may require more intimate knowledge of their gut microbial ecology and the interrelationships between their native microbiota, diet and nutrient harvest.
Our human cultures are also undergoing rapid changes with globalization. Changes in human ecology have been proposed to alter human-associated microbes and diseases76,77. Globalization and the increased movement around the globe is thought to be increasing rates of microbial transmission78. Changes in human ecology may lead to the homogenization of human-associated microbial communities, with resulting erasure of key features of the evolutionary histories of our microbiotas. Therefore, it is imperative that our human microbiome be sampled as thoroughly and as rapidly as possible, particularly in societies that are undergoing dramatic cultural, socioeconomic and technological transformations.
Supplementary Material
BOX: Glossary of terms
- Microbiota
In this report, we define microbiota as the complete set of microbial lineages living in a given environment.
- Microbiome
This term and the term `metagenome' have been used in a variety of ways. Historically, `microbiome' is a word coined by Joshua Lederberg, who defined it as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space and have been all but ignored as determinants of health and disease.”85 Here we define microbiome as the complete collection of genes contained in the genomes of microbes living in a given environment (i.e. the set of genes contained in a microbiota). Some have used the term `metagenome' as the sum of all of the genomes of organisms in an environment.
- Metagenomics
Defined here quite broadly as culture-independent analyses of the composition and dynamic operations of microbial communities. This includes community characterization at the level of DNA (microbiome), RNA (`meta'-transcriptome), protein (meta-proteome) and metabolic network (`metabolome').
- Co-evolution
a process in which the evolution of one taxon or trait reciprocally influences the evolution of another.
- Co-diversification
a process in which two lineages speciate in concert with one another: for example, when pathogens or commensals speciate into specialist lineages when their host lineage speciates.
- Horizontal gene transfer
a process by which a gene moves between two genomes rather than being “vertically” transmitted from parent to offspring.
- Co-metabolites
metabolites that both the host and one or more microbial species are involved in synthesizing or degrading.
- Operational taxonomic units (OTUs)
groups organisms defined by their sequence similarity (in the case of Bacteria and Archaea, typically using their 16S rRNA genes). For example, OTUs at the 97% level (i.e. all 16S rRNA gene sequences are at least 97% identical to one another) are often considered to define a `species'.
- OTU-based analysis
an analysis based on the counts of each type of OTU in each sample. Unlike in phylogenetic analyses, in this approach all OTUs are considered to be equal, independent units: i.e., two OTUs that are closely related to one another will be treated the same as two OTUs that are distantly related.
- Phylogenetic analysis
an analysis based on a phylogenetic tree that treats some sequences as more or less similar to others based on the number of changes that they have accumulated over evolutionary time.
- Principal Coordinates Analysis (PCoA)
an analysis based on a matrix of distances between samples, which finds a few dimensions that explain as much of the variation in the samples as possible using linear algebra techniques.
- UniFrac
a phylogenetic analysis technique that measures the distance between two community samples in terms of the amount of sequence divergence on a phylogenetic tree that is unique to each of the samples. In practice, a `master' phylogenetic tree is constructed using all (16S rRNA) sequences from all biological samples characterized in a study. Pairwise comparisons of samples (communities) are then performed with similarity defined (UniFrac metric) based on the degree of branch length that they share on the tree.
References
- 1.Woese CR. On the evolution of cells. Proc. Natl. Acad. Sci. U S A. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- 3.Martiny JB, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 4.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U S A. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. This paper combines data from hundreds of bacterial communities to show that phylogenetic tree-based measures of diversity (i.e.,UniFrac) can reveal large-scale trends influencing these communities: for example, the unexpectedly large effect of salinity.
- 6.Desnues C, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 7.Butterfield NJ, Knoll AH, Swett K. A bangiophyte red alga from the Proterozoic of arctic Canada. Science. 1990;250:104–107. doi: 10.1126/science.11538072. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 9.Robosky LC, et al. Metabonomic Identification of Two Distinct Phenotypes in Sprague-Dawley (Crl: CD (SD)) Rats. Toxicological Sciences. 2005;87:277–284. doi: 10.1093/toxsci/kfi214. Soc Toxicology. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414.This comparative metagenomic study shows that the gut microbiomes of genetically obese (ob/ob) mice have substantially different representation of different functional categories of genes than those in lean wild-type littermates, especially those genes involved in carbohydrate metabolism. Transplantation of gut microbiotas from obese donors into wild-type germ-free mouse recipients produces a greater gain in adiposity than does transplantation of gut microbiotas from lean donors. These results demonstsrate how comparative metagenomics can be used to link host physiologic states to the microbiome, as well as the power of testing predictions made from such comparative studies with direct experimental tests in gnotobiotic animals.
- 11.Rohde CM, et al. Metabonomic Evaluation of Schaedler Altered Microflora Rats. Chem. Res. Toxicol. 2007;10:1388–1392. doi: 10.1021/tx700184u. [DOI] [PubMed] [Google Scholar]
- 12.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U S A. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teaford MF, Ungar PS. Diet and the evolution of the earliest human ancestors. Proc. Natl. Acad. Sci. U S A. 2000;97:13506–13511. doi: 10.1073/pnas.260368897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connell JF, Hawkes K, Blurton Jones NG. Grandmothering and the evolution of Homo erectus. J. Hum. Evol. 1999;36:461–485. doi: 10.1006/jhev.1998.0285. [DOI] [PubMed] [Google Scholar]
- 16.Ungar PS. In: Evolution of the Hominid Diet: The Known, the Unknown, & the Unknowable. Ungar PS, editor. Oxford University Press; US: 2006. pp. 39–55. [Google Scholar]
- 17.Ungar PS, Grine FE, Teaford MF. Diet in Early Homo: A Review of the Evidence and a New Model of Adaptive Versatility. Ann. Rev. Anthro. 2006;35:209–228. [Google Scholar]
- 18.Yeakel JD, Bennett NC, Koch PL, Dominy NJ. The isotopic ecology of African mole rats informs hypotheses on the evolution of human diet. Proc. Royal Soc. B: Biolog. Sciences. 2007;274:1723–1730. doi: 10.1098/rspb.2007.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiello LC, Wheeler P. The Expensive-Tissue Hypothesis: The Brain and the Digestive System in Human and Primate Evolution. Curr. Anthro. 1995;36:199–221. [Google Scholar]
- 20.Wood B, Brooks A. Human evolution: We are what we ate. Nature. 1999;400:219–220. doi: 10.1038/22227. [DOI] [PubMed] [Google Scholar]
- 21.Kehrer-Sawatzki H, Cooper DN. Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons. Hum. Mutat. 2007;28:99–130. doi: 10.1002/humu.20420. [DOI] [PubMed] [Google Scholar]
- 22.Samuelson LC, Wiebauer K, Snow CM, Meisler MH. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Mol. Cell. Biol. 1990;10:2513–2520. doi: 10.1128/mcb.10.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beja-Pereira A, et al. Gene-culture co-evolution between cattle milk protein genes and human lactase genes. Nat. Genet. 2003;35:311–313. doi: 10.1038/ng1263. This paper demonstrates genetic changes in both humans and cattle that are associated with agriculture. These types of studies emphasize the importance of examining how changes in human culture, technology, and cookery have and are shaping our microbial ecology and microbiomes (the `micro'-evolution of humans).
- 25.Hollox EJ, et al. Lactase haplotype diversity in the Old World. Amer. J. Hum. Gen. 2001;68:160–172. doi: 10.1086/316924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 27.Holmes E, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U S A. 2008;105:2117–2123. doi: 10.1073/pnas.0712038105. This important paper provides a crucial link between human physiology and human microbial ecology by demonstrating that the abundance of certain metabolites correlates with the abundance of particular kinds of gut bacteria, rather with than ancestry.
- 29.Suau A, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 32.Ley R, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725.This paper provides a 16SrRNA-based survey of the gut bacterial communities in 60 species of mammals. The results show that diet and host phylogeny have large influences on which bacteria live in which hosts.
- 33.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hladik CM, Pasquet P. The human adaptations to meat eating: a reappraisal. Hum. Evol. 2002;17:199–206. [Google Scholar]
- 35.Collinson ME, et al. Fossil Evidence of Interactions between Plants and Plant-Eating Mammals. Phil.Trans. Biol. Sci. 1991;333:197–208. doi: 10.1098/rstb.1991.0068. [DOI] [PubMed] [Google Scholar]
- 36.Mackie RI. Mutualistic Fermentative Digestion in the Gastrointestinal Tract: Diversity and Evolution 1. Integrative Comparative Biol. 2002;42:319–326. doi: 10.1093/icb/42.2.319. This helpful review outlines the major carbohydrate fermentation pathways operating in the rumen, and discusses how digestive tract anatomy and diet affect the function of the rumen, as mediated by symbiotic microbes.
- 37.Russell JB, Rychlik JL. Factors That Alter Rumen Microbial Ecology. Science. 2001;292:1119–1122. doi: 10.1126/science.1058830. [DOI] [PubMed] [Google Scholar]
- 38.Stevens C, Hume I. Comparative physiology of the vertebrate digestive system. Cambridge University Press; Cambridge, New York: 2004. [Google Scholar]
- 39.Hackstein JHP, Stumm CK. Methane production in terrestrial arthropods. Proc. Natl. Acad. Sci. USA. 1994;91:5441–5445. doi: 10.1073/pnas.91.12.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackstein JHP, van Alen TA. Fecal Methanogens and Vertebrate Evolution. Evolution. 1996;50:559–72. doi: 10.1111/j.1558-5646.1996.tb03868.x. [DOI] [PubMed] [Google Scholar]
- 41.Dierenfeld ES, Hintz HF, Robertson JB, Van Soest PJ, Oftedal OT. Utilization of bamboo by the giant panda. J. Nutr. 1982;112:636–641. doi: 10.1093/jn/112.4.636. [DOI] [PubMed] [Google Scholar]
- 42.Egert M, Wagner B, Lemke T, Brune A, Friedrich MW. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae) Appl. Environ. Microbiol. 2003;69:6659–6668. doi: 10.1128/AEM.69.11.6659-6668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egert M, Stingl U, Bruun LD, Pommerenke B, Brune A, Friedrich MW. Structure and topology of microbial communities in the major gut compartments of Melolontha melolontha larvae (Coleoptera: Scarabaeidae) Appl. Environ. Microbiol. 2005;71:4556–4566. doi: 10.1128/AEM.71.8.4556-4566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss PD, Delaibera I, Handelsman J, Raffa KF. Bacteria associated with the guts of two wood-boring beetles: Anoplophora glabripennis and Saperda vestita (Cerambycidae) Physiol. Ecol. 2006;35:625–629. [Google Scholar]
- 45.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. Divergence and Redundancy of 16S rRNA Sequences in Genomes with Multiple rrn Operons. J. Bacteriol. 1862004:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner M, Goebel BM, Dojka MA, Pace NR. Specific rDNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 1998;8:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol. Evol. 2006;21:517–523. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 50.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sloan WT, et al. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- 52.Woodcock S, et al. Neutral assembly of bacterial communities. FEMS Microbiol. Ecol. 2007;62:171–180. doi: 10.1111/j.1574-6941.2007.00379.x. This theoretical paper makes an important contribution to our understanding of what would be expected by chance if bacterial communities are assembled according to neutral processes (i.e. those without selection), and shows that many microbial communities appear to fit this model.
- 53.Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press; 2001. [DOI] [PubMed] [Google Scholar]
- 54.Hubbell SP. Neutral theory and the evolution of ecological equivalence. Ecology. 2006;87:1387–1398. doi: 10.1890/0012-9658(2006)87[1387:ntateo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 55.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 56.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Fagarasan S, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgAdeficient gut. Proc. Natl. Acad. Sci. U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanyon CV, et al. Murine scent mark microbial communities are genetically determined. FEMS Micro. Eco. 2007;59:576–583. doi: 10.1111/j.1574-6941.2006.00252.x. This paper demonstrates a strong host genetic influence on the microbial communities that alter the smell of urine, which is used for a range of functions including marking territory, signaling condition, choosing mates, etc. It also shows a specific effect of the major histocompatibility complex (MHC),
- 60.Lombardo MP. Access to mutualistic endosymbiotic microbes: an underappreciated benefit of group living. Behav. Ecol. Sociobiol. 2008:1–19. [Google Scholar]
- 61.Ley RE, et al. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lucas FS, Heeb P. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J. Avian Biol. 2005;36:510–516. [Google Scholar]
- 63.Zoetendal E.G.J.A.G.M.d.V., et al. The Host Genotype Affects the Bacterial Community in the Human Gastrointestinal Tract. Micro. Eco. Health Dis. 2001;13:129–134. [Google Scholar]
- 64.Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J. Med. Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 65.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. Culture independent methods are used to show that establishment of the gut micobial community during the postnatal period follows a highly varied course in infants with different mothers. Dizygotic twins have the most similar pattern of community assembly, suggesting the importance of early environmental exposures on this process.
- 66.Perez PF, Doré J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ, Donnet-Hughes A. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–32. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 67.Frankenfeld CL, et al. Familial correlations, segregation analysis, and nongenetic correlates of soy isoflavone-metabolizing phenotypes. Exp. Biol. Med. 2004;229:902–913. doi: 10.1177/153537020422900906. [DOI] [PubMed] [Google Scholar]
- 68.Moran NA. Symbiosis. Curr. Biol. 2006;16:866–871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 69.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 70.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 71.Borneman J. What Is the Evidence for the Loss of Microbial Diversity? In: Bull AT, editor. Microbial Diversity and Bioprospecting. ASM Press; Herndon, VA: 2004. pp. 421–428. [Google Scholar]
- 72.Allison SD, Martiny JBH. Resistance, resilience and redundancy in microbial communities. Proc. Natl. Acad. Sci. U S A. 2008 doi: 10.1073/pnas.0801925105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fuerst JA, Hugenholtz P. Microorganisms Should Be High on DNA Preservation List. Science. 2000;290:1503. doi: 10.1126/science.290.5496.1503b. [DOI] [PubMed] [Google Scholar]
- 74.Turrini A, Avio L, Bedini S, Giovannetti M. In situ collection of endangered arbuscular mychorrhizal fungi in a Mediterranean UNESCO Biosphere Reserve. Biodiver. Conserv. 2008;17:643–657. [Google Scholar]
- 75.Staley JT. Biodiversity: are microbial species threatened? Commentary. Curr. Opin. Biotech. 1997;8:340–345. doi: 10.1016/s0958-1669(97)80014-6. [DOI] [PubMed] [Google Scholar]
- 76.Ceballos G, Ehrlich PR, Soberon J, Salazar I, Fay JP. Global Mammal Conservation: What Must We Manage? Science. 2005;309:603–607. doi: 10.1126/science.1114015. [DOI] [PubMed] [Google Scholar]
- 77.Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 2006;7:956–960. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pirages DC. Nature, Disease, and Globalization: An Evolutionary Perspective. Intnl. Studies Rev. 2007;9:616–628. [Google Scholar]
- 79.Beard AS, Blaser MJ. The ecology of height: the effect of microbial transmission on human height. Perspect. Biol. Med. 2002;45:475–498. doi: 10.1353/pbm.2002.0064. [DOI] [PubMed] [Google Scholar]
- 80.Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z, Lozupone C, McDonald D, Robeson M, Sammut R, Smit S, Wakefield MJ, Widmann J, Wikman S, Wilson S, Ying H, Huttley GA. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hengstmann U, Chin KJ, Janssen PH, Liesack W. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 1999;65:5050–5058. doi: 10.1128/aem.65.11.5050-5058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeSantis TZ, Jr., et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DeSantis TZ, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. App. Environ. Micro. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ludwig W, et al. ARB: a software environment for sequence data. Nuc. Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lederberg J, McCray AT. 'Ome Sweet 'Omics-- A Genealogical Treasury of Words. The Scientist. 2001;15:8–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.