Abstract
Over 22,000 cases of ovarian cancer were diagnosed in 2007 in the United States but only a fraction of them can be attributed to mutations in highly penetrant genes such as BRCA1. To determine whether low penetrance genetic variants contribute to ovarian cancer risk, we genotyped 1,536 single nucleotide polymorphisms (SNPs) in several candidate gene pathways in 848 epithelial ovarian cancer cases and 798 controls in the North Carolina Ovarian Cancer Study (NCO) using a customized Illumina array. The inflammation gene interleukin-18 (IL18) showed the strongest evidence for association with epithelial ovarian cancer in a gene-by-gene analysis (p=0.002) with a <25% chance of being a false positive finding (q-value=0.240). Using a multivariate model search algorithm over eleven IL18 tagging SNPs, we found the association was best modeled by rs1834481. Further, this SNP uniquely tagged a significantly associated IL18 haplotype and there was an increased risk of epithelial ovarian cancer per rs1834481 allele (OR=1.24, 95% CI: 1.06, 1.45). In a replication stage, twelve independent studies from the Ovarian Cancer Association Consortium (OCAC) genotyped rs1834481 in an additional 5,877 cases and 7,791 controls. The fixed effects estimate per rs1834481 allele was null (OR=0.99, 95% CI: 0.94, 1.05) when data from the twelve OCAC studies were combined. The effect estimate remained unchanged with the addition of the initial NCO data. This analysis demonstrates the importance of consortia, like the OCAC, in either confirming or refuting the validity of putative findings in studies with smaller sample sizes.
Keywords: ovarian cancer, inflammation, consortia, SNPs, IL18
INTRODUCTION
Ovarian cancer is the leading cause of death among cancers of the female reproductive tract and is the fifth leading cause of cancer death in women in the United States (1). The most commonly reported factors that increase ovarian cancer risk are germline mutations in the BRCA1 or BRCA2 genes, family history of ovarian cancer and endometriosis while characteristics such as increasing parity, oral contraceptive use and tubal ligation reduce risk (2–4). Although the mechanisms underlying these reproductive risk factors have yet to be fully elucidated, it has been hypothesized that they may relate to inflammation and DNA damage (5–7). Ovulation involves disruption of the ovarian surface and is associated with an inflammatory response that involves prostaglandins (8), cytokines (9) and reactive oxygen species (10, 11), all of which have been implicated in carcinogenesis (11–14). The inflammation hypothesis is further supported by the observations that endometriosis may cause inflammation, while tubal ligation may reduce inflammation by blocking external agents from coming in contact with the ovary (5). Repair of the ovary after ovulation involves cellular replication that may be prone to DNA damage; thus errors in DNA repair could also contribute to ovarian cancer. In view of the potential importance of inflammation and DNA repair pathways in ovarian carcinogenesis, inherited variation in genes in these pathways could affect ovarian cancer susceptibility.
To address this hypothesis, we used a candidate gene approach to examine the associations between genes involved in inflammation and DNA repair in a population-based, case-control study of ovarian cancer. When examining numerous single nucleotide polymorphisms (SNPs) in multiple genes, the risk of identifying false positives is present, even when statistically adjusting for multiple comparisons. Replication of significant findings using additional independent studies is critical to establish whether true associations exist. In this paper we describe our evaluation of candidate genes related to DNA repair and inflammation, followed by replication of putative significant findings within a large international consortium of ovarian cancer case-control studies.
MATERIALS AND METHODS
Hypothesis generating study
The North Carolina Ovarian Cancer Study (NCO) is a population-based case-control study of epithelial ovarian cancer (borderline and invasive) among women in 48 North Carolina (NC) counties. Eligible cases, aged 20 to 74, were diagnosed between January 1, 1999 and March 31, 2007 and were identified via rapid case ascertainment through the NC Central Cancer Registry. Population-based controls were identified using list-assisted random digit dialing and were frequency-matched to the cases by age and race. DNA was obtained from over 98% of participants. Further details of the study have been described elsewhere (15). All participants signed informed consent forms and approval for the study was obtained from the Duke University Medical Center Institutional Review Board, participating hospitals, and the human subjects committee at the NC Central Cancer Registry.
Replication studies
The Ovarian Cancer Association Consortium (OCAC) was formed to provide a forum for researchers to evaluate genetic associations with ovarian cancer with increased power. A major aim of the OCAC is to follow up on promising genetic associations while addressing the problem of multiple comparisons and false discoveries that are inherent to studies using high-throughput genotyping technologies. Twelve OCAC studies from the United States (DOV, HAW, HOP, MAY, STA, UCI and USC), Europe (GER, MAL, SEA, and UKO) and Australia (AUS) contributed data to the current analysis for a total of 5,877 cases and 7,791 controls of self-reported Caucasian ancestry. These studies have been described in detail previously and are summarized in Table 1 (6, 16–26). All studies obtained approval from their respective human subjects ethics committees and all participants provided signed informed consent.
Table 1.
Description of the NCO and 12 OCAC replication studies with IL18 rs1834481 genotype frequencies (G>C) by case status (listed alphabetically).
| Study | Location | Case ascertainment | Cases (No.) |
Control ascertainment* | Controls (No.) |
||||
|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | GG | GC | CC | ||||
| AUS (16) | Multiregional,Australia | Cancer registries,surgical treatment centres | 475 | 359 | 85 | P: Electoral roll | 573 | 410 | 71 |
| DOV (17) | Washington,USA | Cancer Surveillance System, SEER | 411 | 272 | 55 | P: RDD | 422 | 271 | 46 |
| GER (18) | Multiregional,Germany | Hospital admissions | 143 | 89 | 13 | P: Population registries | 172 | 81 | 13 |
| HAW (19) | Hawaii, USA | Hawaii Tumor Registry, SEER | 56 | 30 | 2 | P: Annual Hawaii Health Survey | 91 | 57 | 8 |
| HOP (20) | Ohio,Pennsylvania and New York,USA | Registries,Physician offices,pathology databases | 174 | 124 | 20 | P: RDD | 367 | 233 | 36 |
| MAL (21) | Multiregional,Denmark | Danish Cancer Registry, 16 Gynecologic departments | 232 | 154 | 29 | P: Danish Central Population Register | 643 | 442 | 89 |
| MAY (22) | Multiregional,USA | Mayo Clinic | 221 | 142 | 13 | C: Women seeking general exams at Mayo Clinic | 245 | 186 | 22 |
| NCO (15) | North Carolina, USA | NC Central Cancer Registry | 450 | 334 | 64 | P: RDD | 470 | 285 | 43 |
| SEA (6) | Cambridge,UK | East Anglia and West Midlands cancer registries | 625 | 395 | 71 | P: EPIC-Norfolk cohort | 672 | 456 | 82 |
| STA (23) | California,USA | Greater Bay Area Cancer Registry, SEER | 201 | 102 | 16 | P: RDD | 244 | 141 | 28 |
| UCI (24) | California,USA | Cancer Surveillance Program of Orange County,Tumor Registry | 268 | 159 | 24 | P: RDD | 264 | 172 | 29 |
| UKO (25) | Multiregional,UK | 10 gynecologic Oncology National Health Service Centres | 148 | 91 | 17 | P: UK Collaborative Trial of Ovarian Cancer Screening & all women followed up for cancers by Office of National Statistics | 302 | 231 | 43 |
| USC (26) | California,USA | LA Cancer Surveillance Program | 413 | 211 | 37 | P: Neighborhood recruits | 408 | 205 | 36 |
| Totals | --- | --- | 3,817 | 2,462 | 446 | --- | 4,873 | 3,170 | 546 |
NCO, North Carolina Ovarian Cancer Study; OCAC, Ovarian Cancer Association Consortium; SEER, Surveillance Epidemiology and End Results; RDD, random digit dialing.
P=Population-based; C=Clinic-based.
SNP selection, genotyping and quality control
The NCO genotyped a total of 1,536 SNPs using the Illumina Golden Gate Assay™ (Supplemental Table 1). SNPs tagging 120 candidate genes and nonsynonymous SNPs from an additional 50 candidate genes were included on the Illumina OPA. Genes were chosen based on previous literature and were defined at 10 kB up and downstream of the gene. While the DNA repair, inflammation and hormone candidate gene pathways were predominantly represented, a limited number of genes from the cell cycle, metabolism, methylation and signal transduction pathways were also included on the OPA. Tagging SNPs were selected using HapMap version 20 (www.hapmap.org) and ldSelect (27); a minor allele frequency (MAF) ≥ 0.05 and pair-wise linkage disequilibrium threshold of r2 ≥0.8 were used for selection. Cases and controls were randomly mixed within each plate and six CEPH-Utah trios-standard by Coriell were distributed across six plates. Only five samples (<1%) failed genotyping. There was one within- and one across-plate duplicate sample on each 96-well DNA plate; the concordance rate for these samples was 99.5%.
Eleven of the 12 OCAC studies used the 5' nuclease TaqMan allelic discrimination assay (Taqman; Applied Biosystems, Foster City, CA, USA) to genotype interleukin-18 (IL18) rs1834481 in seven laboratories using a common batch of reagents. One study (AUS) used the iPLEX Sequenom MassArray system (Sequenom Inc., San Diego, CA, USA). To ensure consistency of genotype calls across laboratories, each site also genotyped a common set of samples from Coriell (http://ccr.coriell.org/Sections/Search/Panel_Detail.aspx?PgId=202&Ref=HAPMAPPT01) consisting of 90 unique DNA samples (30 CEPH trios), five duplicate samples, and a negative template control. Genotype concordance on these plates was >98%.
Statistical Analyses: NCO
We restricted our analysis to white, non-Hispanic NCO participants for whom genotyping data were available. We performed tests for Hardy-Weinberg equilibrium (HWE) among the controls for all SNPs using chi-square goodness-of-fit tests and carried out a two-staged analysis. First, for each gene, we fitted an unconditional logistic regression model for disease given age and indicator variables for both the dominant and recessive genotypes of each SNP in the gene. We used a likelihood ratio test of this model against the model containing only age to assess the gene’s degree of association. We prioritized genes according to the p-values of these tests and calculated a q-value for each gene to provide estimates of false discovery rates (28). Second, within the top genes, we carried out a Bayesian model selection analysis of the SNP genotype variables using the bic.glm algorithm in the BMA package (29) for the R statistical language to determine the combination(s) giving the best fit. The two-stage analysis was completed using the statistical software package R (30). A haplotype analysis for the gene with the strongest evidence for an association with epithelial ovarian cancer was conducted using Haploview, version 3.32 (31).
We completed a full analysis of known and suspected risk factors for epithelial ovarian cancer to determine the extent of confounding; covariates that changed the estimate of effect by 10% or greater were considered to be confounders (there were none).
Statistical Analyses: OCAC
IL18 rs1834481 was evaluated by 12 OCAC studies. Among controls, HWE was tested and MAFs were compared to ensure there were no important population differences with respect to the SNPs’ allele frequencies. For each OCAC study, we fitted unconditional logistic regression models, adjusting for age, to estimate odds ratios (ORs) and 95% confidence intervals (CIs). Using the same approach to confounding described above, we evaluated the final models for confounding (there was none).
Fixed and random effects models accounting for study site were fitted using inverse variance weighting and the DerSimonian-Laird (32) methods, respectively, to estimate summary ORs and 95% CIs for the association between rs1834481 and epithelial ovarian cancer risk. We used Cochran’s Q statistic (33) and I2 (34) to evaluate heterogeneity of results. Data were analyzed with and without the NCO data and were restricted to subgroups of cases (i.e., all, invasive only, serous invasive only) since there may be etiologic heterogeneity of epithelial ovarian cancer by histologic type.
The unconditional logistic regression models for the OCAC studies were fitted using SAS (Version 9.1.3, Cary, NC, USA) and the fixed and random effects analyses were fitted using STATA (Version 9, StataCorp, College Station TX, USA).
RESULTS
Hypothesis generating study: NCO
The initial NCO analysis included 848 epithelial ovarian cancer cases, with a mean age at diagnosis of 55 years, and 798 controls; all participants were white, non-Hispanic. The majority of cases (79%) were invasive and more than 65% of invasive cases were FIGO stage III or IV. Serous histology was the most common subtype (61%).
The gene-by-gene analysis identified IL18 as the gene with the strongest evidence for association (i.e., smallest p-value) with epithelial ovarian cancer (p=0.002, q-value=0.240), compared to all other genes. Eleven SNPs tagging IL18 had been genotyped in the NCO: rs243908; rs1293344; rs1834481; rs1946519; rs2043055; rs549908; rs5744247; rs5744258; rs5744280; rs4937113; and rs11214109. The Bayesian model selection analysis of the IL18 SNPs identified models that included only rs1834481 genotypes as being the most likely, a posteriori. In a subsequent haplotype analysis of IL18 (Supplemental Table 2), four haplotypes had estimated frequencies > 5%. Only one was significantly associated with epithelial ovarian cancer (p=0.0007) and was uniquely tagged by rs1834481, which was in HWE (p=0.981) and had a MAF of 0.23 in NCO controls.
Using Akaike’s Information Criterion, we determined that a log-additive genetic model for rs1834481 best fit the data. There was an increased risk of epithelial ovarian cancer (per allele) for all cases (OR=1.24, 95% CI: 1.06, 1.45), invasive cases (OR=1.25, 95% CI: 1.06, 1.28), and serous invasive cases (OR=1.31, 95% CI: 1.08, 1.60).
Replication studies: OCAC
Twelve OCAC studies genotyped IL18 rs1834481 in an additional 5,877 cases and 7,791 controls. The SNP was in HWE in all studies (alpha=0.10). The MAFs among controls ranged from 0.20 to 0.28 in these studies with no statistically significant differences between them.
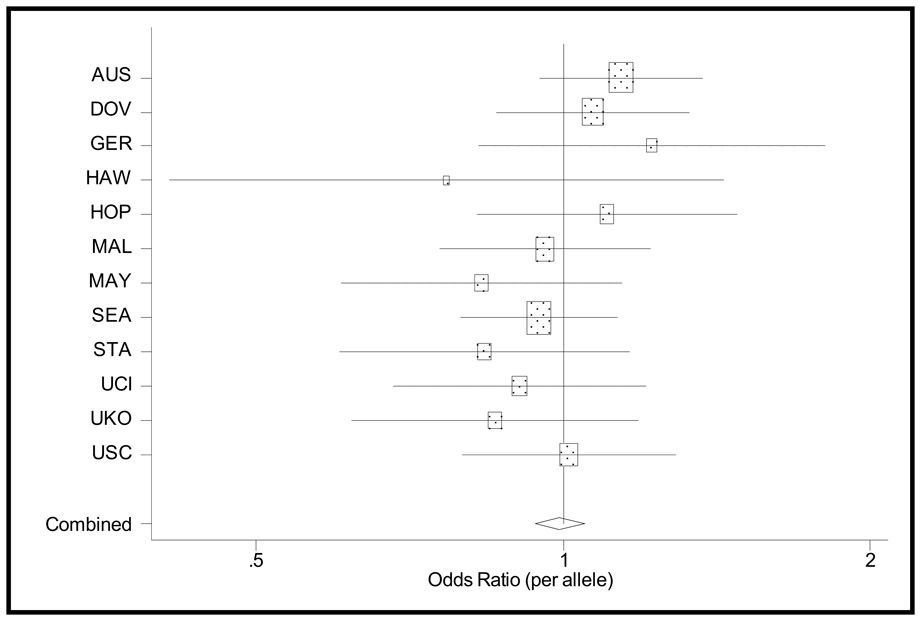
For all of the OCAC studies, the age-adjusted ORs and 95% CIs were not statistically different from a null association (Table 2). In fitting fixed and random effects models to the 12 OCAC studies there was no significant heterogeneity, thus we report fixed effects here. The fixed effects estimates did not change when the NCO data were included; there was, however, significant heterogeneity. In a meta-regression, we did not find evidence that the following study characteristics explained this heterogeneity: U.S. versus non-U.S. populations (p=0.87), incident versus prevalent cases (p=0.38), and population-based versus clinic-based controls (p=0.16). Overall, the fixed effects estimates indicate a null association between IL18 rs1834481 and epithelial ovarian cancer (Table 2, Figure 1).
Table 2.
Site-specific and fixed effects summary ORs (per allele), 95% CIs, and heterogeneity statistics for IL18 rs183441 among 12 OCAC studies, excluding NCO.
| Site | All Cases* | Invasive Cases* | Serous Invasive Cases* | |||
|---|---|---|---|---|---|---|
| OR† | 95% CI | OR† | 95% CI | OR† | 95% CI | |
| AUS | 1.14 | (0.99, 1.31) | 1.08 | (0.93, 1.26) | 1.17 | (0.98, 1.39) |
| DOV | 1.07 | (0.91, 1.26) | 1.04 | (0.87, 1.25) | 1.06 | (0.86, 1.32) |
| GER | 1.22 | (0.91, 1.63) | 1.17 | (0.86, 1.58) | 0.93 | (0.62, 1.39) |
| HAW | 0.77 | (0.48, 1.23) | 0.80 | (0.48, 1.33) | 0.68 | (0.34, 1.33) |
| HOP | 1.10 | (0.89, 1.38) | 1.16 | (0.92, 1.45) | 1.19 | (0.89, 1.58) |
| MAL | 0.96 | (0.80, 1.15) | 0.96 | (0.80, 1.15) | 0.99 | (0.80, 1.23) |
| MAY | 0.83 | (0.66, 1.06) | 0.85 | (0.66, 1.10) | 0.91 | (0.68, 1.24) |
| SEA | 0.95 | (0.83, 1.08) | 0.94 | (0.82, 1.08) | 0.88 | (0.72, 1.08) |
| STA | 0.84 | (0.66, 1.07) | 0.84 | (0.65, 1.07) | 0.92 | (0.68, 1.24) |
| UCI | 0.91 | (0.73, 1.12) | 0.86 | (0.67, 1.11) | 0.93 | (0.68, 1.26) |
| UKO | 0.86 | (0.67, 1.09) | 0.86 | (0.68, 1.10) | 0.85 | (0.62, 1.18) |
| USC | 1.01 | (0.85, 1.22) | 1.08 | (0.89, 1.31) | 1.10 | (0.88, 1.38) |
| Combined‡ | 0.99 | (0.94, 1.05) | 0.99 | (0.93, 1.05) | 1.01 | (0.94, 1.09) |
OR, odds ratio; CI, confidence interval; OCAC, Ovarian Cancer Association Consortium; NCO,North Carolina Ovarian Cancer Study.
Total cases=5877, total invasive cases=4774 and total serous invasive cases=2583 among 12 OCAC studies, excluding NCO.
ORs are age-adjusted.
For all cases, Cochran’s Q=15.147, p=0.18, and I2=27. For invasive cases, Cochran’s Q=12.173,p=0.35, and I2=10. For serous invasive cases, Cochran’s Q=10.053, p=0.53, and I2=0. Low,moderate, and high levels of heterogeneity correspond to I2 values of 25, 50 and 75, respectively (36).
Figure 1.
Estimated age-adjusted odds ratios (boxes) and 95% confidence intervals (lines) from unconditional logistic regression models, for all cases and controls for each of the twelve OCAC replication studies. The combined estimate is from a fixed effects model. The size of each box is proportionate to the study’s size.
DISCUSSION
This is the first report to describe a comprehensive assessment of the relationship between common genetic variation in the IL18 gene and ovarian cancer risk among white, non-Hispanic women. One prior small study investigated a single IL18 polymorphism, not in LD with IL18 rs1834481, and reported null results (19). In the NCO study of 1,536 SNPs from several candidate gene pathways, the IL18 gene showed the strongest evidence for association with epithelial ovarian cancer risk. One of the eleven IL18 SNPs (rs1834481) was significantly associated with epithelial ovarian cancer and uniquely tagged a significant IL18 haplotype, but further studies of this SNP in 12 independent studies by OCAC did not replicate this finding. The most likely explanation for the initial finding is that it represents chance (i.e., a type I error). The magnitude of the q-value (0.240) for IL18 was moderate, suggesting that the association between IL18 and epithelial ovarian cancer could possibly be a false-positive association. However, it is also important to consider that although our results suggest that IL18 rs1834481 is not associated with ovarian cancer, we cannot rule out the possibility that other IL18 polymorphisms may be associated with ovarian cancer risk.
When we fitted the fixed and random effects models in the replication stage, we did so by first excluding the initial NCO findings. Without NCO, there was no heterogeneity of results and the overall effect of the IL18 SNP was null. With the NCO included, the overall effect of IL18 rs1834481 remained null, but there was significant heterogeneity. Since the NCO study was the first to report a positive finding for IL18 rs1834481, it may have exhibited the “winner’s curse phenomenon” (35), whereby the initial finding is often the strongest and most significant, thereby making it statistically different from subsequent studies.
The current study demonstrates the important role of consortia, such as the OCAC, in replication of initial positive findings. Through the OCAC, we were able to refute an initial positive association between a SNP and epithelial ovarian cancer risk via coordinated genotyping and centralized analysis rather than subsequent, potentially conflicting reports from individual studies. Due to the large sample size, both in the number of studies (i.e., 13) and the total number of subjects (i.e., 15,314), it is unlikely that IL18 rs1834481 is associated with epithelial ovarian cancer. Consortia such as the OCAC fill an important role for achieving the sample size and power needed to detect modest associations with common SNPs and aid in confirmation of initial findings from smaller studies.
Supplementary Material
Acknowledgements
ACS/AOCS (Australia): The AOCS Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb) gratefully acknowledges the contribution of all the clinical and scientific collaborators (see http://www.aocstudy.org/). AOCS and the ACS Management Group (A Green, P Parsons, N Hayward, P Webb, D Whiteman) thank all of the project staff, collaborating institutions and study participants.
The authors would like to express their profound thanks to all the study participants who contributed to this research.
Financial Support: Genotyping was funded by a grant from the Ovarian Cancer Research Fund provided by the family and friends of Kathryn Sladek Smith (PI: Andrew Berchuck). Additional support provided by: U.S. Army Medical Research and Materiel Command under DAMD17-01-1-0729, the Cancer Council Tasmania and Cancer Foundation of Western Australia and The National Health and Medical Research Council of Australia (199600) (AUS); R01CA112523 (DOV); Federal Ministry of Education and Research, Programme of Clinical Biomedical Research, Grant 01 GB 9401 (GER); Public Health Service grant R01-CA-58598 and NIH Department of Health and Human Services contracts N01-CN-67001 and NO1-CN-25403 (HAW); DAMD 17-02-1-0669 and R01CA095023 (HOP); Mermaid (MAL); NIH grant 1-R01-CA122443 (MAY); NIH grant 1-R01-CA76016 and Department of Defense grant DAMD17-02-1-0666 (NCO); Cancer Research UK (SEA); Roswell Park Alliance and the National Cancer Institute (CA71766 and Core Grant CA16056) (STA); National Institutes of Health, National Cancer Institute grants CA-58860, CA-92044 and the Lon V Smith Foundation grant LVS-39420 (UCI); Cancer Research UK project grant (no. C8804/A7058), Oak Foundation, Eve Appeal, and Department of Health’s NIHR Biomedical Research Centres funding scheme (UKO); California Cancer Research Program Grants 00-01389V-20170 and 2110200, US Public Health Service Grants CA14089, CA17054, CA61132, CA63464, N01-PC-67010 and R03-CA113148, and California Department of Health Services subcontract 050-E8709 as part of its statewide cancer reporting program (USC); RTP is supported by NIH/NCI grant T32 CA009330-26; DFE is Principal Research Fellow of Cancer Research UK; PDPP is a Cancer Research UK Senior Clinical Research Fellow; SJR is funded by the Mermaid component of the Eve Appeal; HS was funded by a grant from Wellbeing; and GCT and PMW are supported by fellowships from the NHMRC.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007 Jan–Feb;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Whittemore AS, Harris R, Itnyre J Collaborative Ovarian Cancer Group. Characteristics relating to ovarian cancer risk:collaborative analysis of 12 US case-control studies. IV. The pathogenesis of epithelial ovarian cancer. Am J Epidemiol. 1992 Nov 15;136(10):1212–1220. doi: 10.1093/oxfordjournals.aje.a116429. [DOI] [PubMed] [Google Scholar]
- 3.Whittemore AS, Harris R, Itnyre J Collaborative Ovarian Cancer Group. Characteristics relating to ovarian cancer risk:collaborative analysis of 12 US case-control studies. II. Invasive epithelial ovarian cancers in white women. Am J Epidemiol. 1992 Nov 15;136(10):1184–1203. doi: 10.1093/oxfordjournals.aje.a116427. [DOI] [PubMed] [Google Scholar]
- 4.Riman T, Nilsson S, Persson IR. Review of epidemiological evidence for reproductive and hormonal factors in relation to the risk of epithelial ovarian malignancies. Acta Obstet Gynecol Scand. 2004 Sep;83(9):783–795. doi: 10.1111/j.0001-6349.2004.00550.x. [DOI] [PubMed] [Google Scholar]
- 5.Ness RB, Grisso JA, Cottreau C, et al. Factors related to inflammation of the ovarian epithelium and risk of ovarian cancer. Epidemiology. 2000 Mar;11(2):111–117. doi: 10.1097/00001648-200003000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Song H, Ramus SJ, Quaye L, et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006 Nov;27(11):2235–2242. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 7.Auranen A, Song H, Waterfall C, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005 Nov 20;117(4):611–618. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 8.Espey LL. Current status of the hypothesis that mammalian ovulation is comparable to an inflammatory reaction. Biol Reprod. 1994 Feb;50(2):233–238. doi: 10.1095/biolreprod50.2.233. [DOI] [PubMed] [Google Scholar]
- 9.Terranova PF, Rice VM. Review: cytokine involvement in ovarian processes. Am J Reprod Immunol. 1997 Jan;37(1):50–63. doi: 10.1111/j.1600-0897.1997.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 10.Behrman HR, Kodaman PH, Preston SL, Gao S. Oxidative stress and the ovary. J Soc Gynecol Investig. 2001 Jan–Feb;8(1 Suppl Proceedings):S40–S42. doi: 10.1016/s1071-5576(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 11.Murdoch WJ, Martinchick JF. Oxidative damage to DNA of ovarian surface epithelial cells affected by ovulation: carcinogenic implication and chemoprevention. Exp Biol Med (Maywood) 2004 Jun;229(6):546–552. doi: 10.1177/153537020422900613. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006 Jan;55(1):115–122. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grisham MB, Jourd'heuil D, Wink DA. Review article: chronic inflammation and reactive oxygen and nitrogen metabolism--implications in DNA damage and mutagenesis. Aliment Pharmacol Ther. 2000 Apr;14 Suppl 1:3–9. doi: 10.1046/j.1365-2036.2000.014s1003.x. [DOI] [PubMed] [Google Scholar]
- 14.Ristimaki A. Cyclooxygenase 2: from inflammation to carcinogenesis. Novartis Found Symp. 2004;256:215–221. discussion 21-6, 59–69. [PubMed] [Google Scholar]
- 15.Schildkraut J, Moorman P, Bland A, et al. Cyclin E overexpression in epithelial ovarian cancer characterizes an etiologic subgroup. Cancer Epidemiol Biomarkers Prev. 2008;17(3):585–593. doi: 10.1158/1055-9965.EPI-07-0596. [DOI] [PubMed] [Google Scholar]
- 16.Merritt MA, Green AC, Nagle CM, Webb PM. Talcum powder, chronic pelvic inflammation and NSAIDs in relation to risk of epithelial ovarian cancer. Int J Cancer. 2008 Jan 1;122(1):170–176. doi: 10.1002/ijc.23017. [DOI] [PubMed] [Google Scholar]
- 17.Rossing MA, Cushing-Haugen KL, Wicklund KG, Doherty JA, Weiss NS. Menopausal hormone therapy and risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2007 Dec;16(12):2548–2556. doi: 10.1158/1055-9965.EPI-07-0550. [DOI] [PubMed] [Google Scholar]
- 18.Royar J, Becher H, Chang-Claude J. Low-dose oral contraceptives: protective effect on ovarian cancer risk. Int J Cancer. 2001 Nov 20;95(6):370–374. doi: 10.1002/1097-0215(20011120)95:6<370::aid-ijc1065>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Bushley AW, Ferrell R, McDuffie K, et al. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol. 2004 Dec;95(3):672–679. doi: 10.1016/j.ygyno.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Pearce CL, Wu AH, Gayther SA, et al. Progesterone receptor variation and risk of ovarian cancer is limited to the invasive endometrioid subtype: results from the ovarian cancer association consortium pooled analysis. Br J Cancer. 2008 Jan 29;98(2):282–288. doi: 10.1038/sj.bjc.6604170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soegaard M, Jensen A, Hogdall E, et al. Different risk factor profiles for mucinous and nonmucinous ovarian cancer: results from the Danish MALOVA study. Cancer Epidemiol Biomarkers Prev. 2007 Jun;16(6):1160–1166. doi: 10.1158/1055-9965.EPI-07-0089. [DOI] [PubMed] [Google Scholar]
- 22.Sellers TA, Huang Y, Cunningham J, et al. Association of single nucleotide polymorphisms in glycosylation genes with risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008 Feb;17(2):397–404. doi: 10.1158/1055-9965.EPI-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuire V, Felberg A, Mills M, et al. Relation of contraceptive and reproductive history to ovarian cancer risk in carriers and noncarriers of BRCA1 gene mutations. Am J Epidemiol. 2004 Oct 1;160(7):613–618. doi: 10.1093/aje/kwh284. [DOI] [PubMed] [Google Scholar]
- 24.Ziogas A, Gildea M, Cohen P, et al. Cancer risk estimates for family members of a population-based family registry for breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2000 Jan;9(1):103–111. [PubMed] [Google Scholar]
- 25.Ramus S, Vierkant R, Johnatty S, et al. Consortium Analysis of 7 Candidate SNPs for Ovarian Cancer. International Journal of Cancer. 2008;123(2):380–388. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pike MC, Pearce CL, Peters R, Cozen W, Wan P, Wu AH. Hormonal factors and the risk of invasive ovarian cancer: a population-based case-control study. Fertil Steril. 2004 Jul;82(1):186–195. doi: 10.1016/j.fertnstert.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004 Jan;74(1):106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003 Aug 5;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raftery AE. Bayesian Model Selection in Social Research. Sociological Methodology. 1995;25:111–163. [Google Scholar]
- 30.R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2008. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986 Sep;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 33.Cochran W. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 34.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002 Jun 15;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 35.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008 Feb;123(1):1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003 Sep 6;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.