Abstract
Two recent studies conducted in our laboratory have demonstrated formation and accumulation of pyridyloxobutyl (POB) and pyridylhydroxybutyl (PHB) adducts in lung and liver total DNA of F344 rats chronically treated with the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and (R)- and (S)-enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL). In this study, we measured POB and PHB adducts in lung and liver mitochondrial DNA (mtDNA), as previous studies suggest a potentially important role of mtDNA in carcinogenesis. Rats were sacrificed after 1, 2, 5, 10, 16, and 20 weeks of treatment with 10 ppm of NNK or (S)-NNAL in drinking water, and mtDNA and nuclear DNA (nDNA) adduct levels in lung and liver were determined by LC-ESI-MS/MS-SRM. The mean levels of individual POB adducts in mtDNA at all time points were slightly higher than those in nDNA for both NNK and (S)-NNAL-treated rats in the lung (P < 0.001 for both treatments), but not in the liver (P > 0.05). Lung mtDNA of both NNK- and (S)-NNAL-treated rats contained higher concentrations of the sum of three POB adducts (P < 0.001 for both treatments) than nDNA, while the levels of mtDNA and nDNA total POB adducts in the liver were not significantly different in either NNK- or (S)-NNAL-treated rats. Analysis of PHB adducts in mtDNA and nDNA produced results similar to those obtained for POB adducts. The steady accumulation of the lung and liver mtDNA adducts over the course of the study indicates inefficient repair of these adducts in mtDNA. This is the first study to examine the formation of NNK- and (S)-NNAL-derived adducts in rat mtDNA. The results support the hypothesis that preferential binding of tobacco carcinogens to mtDNA of the lung might be functionally important in the development of smoking-induced lung cancer.
Keywords: NNK, NNAL, mitochondria, DNA adducts
INTRODUCTION
Despite the certainty of the causal relationship between exposure to tobacco smoke carcinogens and lung cancer (1–4), and a great deal of knowledge about genetic and epigenetic abnormalities accompanying this complex multistep process, some aspects of the mechanism of lung cancer development following exposure remain unclear. Tobacco smoke is a complex mixture containing over 60 compounds rated by the International Agency for Research on Cancer as having sufficient evidence for carcinogenicity in laboratory animals or humans (1). Of these, polycyclic aromatic hydrocarbons (PAH), tobacco-specific N-nitrosamines (TSNA), and aromatic amines have been most thoroughly studied due to their established carcinogenicity to humans (1,5–7). Since TSNA are formed from nicotine during tobacco processing, the unique attribute of these carcinogens is their specificity to tobacco products (5,8). One of the most important representatives of this group is 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a remarkably effective systemic lung carcinogen in laboratory animals (5). These two qualities – specificity of its origin and preferential targeting of the lung – make NNK an excellent tool in the investigations of the mechanisms of tobacco-attributed lung cancer.
Formation of DNA adducts is accepted as a critical factor in the induction of lung cancer by NNK. An established concept is that cytochrome P450-mediated NNK metabolic activation leads to formation of electrophiles which bind to DNA (1,4,5,9–11). The DNA adducts thus formed, if not repaired, can cause miscoding upon DNA replication, leading to mutational activation of oncogenes and/or inactivation of tumor suppressor genes, followed by the loss of normal cellular growth control functions, ultimately resulting in cellular proliferation and cancer (4). This concept implies processes that take place in nuclear DNA (nDNA), the key player in the control of major cellular functions. However, in addition to the nuclear genome, mammalian cells contain mitochondrial DNA (mtDNA), and its potential role in tobacco smoke-induced carcinogenesis cannot be neglected.
Mitochondria are ubiquitous semi-autonomously functioning intracellular organelles that generate approximately 90% of cellular adenosine triphosphate (ATP), thus being the major source of energy needed by the normal human cell. Mitochondria also play a critical and complex role in mammalian cell apoptosis via the release into the cytosol of several apoptogenic proteins, such as cytochrome c, AIF, Smac/Diablo, and EndoG, that directly activate cellular apoptotic programs (12–16). The discovery that most cancer cells produce energy predominantly via aerobic glycolysis (Warburg effect (17)), followed by the identification of a number of cancer-related mitochondrial alterations (reviewed in (18)), led to the hypothesis that mutations in mtDNA may be involved in the carcinogenic process (19,20). The observations of preferential accumulation of some carcinogens in mitochondria and their effective binding to mtDNA (reviewed in (19,20)), along with inefficient mtDNA repair mechanisms (21,22), provided additional support for this hypothesis. Furthermore, it has been demonstrated that smoking inhibits mitochondrial enzyme activity in platelets (23) and damages respiratory chain function in lymphocytes (24). Such symptoms as fatigue and poor gastrointestinal motility, leading to the loss of appetite, are associated with both smoking (25–27) and systemic mitochondrial dysfunction (reviewed in (28)). Increased levels of mtDNA damage have been demonstrated in lung tissues (29) and buccal cells (30) from smokers compared to nonsmokers. Decrease in mitochondrial function in smokers is accompanied by an increase in mitochondrial content that persists for an average of 2 decades after smoking cessation (31).
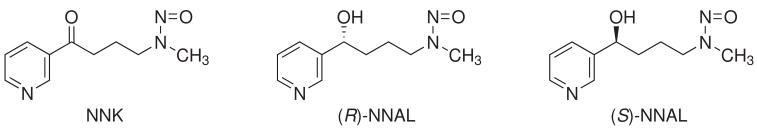
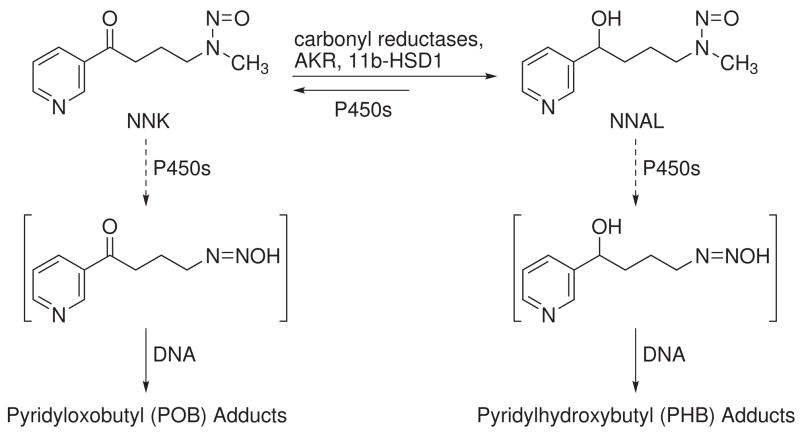
Two recent studies conducted in our laboratory have demonstrated formation and accumulation of pyridyloxobutyl (POB) (11) and pyridylhydroxybutyl (PHB) (32) DNA adducts in F344 rats chronically treated with NNK or (R)- and (S)-enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL). The pulmonary carcinogenicity of NNAL is similar to that of NNK (5). Structures of NNK, (R)-NNAL, and (S)-NNAL are illustrated in Chart 1. POB-DNA adducts are formed via a-methyl hydroxylation of NNK, while hydroxylation of the NNAL methyl group results in the formation of PHB adducts (Scheme 1) (33,34). However, in vivo experiments in rats demonstrate that PHB-DNA adducts are formed primarily from (R)-NNAL, while (S)-NNAL produces mainly POB-DNA adducts, similar to NNK (11,32).
Chart 1.
Structures of NNK, (R)-NNAL, and (S)-NNAL.
Scheme 1.
DNA adduct formation from NNK and NNALa
a For more detailed scheme, see ref. 11
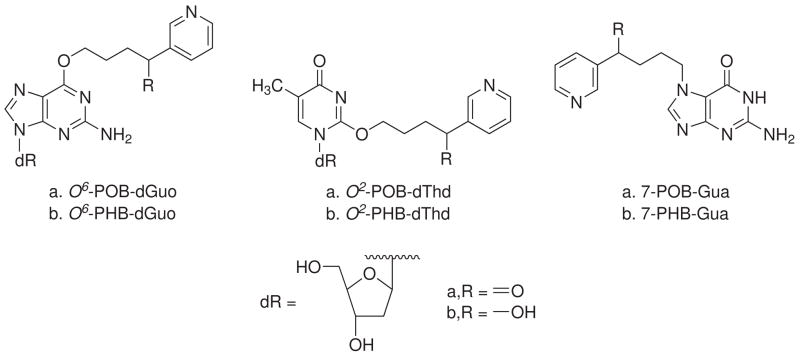
In this study, we further investigate NNK and (S)-NNAL-induced damage to DNA of lung and liver cells of rats chronically treated with these carcinogens. Since the DNA analyzed in the previous studies was isolated by a method that did not include subcellular fractionation, it contained mostly nDNA and probably some mtDNA (11,32). In this study, we modified our DNA isolation procedure to separate mtDNA from nDNA. POB and PHB adducts were measured in mtDNA, and the results were compared to nDNA levels in the same tissue. Structures of POB- and PHB-DNA adducts analyzed here are illustrated in Chart 2.
Chart 2. Structures of POB- and PHB-DNA adducts analyzed in this studya.
a dR: 2′-deoxyribosyl
MATERIALS AND METHODS
Caution
NNK and NNAL are potent carcinogens and should be handled with extreme care, in a well-ventilated hood and with personal protective equipment
Chemicals and Enzymes
The following were synthesized as previously described: POB-DNA adducts (35), deuterated POB-DNA adducts (10), PHB-DNA adducts (34,36), and deuterated PHB-DNA adducts (32). Micrococcal nuclease (LS004797, 15 kU) and phosphodiesterase II (LS003603, 10 U) were purchased from Worthington Biochemical Corp. (Lakewood, NJ), and alkaline phosphatase (567752, 30 U/mL) was obtained from Roche Molecular Biochemicals (Indianapolis, IN). Reagents and enzymes for DNA isolation were obtained from QIAGEN Sciences (Germantown, MD). All other chemicals and solvents were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI) or Fisher Scientific (Fairlawn, NJ).
Animal Experiment
Lung and liver tissues analyzed here were those produced in a previous study in which rats were treated with 10 ppm of NNK, (R)-NNAL, or (S)-NNAL in the drinking water (11). Tissues harvested from rats sacrificed at 1, 2, 5, 10, 16, and 20 weeks of treatment were stored at −80 °C until DNA isolation. Lung tissues from rats treated with NNK for 16 weeks were not available for this study. For the rest of the time points, 1 to 3 tissue samples per time point were available.
Isolation of mtDNA and nDNA
Frozen lung and liver tissue samples were cut into small pieces and further subjected to homogenization and differential centrifugation by a slight modification of the method described by Balansky et al. (37). Briefly, whole lung or ~ 1 g liver samples were homogenized in 10 mL or 5 mL, respectively, of ice-cold 10 mM Tris-EDTA buffer (pH 7.4) containing 0.25 M sucrose. The homogenates were centrifuged at 1000 × g for 30 min, and the supernatants containing mitochondria were transferred into polypropylene tubes. The pellets containing nuclei and cell debris were lysed in 6 mL cell lysis solution from QIAGEN, and after treating with proteinase K, RNase A, and precipitating proteins, nDNA was isolated and purified as previously described (38,39). The mitochondria were sedimented by supernatant centrifugation at 10,000 × g for 15 min. The mitochondrial pellets were washed with 1 mL ice-cold 10 mM Tris-EDTA buffer (pH 7.4), resuspended in 0.25 mL of mitochondrial buffer (50 mM glucose, 10 mM EDTA, and 25 mM Tris (pH 8), lysed by adding 0.5 mL alkaline SDS solution (0.2 M NaOH, 1% SDS), and neutralized by adding 0.375 mL of ice-cold 3 M NaOAc in 2 M acetic acid (pH 4.8). The mix was centrifuged at 10,000 × g for 10 min, the supernatant was transferred to a clean 2-mL Eppendorf tube, and mtDNA was isolated and purified as described for nDNA. Balansky et al. (37) have demonstrated that mtDNA preparations produced by this method are free from nDNA contamination. Contents of dGuo and dThd in isolated mtDNA and nDNA were determined by HPLC (10), and the amount of DNA was calculated as described (11).
Analysis of POB-DNA and PHB-DNA Adducts
The adducts were analyzed by HPLC-ESI-MS/MS-SRM essentially as described previously (11,32). We used all isolated DNA of each mtDNA sample, and 0.5–1.0 mg of each nDNA sample. Briefly, six deuterated internal standards (3 for POB and 3 for PHB adducts) were added to the DNA sample dissolved in 10 mM sodium succinate buffer (pH 7.0), and the mix was subjected to neutral thermal hydrolysis (100 °C, 30 min) followed by enzymatic hydrolysis with micrococcal nuclease, phosphodiesterase II, and alkaline phosphatase. The hydrolysate was purified on a solid-phase extraction cartridge (Strata-X-cartridge, Phenomenex, CA), concentrated to dryness, and the resulting sample was stored at −20 °C until analysis. Prior to analysis by LC-ESI-MS/MS-SRM, the samples were dissolved in 20 μL of 2% NH4OAc; 8 μL of each sample was injected on LC-MS/MS to analyze POB-DNA adducts and another 8 μL was injected for PHB-DNA adduct analysis. LC-ESI-MS/MS-SRM analysis was performed as described (11,32).
Statistical Analyses
Repeated measures analysis of variance was used with nDNA or mtDNA as the repeated factors and weeks as a grouping factor. Linear regression was used to identify adducts that had a significant rate of increase with time. Due to distributions skewed to higher values, the adducts were analyzed on the natural logarithmic scale. No adjustments were made for multiple comparisons.
RESULTS
The method used here allowed us to isolate both mtDNA and nDNA from the same tissue sample. The amount of nDNA taken for analysis was determined based on the 22% guanine contribution to the nuclear genome, as previously determined in our laboratory. The amount of isolated mtDNA was calculated based on the 13% guanine contribution to the rat mitochondrial genome determined in this study, which is in agreement with the data reported elsewhere (40). The average (± SD) amounts of mtDNA isolated from lung and liver samples were 9.21 (± 4.40) μg and 14.3 (± 7.41) μg, respectively. The yield of mtDNA, expressed as μg per g wet tissue, averaged 4.97 (± 2.37) for lungs and 12.7 (± 7.23) for the liver.
POB Adducts in mtDNA and nDNA of NNK- and (S)-NNAL-Treated Rats
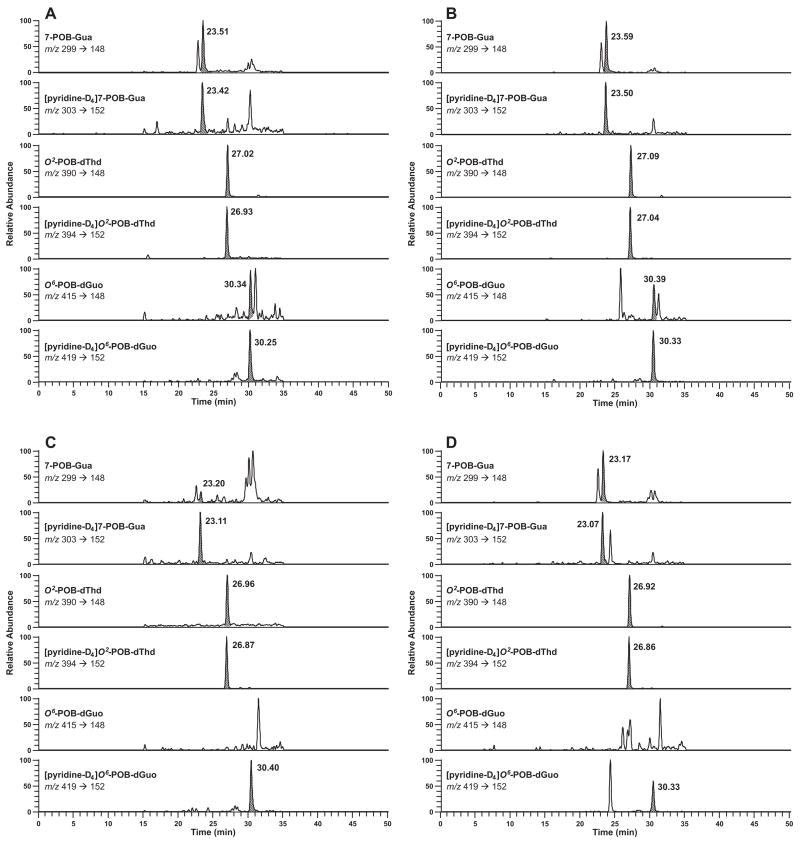
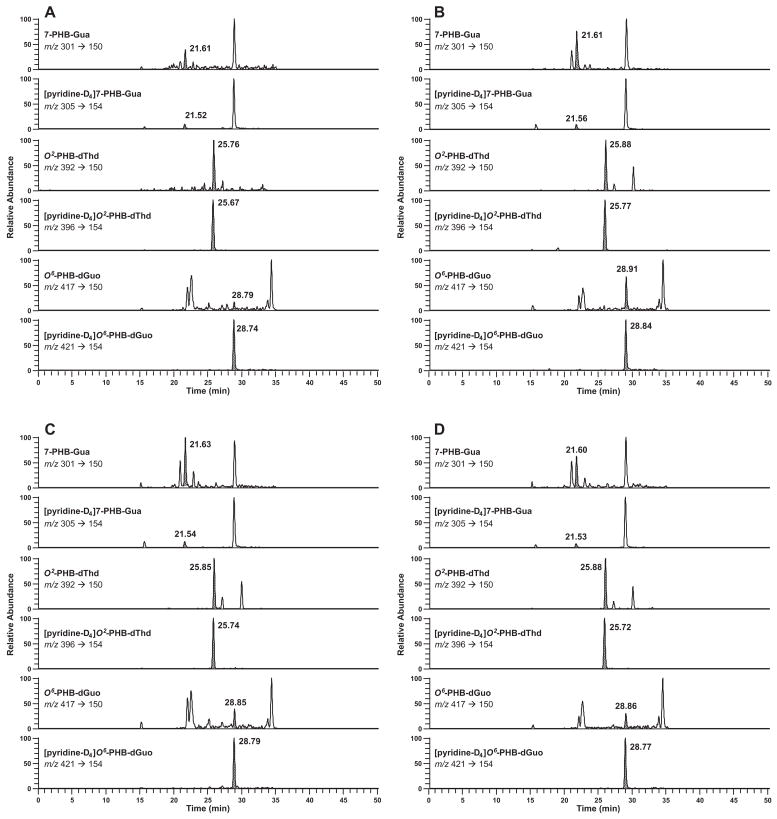
POB adducts were detectable in most samples, with the exception of O6-POB-dGuo in liver of both NNK and (S)-NNAL-treated rats. Typical chromatograms obtained upon POB-DNA adduct analysis of mtDNA and nDNA from NNK-treated rats are shown in Figure 1A-D. Analysis of mtDNA and nDNA from (S)-NNAL-treated rats produced similar LC-ESI-MS/MS-SRM traces.
Figure 1.
Typical SRM chromatograms obtained upon analysis of POB adducts in (A) lung mtDNA; (B) lung nDNA; (C) liver mtDNA; (D) liver nDNA from NNK-treated rat after 10 weeks of treatment. For each transition, the peak corresponding to the adduct is shaded.
Levels of mitochondrial and nuclear POB-DNA adducts in the lung of NNK- and (S)-NNAL-treated rats, expressed per mg DNA, are summarized in Table 1, and for liver in Table 2. In the lung, mean levels of 7-POB-G and O2-POB-dThd in mtDNA were slightly higher than those in nDNA at all time points for both NNK-treated rats (P < 0.001 for both adducts) and (S)-NNAL-treated rats (P < 0.001 for both adducts). This was generally true for O6-POB-dGuo in the lung mtDNA and nDNA (P < 0.001 for both NNK and (S)-NNAL treatment), with the exception of the 1-week time-point in (S)-NNAL-treated rats (Table 1). In the liver of NNK-treated rats, the differences between mtDNA and nDNA levels of 7-POB-Gua and O2-POB-dThd were not statistically significant (P = 0.73 and P = 0.11, respectively). In the liver of (S)-NNAL-treated rats, 7-POB-Gua in mtDNA was not different from nDNA (P = 0.24), and O2-POB-dThd was slightly higher in mtDNA than in nDNA (P < 0.001).
Table 1.
POB-DNA adducts in lung nuclear and mitochondrial DNA of rats treated with NNK and (S)-NNAL
| Week of treatment | fmol/mg DNA (mean ± S.D.)a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7-POB-Gua | O2-POB-dThd | O6-POB-dGuo | ||||||||||
| NNK | (S)-NNAL | NNK | (S)-NNAL | NNK | (S)-NNAL | |||||||
| nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | |
| 1 | 1310 (1) | 1770 (1) | 949±153 | 1730±196 | 1230 (1) | 2400 (1) | 1220±173 | 1900±143 | 44 (1) | 46 (1) | 38±7 | 32±10 |
| 2 | 1710 (2) | 2990 (2) | 1430±452 | 2390±40 | 1940 (2) | 4770 (2) | 2150±580 | 3170±141 | 41 (2) | 48 (2) | 35±8 | 38±13 |
| 5 | 2530±692 | 2900±692 | 1440±170 | 1980±48 | 3790±398 | 6260±947 | 3380±733 | 5680±268 | 48±6 | 62±3 | 25±2 | 62±5 |
| 10 | 2810±816 | 2920±175 | 1770±1270 | 2400±467 | 6990±2030 | 8080±205 | 6030±810 | 7170±2010 | 42±4 | 97±4 | 38±13 | 69±13 |
| 16 | NAb | NA | 1820±351 | 2250±112 | NA | NA | 4730±816 | 6790±563 | NA | NA | 40±12 | 99±10 |
| 20 | 1610±218 | 3350±136 | 1510 (2) | 2580 (2) | 5050±1600 | 11200±1820 | 3990 (2) | 9010 (2) | 31±7 | 141±43 | 19 (2) | 102 (2) |
N = 3, unless stated otherwise (N in parentheses); when N = 2, the cell contains mean of two analyses; when N = 1, the cell contains the result of a single analysis
NA, not analyzed (no samples available for this time-point).
Table 2.
POB-DNA adducts in liver nuclear and mitochondrial DNA of rats treated with NNK and (S)-NNAL
| Week of treatment | fmol/mg DNA (mean ± S.D., N = 3) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7-POB-Gua | O2-POB-dThd | O6-POB-dGuo | ||||||||||
| NNK | (S)-NNAL | NNK | (S)-NNAL | NNK | (S)-NNAL | |||||||
| nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | nDNA | mtDNA | |
| 1 | 1030±252 | 1000±346 | 803±189 | 568±242 | 830±76 | 897±286 | 806±48 | 831±164 | NAb | NDc | NA | ND |
| 2 | 1220±214 | 1010±52 | 948±154 | 1100±105 | 1070±134 | 1350±570 | 841±131 | 1340±84 | NA | ND | NA | ND |
| 5 | 1390±154 | 1160±48 | 1150±292 | 994±88 | 1620±268 | 2320±579 | 1740±263 | 2050±429 | NA | ND | NA | ND |
| 10 | 1220±479 | 1300±208 | 1210±80 | 1200±123 | 3720±152 | 2780±860 | 2790±258 | 2460±435 | NA | ND | NA | ND |
| 16 | 1530±408 | 1480±156 | 869±130 | 1260±345 | 2740±544 | 3260±628 | 1920±349 | 3060a | NA | ND | NA | ND |
| 20 | 1430±199 | 1670±168 | 751±40 | 1350±202 | 2710±528 | 3540±112 | 2000±169 | 3450±276 | NA | ND | NA | ND |
This value is the amount detected in one out of 3 samples analyzed. The adduct was not detected in the other two samples, probably due to low yield of mtDNA
NA, not analyzed
ND, not detected (the limit of detection for this adduct is 1 fmol/mg DNA (10)).
One common feature of all mtDNA adducts in both the lung and liver was their steady accumulation over the period of the study (Tables 1 and 2). Thus, 7-POB-Gua in the lung mtDNA increased from 1770 fmol/mg DNA after 1 week of treatment with NNK to 3350 fmol/mg DNA after 20 weeks of treatment (P = 0.034). These amounts in (S)-NNAL-treated rats were 1730 fmol/mg DNA and 2580 fmol/mg DNA, respectively (P < 0.001). Similarly, 7-POB-Gua in the liver mtDNA increased about 2-fold over the period studied in both NNK- and (S)-NNAL-treated rats (P < 0.001 and P = 0.011, respectively). The levels of O2-POB-dThd increased about 5-fold in the lung of NNK- and (S)-NNAL-treated rats (P < 0.001 for both treatments), and about 4-fold in the liver (P < 0.001 for both treatments). The levels of O6-POB-dGuo in the lung mtDNA increased from 46 fmol/mg DNA after 1 week of treatment with NNK to 141 fmol/mg DNA after 20 weeks of treatment (P < 0.001), and from 32 fmol/mg DNA after 1 week of treatment with (S)-NNAL to 102 fmol/mg DNA after 20 weeks of treatment (P < 0.001).
In the nDNA of both the lung and liver, 7-POB-Gua and O2-POB-dThd increased during the initial period of either NNK or (S)-NNAL treatment and reached a maximum at 10 or 16 weeks, which was followed by a slight decline in the adduct levels. Thus, after the first week of treatment with NNK, the mean levels of 7-POB-Gua in nDNA were 1310 fmol/mg DNA in the lung and 1030 fmol/mg DNA in the liver. These values increased to 2810 fmol/mg DNA in the lung after 10 weeks of treatment, and 1530 fmol/mg DNA in the liver after 16 weeks of treatment, and subsequently declined to 1610 fmol/mg DNA and 1430 fmol/mg DNA, respectively, after 20 weeks of treatment. The mean levels of O2-POB-dThd in nDNA after the first week of treatment with NNK were 1230 fmol/mg DNA in the lung and 830 fmol/mg DNA in the liver, then increased to 6990 fmol/mg DNA and 3720 fmol/mg DNA, respectively, after 10 weeks of treatment, and declined to 5050 fmol/mg DNA and 2710 fmol/mg DNA, respectively, after 20 weeks of treatment. Analysis of lung and liver nDNA from rats treated with (S)-NNAL produced results similar to NNK treatment. The levels of O6-POB-dGuo in lung nDNA of NNK- and (S)-NNAL-treated rats also followed a similar pattern (Table 1).
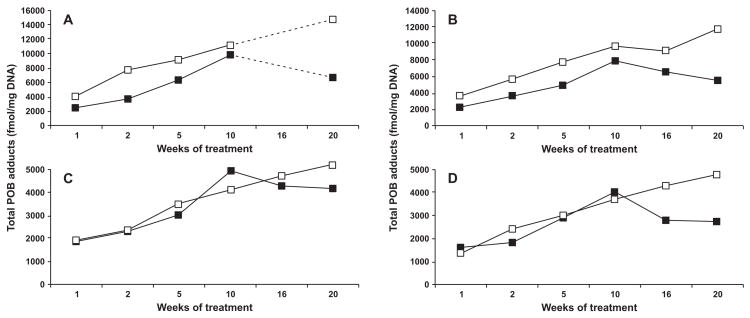
The sum of 7-POB-Gua, O2-POB-dThd, and O6-POB-dGuo, referred to as total POB-DNA adducts, was calculated for each time point of treatment with both NNK and (S)-NNAL, and the results were compared for mtDNA and nDNA (Figure 2A–D). Lung mtDNA of both NNK- and (S)-NNAL-treated rats contained higher concentrations of total POB adducts (P < 0.001 for both treatments) than nDNA, while the levels of mtDNA and nDNA total POB adducts in the liver were not significantly different in either NNK- or (S)-NNAL-treated rats (P = 0.40 and P = 0.81, respectively).
Figure 2.
Plots of total POB adducts (fmol/mg DNA) vs time (weeks) in the (A) lung of NNK-treated rats; (B) lung of (S)-NNAL-treated rats; (C) liver of NNK-treated rats; (D) liver of (S)-NNAL-treated rats. There were no lung samples available for 16 weeks of treatment with NNK (figure 2A). Symbol designations are ■, nDNA; and □, mtDNA.
PHB Adducts in mtDNA and nDNA of NNK- and (S)-NNAL-Treated Rats
PHB-DNA adducts were detectable in most of the lung samples, and were below the limit of detection in the liver mtDNA of both NNK and (S)-NNAL-treated rats. Typical chromatograms obtained upon PHB adduct analysis of mtDNA and nDNA from NNK- and (S)-NNAL-treated rats are shown in Figure 3A–D.
Figure 3.
Typical SRM chromatograms obtained upon analysis of PHB adducts in (A) lung mtDNA from NNK-treated rat; (B) lung nDNA from NNK-treated rat; (C) lung mtDNA from (S)-NNAL-treated rat; (D) lung nDNA from (S)-NNAL-treated rat after 10 weeks of treatment. For each transition, the peak corresponding to the adduct is shaded.
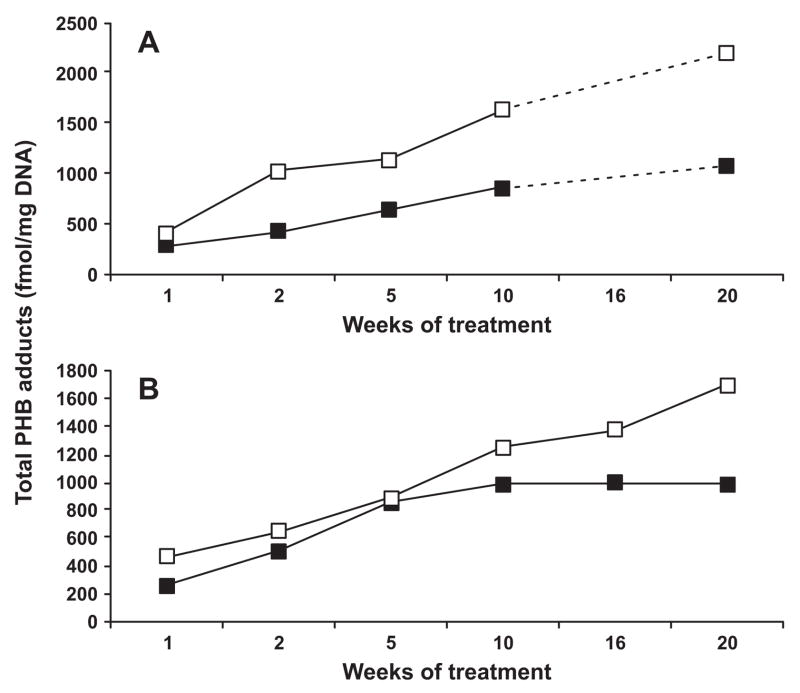
Overall, the levels of PHB adducts in mtDNA were slightly higher than those in nDNA for both NNK-treated rats and (S)-NNAL-treated rats. Similar to mitochondrial POB-DNA adducts, the common feature of mtDNA PHB adducts was their steady accumulation over the whole period of the study (Figure 4 A–B). Thus, total PHB-DNA adducts (the sum of 7-PHB-Gua, O2-PHB-dThd, and O6-PHB-dGuo) in the lung mtDNA increased from 395 fmol/mg DNA after 1 week of treatment with NNK to 2220 fmol/mg DNA after 20 weeks of treatment (P = 0.002). These amounts in (S)-NNAL-treated rats were 469 fmol/mg mtDNA and 1690 fmol/mg mtDNA, respectively (P < 0.001).
DISCUSSION
The findings that genotoxic carcinogens preferentially accumulate in mitochondria and effectively bind to mtDNA, the observations that mitochondria are altered in cancer cells, and the knowledge of the crucial role of mitochondria in apoptosis, suggest that mtDNA mutations may play an important role in the carcinogenic process. The purpose of this study was to investigate the formation of mtDNA adducts in rats chronically treated with the tobacco-specific carcinogen NNK and its carcinogenic metabolite (S)-NNAL, and to compare the levels of mtDNA and nDNA adducts in the same tissue samples. This is the first study to demonstrate effective binding of NNK and (S)-NNAL metabolites to the mitochondrial genome.
We focused on NNK and (S)-NNAL-treated animals because of a number of indications that (S)-NNAL, but not (R)-NNAL, is important in lung tumorigenesis by NNK. These include predominant formation of (S)-NNAL from NNK in lung and liver microsomes and cytosol (41,42), stereoselective retention of (S)-NNAL in the lung (43,44), and a striking similarity between NNK and (S)-NNAL in the formation of POB-DNA adducts (11) – the pathway believed to be an important mechanism of NNK carcinogenesis in rodents (45,46) and likely in smokers (47,48).
Overall, the levels of mtDNA and nDNA adducts were higher in the lung than in the liver of both NNK and (S)-NNAL-treated rats, reproducing the results that we obtained previously upon analysis of total POB- and PHB-DNA adducts in the same animals (11,32). When compared in the same samples, mtDNA adducts were higher than nDNA adducts in the lung, but not in the liver. Since lung is the primary target for NNK- and (S)-NNAL-induced carcinogenesis, our findings are consistent with the results of a study in which preferential binding of N-nitrosodimethylamine and N-nitrosodiethylamine to mtDNA was demonstrated only for tumor susceptible tissues, while there was no difference in binding of these carcinogens to mtDNA or nDNA in non-tumor susceptible tissues (49). The difference between the levels of mtDNA and nDNA adducts in the lung observed in our study is also in agreement with reports on the relative efficiency of nDNA and mtDNA binding for nitroso compounds and benzo[a]pyrene (BaP). Thus, we found that after 20 weeks of treatment with NNK, levels of 7-POB-Gua, O6-POB-dGuo, and O2-POB-dThd in lung mtDNA were 2.08, 4.54, and 2.23 times, respectively, higher than the levels of these adducts in nDNA. In other studies, treatment of laboratory animals with N-methyl-N-nitrosourea (50) and N-nitrosodimethylamine (51) led to 3–7 times more efficient methylation of liver and kidney mtDNA compared to nDNA. Some in vitro experiments on preferential accumulation of carcinogens in mitochondria demonstrated that BaP and its carcinogenic dihydrodiol epoxide derivative bind to mtDNA of animal cells about 50 times more than to nDNA (52,53). An in vivo study, however, demonstrated that the levels of mtDNA adducts in the liver, lung, and kidney of rats treated with BaP were only 2 times higher than those of nDNA adducts (37). In the same study, treatment of rats with cigarette smoke produced results comparable to BaP treatment.
The steady accumulation of mtDNA adducts observed in the rat lung and liver over the 20-week period of NNK- and (S)-NNAL-treatment is in contrast with the results obtained upon analysis of nDNA adducts. The fact that nDNA adducts reach maximum values after 10–16 weeks of treatment and subsequently decline is indicative of a shift of balance between adduct formation and DNA repair (11). In the case of mtDNA, continuous accumulation of adducts points to the inefficiency or lack of appropriate mtDNA repair mechanisms. Thus, nuclear O6-POB-dGuo is efficiently repaired by AGT (54,55), while in mitochondria AGT repairs methyl and ethyl adducts, but not bulky ones (56). Similarly, O2-POB-dThd is probably a nucleotide excision repair (NER) substrate in nDNA, while mitochondria are deficient in the NER pathway (reviewed in (21,22)). Similarly, the mechanism of 7-POB-Gua repair, yet unknown in nDNA, is apparently not effective in mitochondria. Of the measured adducts, O6-POB-dGuo has been shown to be highly mutagenic (55). Accumulation of this adduct in lung mtDNA might play an important role in the induction of lung tumors by NNK in laboratory animals, and potentially in smokers.
The biological plausibility of the role of mtDNA mutations in the carcinogenic process is supported by the critical role of mitochondria in cellular energy production, apoptosis, and cellular growth and differentiation. In addition to these theoretical considerations, there is growing experimental evidence of a relationship between mtDNA mutations and cancer. Such evidence includes propagation of mtDNA mutations in cancer cells, changes in the cell surface produced by mtDNA mutations, suppression of the tumorigenic phenotype by the fusion of cancer cells with cytoplasts from non-tumorigenic cells, and appearance of rearranged and normal segments of mtDNA in nuclear genomes of various cell species (reviewed in (20)). Furthermore, mathematical models demonstrate that both random distribution of mutated mtDNA among daughter cells and relaxed replication independent of the cell cycle, can lead to clonal expansion of a single mtDNA mutation during biological time scales (57,58). Each of 2 to 10 mtDNA molecules present in a mitochondrion encodes 2 ribosomal RNA, 22 mitochondrial transfer RNA, and 13 mitochondrial proteins (subunits of respiratory complexes I, III, IV, and V) (59,60). Many mutational hotspots occur in the non-coding D-loop of mtDNA, which serves as the main site for mtDNA replication and transcription. On the other hand, mtDNA lacks introns, and all non-D-loop mutations will occur in sequences coding for functionally important enzymes in the respiratory chain complex.
In summary, we demonstrate here that the chronic treatment of rats with NNK and (S)-NNAL at low doses gives higher levels of POB and PHB adducts in mtDNA than in nDNA of the lung, but not the liver. In both the lung and the liver, mtDNA adducts accumulated over the course of treatment, indicating inefficient repair of these adducts in mtDNA. This is the first study to examine the formation of NNK- and (S)-NNAL-derived adducts in rat mtDNA. The results support the hypothesis that preferential binding of tobacco carcinogens to mtDNA of the lung might be functionally important in the development of smoking-induced lung cancer.
Figure 4.
Plots of total PHB adducts (fmol/mg DNA) vs time (weeks) in the (A) lung of NNK-treated rats; (B) lung of (S)-NNAL-treated rats. There were no lung samples available for 16 weeks of treatment with NNK (figure 4A). Symbol designations are ■, nDNA; and □, mtDNA.
Acknowledgments
We thank Mingyao Wang and Pramod Upadhyaya for valuable advice, Siyi Zhang and Steve Kalscheuer for help with DNA isolation, Peter Villalta for help with mass spectrometry, Bruce Lindgren for statistical analyses, and Bob Carlson for editorial assistance. This study was supported by NCI grant CA-81301.
Footnotes
Abbreviations: AKR, aldo-keto reductases; ATP, adenosine triphosphate; BaP, benzo[a]pyrene; HPLC-ESI-MS/MS, high performance liquid chromatography-electrospray ionization-tandem mass spectrometry; 11β-HSD1, 11β-hydroxysteroid dehydrogenase type 1; mtDNA, mitochondrial DNA; nDNA, nuclear DNA; NNK, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone; NNAL, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol; O2-POB-dThd, O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine; O6-POB-dGuo, O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine; 7-POB-Gua, 7-[4-(3-pyridyl)-4-oxobut-1-yl]guanine; PAH, polycyclic aromatic hydrocarbons; PHB, 4-(3-pyridyl)-4-hydroxybut-1-yl; POB, 4-(3-pyridyl)-4-oxobut-1-yl; SRM, selected reaction monitoring; TSNA, tobacco-specific nitrosamines.
Reference List
- 1.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon, FR: 2004. Tobacco Smoke and Involuntary Smoking; pp. 35–102. [PMC free article] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 89. IARC; Lyon, FR: 2008. Smokeless tobacco and tobacco-specific nitrosamines. [PMC free article] [PubMed] [Google Scholar]
- 3.www.deathsfromsmoking.net. (2007) International Union Against Cancer.
- 4.Hecht SS. Progress and challenges in selected areas of tobacco carcinogenesis. Chem Res Toxicol. 2008;21:160–171. doi: 10.1021/tx7002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 6.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 7.Vineis P, Pirastu R. Aromatic amines and cancer. Cancer Causes Control. 1997;8:346–355. doi: 10.1023/a:1018453104303. [DOI] [PubMed] [Google Scholar]
- 8.Wiley JC, Chien DHT, Nungesser NA, Lin D, Hecht SS. Synthesis of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, 4-(carbethoxynitrosamino)-1-(3-pyridyl)-1-butanone, and N’-nitrosonornicotine labelled with tritium in the pyridine ring. J Label Comp Radiopharm. 1988;25:707–716. [Google Scholar]
- 9.Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 10.Lao Y, Villalta PW, Sturla SJ, Wang M, Hecht SS. Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:674–682. doi: 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lao Y, Yu N, Kassie F, Villalta PW, Hecht SS. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 13.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 14.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 15.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connoly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 16.Li LY, Luo X, Wang X. Endonuclease G (EndoG) is an apoptotic DNAse when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 17.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 18.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shay JW, Werbin H. Are mitochondrial DNA mutations involved in the carcinogenic process? Mutat Res. 1987;186:149–160. doi: 10.1016/0165-1110(87)90028-5. [DOI] [PubMed] [Google Scholar]
- 20.Bandy B, Davidson AJ. Mitochondrial mutations may increase oxidative stress: Implications for carcinogenesis and aging? Free Radic Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 21.Croteau DL, Stierum RH, Bohr VA. Mitochondrial DNA repair pathways. Mutat Res. 1999;434:137–148. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- 22.Sawyer DE, Van Houten B. Repair of DNA damage in mitochondria. Mutat Res. 1999;434:161–176. doi: 10.1016/s0921-8777(99)00027-0. [DOI] [PubMed] [Google Scholar]
- 23.Smith PR, Cooper JM, Govan GG, Harding AE, Schapira AH. Smoking and mitochondrial function: a model for environmental toxins. Q J Med. 1993;86:657–660. doi: 10.1093/qjmed/86.10.657. [DOI] [PubMed] [Google Scholar]
- 24.Miro O, Alonso JR, Jarreta D, Casademont J, Urbano-Marquez A, Cardellach F. Smoking disturbs mitochondrial respiratory chain function and enhances lipid peroxidation on human circulating lymphocytes. Carcinogenesis. 1999;20:1331–1336. doi: 10.1093/carcin/20.7.1331. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Hansen MJ, Jones JE, Vlahos R, Anderson GP, Morris MJ. Long-term cigarette smoke exposure increases uncoupling protein expression but reduces energy intake. Brain Res. 2008;1228:81–88. doi: 10.1016/j.brainres.2008.06.067. [DOI] [PubMed] [Google Scholar]
- 26.Hughes JR, Crow RS, Jacobs DR, Jr, Mittelmark MB, Leon AS. Physical activity, smoking, and exercise-induced fatigue. J Behav Med. 1984;7:217–230. doi: 10.1007/BF00845388. [DOI] [PubMed] [Google Scholar]
- 27.Corwin EJ, Klein LC, Rickelman K. Predictors of fatigue in healthy young adults: Moderating effects of cigarette smoking and gender. Biological Research for Nursing. 2002;3:222–233. doi: 10.1177/109980040200300407. [DOI] [PubMed] [Google Scholar]
- 28.Cohen BH, Gold DR. Mitochondrial cytopathy in adults: what we know so far. Cleve Clin J Med. 2001;68:625–642. doi: 10.3949/ccjm.68.7.625. [DOI] [PubMed] [Google Scholar]
- 29.Ballinger SW, Bouder TG, Davis GS, Judice SA, Nicklas JA, Albertini RJ. Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 1996;56:5692–5697. [PubMed] [Google Scholar]
- 30.Tan D, Goerlitz DS, Dumitrescu RG, Han D, Seillier-Moiseiwitsch F, Spernak SM, Orden RA, Chen J, Goldman R, Shields PG. Associations between cigarette smoking and mitochondrial DNA abnormalities in buccal cells. Carcinogenesis. 2008;29:1170–1177. doi: 10.1093/carcin/bgn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masayesva BG, Mambo E, Taylor RJ, Goloubeva OG, Zhou S, Cohen Y, Minhas K, Koch W, Sciubba J, Alberg AJ, Sidransky D, Califano J. Mitochondrial DNA content increase in response to cigarette smoking. Cancer Epidemiol Biomarkers Prev. 2006;15:19–24. doi: 10.1158/1055-9965.EPI-05-0210. [DOI] [PubMed] [Google Scholar]
- 32.Upadhyaya P, Kalscheuer S, Hochalter JB, Villalta PW, Hecht SS. Quantitation of pyridylhydroxybutyl-DNA adducts in liver and lung of F-344 rats treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol. 2008;21:1468–1476. doi: 10.1021/tx8001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Upadhyaya P, Sturla S, Tretyakova N, Ziegel R, Villalta PW, Wang M, Hecht SS. Identification of adducts produced by the reaction of 4-(acetoxymethynitrosamino)-1-(3-pyridyl)-1-butanol with deoxyguanosine and DNA. Chem Res Toxicol. 2003;16:180–190. doi: 10.1021/tx0256376. [DOI] [PubMed] [Google Scholar]
- 34.Hecht SS, Villalta PW, Sturla SJ, Cheng G, Yu N, Upadhyaya P, Wang M. Identification of O2-substituted pyrimidine adducts formed in reactions of 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-1butanone and 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanol with DNA. Chem Res Toxicol. 2004;17:588–597. doi: 10.1021/tx034263t. [DOI] [PubMed] [Google Scholar]
- 35.Sturla SJ, Scott J, Lao Y, Hecht SS, Villalta PW. Mass spectrometric analysis of relative levels of pyridyloxobutylation adducts formed in the reaction of DNA with a chemically activated form of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol. 2005;18:1048–1055. doi: 10.1021/tx050028u. [DOI] [PubMed] [Google Scholar]
- 36.Wang M, Cheng G, Sturla SJ, Shi Y, McIntee EJ, Villalta PW, Upadhyaya P, Hecht SS. Identification of adducts formed by pyridyloxobutylation of deoxyguanosine and DNA by 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone, a chemically activated form of tobacco-specific carcinogens. Chem Res Toxicol. 2003;16:616–626. doi: 10.1021/tx034003b. [DOI] [PubMed] [Google Scholar]
- 37.Balansky R, Izzotti A, Scatolini L, D’Agostini F, De Flora S. Induction by carcinogens and chemoprevention by N-acetylcysteine of adducts to mitochondrial DNA in rat organs. Cancer Res. 1996;56:1642–1647. [PubMed] [Google Scholar]
- 38.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem Res Toxicol. 2006;19:319–324. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S, Villalta PW, Wang M, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1, N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography-electrsopray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:1386–1392. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- 41.Breyer-Pfaff U, Martin HJ, Ernst M, Maser E. Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab Dispos. 2004;32:915–922. [PubMed] [Google Scholar]
- 42.Upadhyaya P, Carmella SG, Guengerich FP, Hecht SS. Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis. 2000;21:1233–1238. [PubMed] [Google Scholar]
- 43.Wu Z, Upadhyaya P, Carmella SG, Hecht SS, Zimmerman CL. Disposition of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in bile duct- cannulated rats: stereoselective metabolism and tissue distribution. Carcinogenesis. 2002;23:171–179. doi: 10.1093/carcin/23.1.171. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman CL, Wu Z, Upadhyaya P, Hecht SS. Stereoselective metabolism and tissue retention in rats of the individual enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), metabolites of the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Carcinogenesis. 2004;25:1237–1242. doi: 10.1093/carcin/bgh120. [DOI] [PubMed] [Google Scholar]
- 45.Trushin N, Rivenson A, Hecht SS. Evidence supporting the role of DNA pyridyloxobutylation in rat nasal carcinogenesis by tobacco specific nitrosamines. Cancer Res. 1994;54:1205–1211. [PubMed] [Google Scholar]
- 46.Staretz ME, Foiles PG, Miglietta LM, Hecht SS. Evidence for an important role of DNA pyridyloxobutylation in rat lung carcinogensis by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone: effects of dose and phenethyl isothiocyanate. Cancer Res. 1997;57:259–266. [PubMed] [Google Scholar]
- 47.Foiles PG, Akerkar SA, Carmella SG, Kagan M, Stoner GD, Resau JH, Hecht SS. Mass spectrometric analysis of tobacco-specific nitrosamine-DNA adducts in smokers and nonsmokers. Chem Res Toxicol. 1991;4:364–368. doi: 10.1021/tx00021a017. [DOI] [PubMed] [Google Scholar]
- 48.Hölzle D, Schlöbe D, Tricker AR, Richter E. Mass spectrometric analysis of 4-hydroxy-1-(3-pyridyl)-1-butanone-releasing DNA adducts in human lung. Toxicology. 2007;232:277–285. doi: 10.1016/j.tox.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Daugherty JP, Clapp NK. Association of nitrosamine-derived radioactivity with nuclear and mitochondrial DNA in mice. Jpn J Cancer Res. 1985;76:197–201. [PubMed] [Google Scholar]
- 50.Wunderlich V, Schutt M, Bottger M, Graffi A. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem J. 1970;118:99–109. doi: 10.1042/bj1180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wunderlich V, Tetzlaff I, Graffi A. Studies on nitrosodimethylamine: preferential methylation of mitochondrial DNA in rats and hamsters. Chem -Biol Interactions. 1971;4:81–89. doi: 10.1016/0009-2797(72)90001-4. [DOI] [PubMed] [Google Scholar]
- 52.Graffi A. Intracellular benzpyrene accumulation in living normal and tumor cells. Z Krebsforschung und Klin Onkol. 1940;50:196–219. [Google Scholar]
- 53.Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 55.Pauly GT, Peterson LA, Moschel RC. Mutagenesis by O6-[4-oxo-4-(3-pyridyl)butyl]guanine in Escherichia coli and human cells. Chem Res Toxicol. 2002;15:165–169. doi: 10.1021/tx0101245. [DOI] [PubMed] [Google Scholar]
- 56.Myers KA, Saffhill R, O’Connor PJ. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9:285–292. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 57.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jiminez P, Thilly WG. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 58.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? The Lancet. 2002;360:1323–1325. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- 59.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 60.Chan DC. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]