Abstract
The well-known fig–fig wasp and yucca–yucca moth mutualisms are classic examples of obligate mutualisms that have been shaped by millions of years of coevolution. Pollination systems involving obligate seed parasites are only expected to evolve under rare circumstances where their positive effects are not swamped by abundant co-pollinators and heavy costs resulting from seed destruction. Here, we show that, in Phyllantheae, specialization to pollination by Epicephala moths evolved at least five times, involving more than 500 Phyllantheae species in this obligate association. Active pollination behaviour evolved once in Epicephala, 10–20 Myr after the initial divergence of their host plants. The pollinating Epicephala moths thus radiated on an already-diverged host lineage and successively colonized new Phyllantheae hosts, thereby giving rise to repeated independent evolution of the specialized pollination system in Phyllantheae. The present evolutionary success of this association rests entirely upon active pollination by Epicephala, making this a distinct example of an evolutionary key innovation. Overall, our findings provide a clear empirical demonstration of how a combination of evolutionary innovation and partner shifts facilitates the spread of mutualism in a coevolving species interaction.
Keywords: active pollination behaviour, Epicephala, key innovation, obligate pollination mutualism, Phyllantheae
1. Introduction
The well-known obligate fig–fig wasp and yucca–yucca moth mutualisms have become principal model systems for the study of coevolution (Weiblen 2002; Cook & Rasplus 2003; Pellmyr 2003). Female fig wasps and yucca moths pollinate the flowers in which they lay eggs, and the plants sacrifice a fraction of their developing seeds to nourish pollinator larvae. Despite a wealth of documented examples of specialized pollination systems in angiosperms, however, pollination by obligate seed parasites is rare. This is because seed parasitism inflicts a heavy cost on plants, while abundant co-pollinators swamp the mutualistic effect of pollination by seed parasites (Thompson & Pellmyr 1992; Thompson & Cunningham 2002). Consequently, there are only a handful of mutualisms wherein obligate seed parasites act as effective pollinators of their host plants (Pellmyr 1989; Pellmyr et al. 1996a; Fleming & Holland 1998).
A novel example of such a coevolved obligate pollination mutualism has recently been reported between Phyllantheae plants (Phyllanthaceae) and Epicephala moths (Gracillariidae) (Kato et al. 2003; Kawakita & Kato 2004a,b). Phyllantheae is a pantropical tribe of more than 1200 species having a remarkable diversity of growth forms, ranging from annual and perennial herbs, shrubs and trees to climbers, succulents, rheophytes and aquatics (Hoffmann et al. 2006; Kathriarachchi et al. 2006). The plants have minute, unisexual flowers that are borne as clusters on leaf axils (figure 1). Production of nectar is variable across taxa, which probably reflects differences in pollination systems. Species belonging to Glochidion (more than 300 spp.), Breynia (35 spp.) and Phyllanthus subgenus Gomphidium (more than 150 spp.) are each pollinated nocturnally by a species-specific Epicephala moth whose larvae feed on the seeds (figure 1). The female moths have evolved to actively collect and transport pollen between flowers using specialized proboscides equipped with numerous sensilla (Kawakita & Kato 2006). They lay eggs in the flowers they pollinate, and the larvae consume a fraction of the developing seeds, leaving others viable for plant reproduction.
Figure 1.
Phyllantheae–Epicephala obligate pollination mutualism. (a) Glochidion acuminatum is pollinated by a species-specific Epicephala moth that infests (b) the fruit as larvae. The adult female moth (approx. 5 mm in body length) (c) actively collects pollen on male flowers and (d) pollinates the female flower in which she lays an egg.
Although the phylogenetic classification of Phyllantheae has been the subject of previous investigations (Kathriarachchi et al. 2005, 2006), virtually nothing is known about pollination biology in the remaining groups of Phyllantheae. We therefore studied pollination systems and associations with Epicephala in 26 species of Phyllantheae during 2002–2007 in Southeast Asia, New Caledonia, Australia, Madagascar, Guinea and North America (table 1). Based on the results of the pollination study, we explored the origin of the Phyllantheae–Epicephala mutualism by reconstructing robust phylogenies for 46 species of Phyllantheae and associated Epicephala moths. While the present study focuses on only a small proportion of the global diversity of Phyllantheae, the sampled species cover the entire range of taxonomic diversity within the tribe (Hoffmann et al. 2006; Kathriarachchi et al. 2006), allowing a sound investigation into broad coevolutionary history of the Phyllantheae–Epicephala association. Overall, our results reveal an unexpectedly complex origin of the Phyllantheae–Epicephala pollination mutualism and provide important general insights into how a combination of evolutionary innovation and partner shifts shapes the evolutionary dynamics of mutualism in coevolving species interactions.
Table 1.
List of species studied.
| species sampleda | abbreviation | study site/sampling locality | Epicephala as the pollinator | criteria for pollinator determinationb | style spreading |
|---|---|---|---|---|---|
| Margaritaria | |||||
| M. discoidea (Baill.) G. L. Webster | Mdis | Guinea: Bossou | no | E (fr), M | 7.07 |
| M. indica (Dalziel) Airy Shaw | Mind | Japan: Okinawa Island | no | M | 5.46 |
| Flueggea | |||||
| F. jullienii (Beille) G. L. Webster | Fjul | Laos: Mahaxai | no | M | 4.99 |
| F. suffruticosa Baill. | Fsuf | Japan: Hyogo/Hiroshima/Amami Islands | no | E (fv, fr, fl), M | 4.82 |
| F. virosa (Willd.) Voigt | Fvir | Laos: Vieng Xai/Taiwan: Fangliao | no | E (fv, fr), M | 4.47 |
| Phyllanthus | |||||
| P. (Is.) ussuriensis Rupr. & Maxim. | Puss | Japan: Tokyo/Kyoto | no | E (fv, fr, fl), M | 7.01 |
| P. (Is.) virgatus G. Forst | Pvir | Laos: Vientiane | no | E (fv, fr), M | 5.97 |
| P. (Er.) liukiuensis Hayata | Pliu | Japan: Okinawa Island | no | E (fr, fl), M | 7.87 |
| P. (Er.) pulcheroides Beille | Ppul | Laos: Mahaxai | no | E (fr, fl), M | 8.69 |
| P. (Ki.) reticulatus Poir. | Pret | Taiwan: Henchun | yes | E (fv, fr, fl), M | 0.45 |
| P. (Ki.) sp. | Psp | Laos: Laksao | yes | E (fl), M | 0.50 |
| P. (Ki.) flexuosus Müll. Arg. | Pfle | Japan: Kyoto/Hyogo/Miyazaki | no | E (fv, fr, fl), M | 4.87 |
| P. (Ki.) oligospermus Hayata | Poli | Japan: Yonaguni Island | no | E (fv, fr, fl), M | 4.96 |
| P. (Ki.) tenellus Roxb. | Pten | Japan: Okinawa Island | no | E (fr, fl), M | 6.41 |
| P. (Ph.) amarus Schumach. & Thonn. | Pama | Japan: Ishigaki Island/Laos: Thakhaek | no | E (fv, fr, fl), M | 4.09 |
| P. (Ph.) debilis Willd. | Pdeb | Japan: Ishigaki Island | no | E (fv, fr, fl), M | 4.32 |
| P. (Ph.) lepidocarpus Siebold & Zucc. | Plep | Japan: Kyoto/Miyako Island/Ishigaki Island | no | E (fv, fr, fl), M | 3.12 |
| P. (Go.) aeneus Baill. | Paen | New Caledonia: Cap Bocage | yes | L, M | 1.08 |
| P. (Go.) gneissicus S. Moore | Pgne | New Caledonia: Mt Panié | yes | L | n.a. |
| P. (Go.) guillauminii Däniker | Pgui | New Caledonia: Tiébaghi | yes | L | n.a. |
| P. (Go.) vulcani Guillaumin | Pvul | New Caledonia: Riviere Bleue | yes | L, M | 0.62 |
| P. (Go.) bourgeoisii Baill. | Pbou | New Caledonia: Cap Bocage | yes | L, M | 0.38 |
| P. (Go.) chamaecerasus Baill. | Pcha | New Caledonia: Chutes de Ba | yes | L | n.a. |
| P. (Go.) caudatus Müll. Arg. | Pcau | New Caledonia: Riviere Bleue | yes | L | n.a. |
| P. (Go.) koniamboensis M. Schmid | Pkon | New Caledonia: Tinip | yes | L | n.a. |
| P. (Go.) mangenotii M. Schmid | Pman | New Caledonia: Cap Bocage | yes | L, M | 0.49 |
| P. (Ci.) acidus (L.) Skeels | Paci | Laos: Vientiane (cultivated) | no | L, E (fr), M | 2.50 |
| P. (Em.) emblica L. | Pemb | Laos: Ban Chomesy | no | L, E (fr) | n.a. |
| P. (Pd.) roseus Beille | Pros | Laos: Phialat | no | E (fv, fr, fl), M | 1.99 |
| P. marojejiensis (Leandri) Petra Hoffm. & McPherson | Pmar | Madagascar: Mt Marojeji | yes | E (fv, fr, fl), M | 0.18 |
| P. humbertii (Leandri) Petra Hoffm. & McPherson | Phum | Madagascar: Mt Marojeji | yes | E (fr), M | 0.39 |
| Reverchonia | |||||
| R. arenaria A. Gray | Rare | USA: New Mexico | no | E (fv, fr), M | 1.87 |
| Sauropus | |||||
| S. androgynus Merr. | Sand | Laos: Thakhaek | no | E (fr, fl), M | 2.03 |
| S. brevipes Müll. Arg. | Sbre | Laos: Vientiane | no | E (fr, fl), M | 2.14 |
| S. granulosus Airy Shaw | Sgra | Laos: Vientiane | no | E (fv, fl), M | 2.04 |
| S. quadrangularis Müll. Arg. | Squa | Laos: Vientiane | no | E (fv, fr, fl), M | 2.53 |
| Breynia | |||||
| B. disticha Forst. | Bdis | New Caledonia: Koumac | yes | M | 0.25 |
| B. fruticosa (Müll. Arg.) Hook.f. | Bfru | Laos: Vientiane | yes | L, M | 1.45 |
| B. oblongifolia Müll. Arg. | Bobl | Australia: Windsor Tableland | yes | M | 0.20 |
| B. retusa Alston | Bret | Laos: Vientiane | no | E (fv, fr, fl), M | 3.02 |
| B. vitis-idaea (Burm.f.) C. E. C. Fisch. | Bvit | Japan: Amami Islands | yes | L, M | 0.43 |
| Glochidion | |||||
| G. acuminatum Müll. Arg. | Gacu | Japan: Amami Islands | yes | L, M | 0.86 |
| G. lanceolatum Hayata | Glan | Japan: Ishigaki Island | yes | L, M | 0.31 |
| G. obovatum Siebold & Zucc. | Gobo | Japan: Wakayama | yes | L, M | 0.93 |
| G. rubrum Blume | Grub | Japan: Ishigaki Island | yes | L, M | 0.87 |
| G. zeylanicum (Gaertn.) A. Juss. | Gzey | Japan: Okinawa Island | yes | L, M | 0.24 |
Phyllanthus subgenera are abbreviated as follows: Is., Isocladus; Er., Eriococcus; Ki., Kirganelia; Ph., Phyllanthus; Ci., Cicca; Em., Emblica; Pd., Phyllanthodendron. Phyllanthus marojejiensis and Phyllanthus humbertii have not been assigned to any subgenus.
Each species was judged as either Epicephala or non-Epicephala pollinated based on literature information (L), ecological data (E) and/or style morphology of the female flower (M). Ecological data consisted of direct observation of flower visitors (fv) and/or examination of Epicephala larvae/eggs in fruits (fr)/flowers (fl), respectively.
2. Material and methods
(a) Determination of pollination systems
We determined whether a species is Epicephala-pollinated or not by examining the presence/absence of Epicephala eggs and larvae in female flowers and fruits, respectively. We judged that a species has a non-Epicephala pollination system if no larvae were found in the sampled fruits (table S1 in the electronic supplementary material). Whenever possible, this was further confirmed by the absence of eggs in pollinated female flowers under a light microscope. Species that had Epicephala larvae in fruits were further examined for eggs in pollinated female flowers (tables S1 and S2 in the electronic supplementary material). If the pollinated status of a female flower and the presence of Epicephala eggs are coupled, it is likely that the species is Epicephala-pollinated because the female moth invariably lays an egg in the flower that she pollinated (Kato et al. 2003; Kawakita & Kato 2004a,b). In turn, a lack of association or absence of eggs in female flowers indicates that Epicephala moths are not involved in pollination. Pollination status was determined by checking for the presence of pollen grains on the stigma under a light microscope. Overall, we dissected 19–461 fruits representing 1–31 plants for each of 19 species to examine the presence of seed feeding by Epicephala larvae. Also, we looked for Epicephala eggs in 25–393 female flowers representing 2–13 plants in 24 species.
Inference of pollination systems based on the above methods was further substantiated by field observations of diurnal and nocturnal flower visitors (table S3 in the electronic supplementary material). We paid special attention to the behaviour of Epicephala to determine whether the moths exhibited pollinating behaviour. In herbaceous species of Phyllanthus that belong to subgenera Isocladus and Phyllanthus, ants were the predominant flower visitors (table S3 in the electronic supplementary material). Because antibiotic substances secreted from ant metapleural glands can inhibit pollen germination (Beattie et al. 1984), we experimentally tested the effectiveness of ant pollination in Phyllanthus lepidocarpus (see the electronic supplementary material).
Although our inference of pollination system does not rely on pollinator exclusion or pollen deposition experiments, which provide a more direct evidence of pollinator contributions, the above approach has proven to be reliable in previous studies (Kato et al. 2003; Kawakita & Kato 2004a,b) and consistently leads to unambiguous inference of whether or not a species is Epicephala-pollinated (table 1; tables S1–S3 in the electronic supplementary material); thus, pollination systems inferred as such can be reliably combined with phylogenetic results to explore an overall evolutionary pattern of Epicephala pollination in Phyllantheae.
(b) Quantification of style spreading
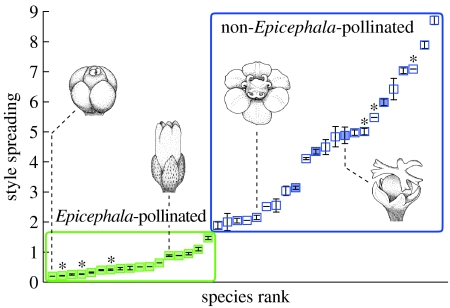
Of the 46 Phyllantheae species sampled in this study, information on the pollination systems for 40 species was available from previous literature and field data collected as described above. To infer pollination systems for the other six species, we quantified the degree of style spreading, which reflects a syndrome associated with Epicephala pollination. In species pollinated by the moths, styles are reduced and fused to form a narrow apical cavity into which moths actively deposit pollen (figure 2; Kato et al. 2003; Kawakita & Kato 2004a,b). By contrast, species diurnally pollinated by various nectar-seeking insects usually have bifid styles that are spread horizontally, which facilitates passive pollen receipt from insect bodies (figure 2). Style spreading is a continuous, quantitative measure that does not by itself provide information about pollination system. However, species with different pollination syndromes had non-overlapping distributions of style spreading, and the six species with unknown pollination systems were nested well within either of the cohorts (see §3); thus, we were able to reliably assign their pollination modes despite lack of ecological data.
Figure 2.
Distribution of style spreading in Phyllantheae. Species pollinated by Epicephala have reduced styles that are medially fused, whereas non-Epicephala-pollinated species have horizontally spread, bifid styles. Filled and open boxes indicate species with and without associations with Epicephala, respectively. Ecological data were not available for species with asterisks. Female flowers are drawn for Phyllanthus marojejiensis, G. acuminatum, Sauropus brevipes and Flueggea suffruticosa (from left to right). Error bars, ±1 s.e.
We quantified style spreading as the ratio of apical-to-basal style width using 3–45 flowers representing 1–10 plants for each species in 40 Phyllantheae taxa. Measurements of style width were done under a light microscope equipped with an eyepiece micrometer using either fresh plant materials or specimens preserved in 70 per cent ethanol. Because flowers of Phyllantheae are trimerous, the apical stylar width was represented by the diameter of the minimum circle circumscribing the triangle formed by the three styles. The average lengths of the three sides were used to calculate the diameter by approximating the triangle to be equilateral. The left arms of the styles were used for measurements in species with bifid styles.
(c) Molecular phylogenetic analysis
DNA extraction and sequencing followed previously described methods (Kawakita et al. 2004; Kawakita & Kato 2006). The phylogenetic analysis of the 46 species of Phyllantheae was based on sequences of the chloroplast matK, ndhF, atpB and nuclear PHYC genes. In addition to the species sampled here, we included 46 sequences of other members of Phyllanthaceae and four outgroup taxa representing Picrodendraceae and Putranjivaceae that were available from a previous study (Kathriarachchi et al. 2005) to allow fossil calibrations in the following divergence time estimation. Species with more than one missing gene in Kathriarachchi et al. (2005) were not included, to avoid potential confounding effects of large amounts of missing data on phylogenetic reconstruction and divergence time estimation (Douzery et al. 2004; Wiens 2006). We also reconstructed the phylogeny of the associated 26 Epicephala species, which were each specific to one, or rarely two, Phyllantheae species. These moths are currently all undescribed but can be clearly distinguished based on male genital morphology. The phylogenetic analysis of Epicephala was based on sequences of the mitochondrial COI, nuclear arginine kinase (ArgK), elongation factor-1 alpha (EF-1α), wingless (Wg) and 18S rDNA genes. For the moth outgroups, we sampled three gracillariid moths: Cuphodes diospyrosella, Stomphastis labyrinthica and Melanocercops ficuvorella. The last species was included as the farthest outgroup based on a larger phylogenetic analysis of the subfamily based on EF-1α (A. Kawakita & M. Kato 2007, unpublished data; also see the electronic supplementary material for details of sampling). Polymerase chain reactions were done using primers and protocols provided previously (Brower & DeSalle 1998; Heraty et al. 2004; Kathriarachchi et al. 2005; Kawakita & Kato 2006) and additional matK primers (listed in the electronic supplementary material, table S4). Available plant and moth voucher specimens are preserved in the Kyoto University Museum, and all newly obtained sequences have been deposited in the GenBank database under accession numbers FJ235206–FJ235514 (see tables S5 and S6 in the electronic supplementary material for the full list of accession numbers).
Phylogenetic analysis was conducted using maximum-parsimony, maximum-likelihood (ML) and Bayesian methods. Maximum-parsimony heuristic searches were done in PAUP* v. 4.0.b10 (Swofford 2002) with 100 random addition analyses and tree bisection–reconnection branch swapping, and the robustness of trees was assessed by non-parametric bootstrapping with 1000 replicates. We also performed ML heuristic searches using PAUP* with 10 random addition analyses and subtree pruning–regrafting branch swapping. The ApproxLim parameter was adjusted to 2 to accelerate the tree search (Morrison 2007). Appropriate models of base substitution were selected using Modeltest v. 3.06 (Posada & Crandall 1998). The robustness of ML trees was validated with non-parametric bootstrapping using PhyML (Guindon & Gascuel 2003) with 500 replicates. We performed a Bayesian analysis using MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003), with unlinked models for each partition. The analysis consisted of two million generations, sampling trees every 1000 generations for a total of 2001 trees. We plotted ln-likelihood of the sampled trees against generation time to identify the region of the analysis in which the parameter estimates were stable, and accordingly discarded the initial 1001 trees as burn-in.
Because Epicephala-pollinated Phyllantheae species were not recovered as monophyletic (see §3), we tested the robustness of this reconstruction using an approximately unbiased test (Shimodaira 2002). Similarly, we tested the monophyly of the actively pollinating Epicephala species, which were also recovered as non-monophyletic (see §3). The tests were conducted using Consel (Shimodaira & Hasegawa 2001), with alternative hypotheses obtained by constraining the monophyly of Epicephala-pollinated Phyllantheae species or that of actively pollinating Epicephala, and performing ML searches as described above.
Major clades of Epicephala were generally associated with well-defined taxonomic groups of Phyllantheae (see §3), but relationships at higher levels were not congruent between the two phylogenies. We therefore tested the cophylogenetic association between plant and moth phylogenies at higher taxonomic levels using ParaFit (Legendre et al. 2002). To focus on phylogenetic relationships at higher levels, terminal taxa in the plant phylogeny were pruned to major plant taxonomic groups, and those in the moth phylogeny to well-defined Epicephala clades having exclusive associations with major host groups (see the electronic supplementary material).
(d) Local ML ancestral state reconstruction
To investigate the evolutionary history of the Phyllantheae–Epicephala mutualism, we conducted local ML ancestral character state reconstructions using BayesMultistate (Pagel et al. 2004) and ML phylogenies of Phyllantheae and Epicephala. We first tested for a difference in transition rates between the two states (presence/absence of Epicephala pollination in Phyllantheae and pollination behaviour in Epicephala) by comparing likelihoods between the one-rate and the two-rate model. We used a likelihood ratio test to determine statistical significance following a Χ2-distribution with one degree of freedom. Because there was no significant difference between the two rates in either the plant or moth phylogeny, the one-rate model was used in ancestral state reconstructions (Pagel 1999). A likelihood ratio of greater than 2 was considered significant evidence for the occurrence of either state at ancestral nodes (Pagel 1999). The common ancestor of Phyllantheae and its sister outgroup clade was assumed to have a non-Epicephala pollination system because none of the plants outside Phyllantheae are known to have associations with Epicephala. Similarly, the common ancestor of Epicephala and its sister clade was assumed to be a non-pollinator, because the rest of the gracillariid moths are generally leaf miners and do not visit flowers. The robustness of the results to phylogenetic uncertainty was validated using 100 subset trees from the Bayesian analysis and performing the reconstructions as described above, which indicated that all significant reconstructions were stable in at least 70 per cent of the trees tested (results not shown).
(e) Divergence time estimation
The above analysis of ancestral character state reconstruction indicated that Epicephala-pollinated Phyllantheae plants evolved multiple times independently (see §3). However, because our taxon sampling was limited to 46 species amid the global diversity of Phyllantheae, results of ancestral state reconstruction might change with the addition of more taxa. We therefore estimated divergence times for the Phyllantheae and Epicephala phylogenies to test whether the multiple-origins hypothesis is in fact the preferred scenario. If the age of the most recent common ancestor (MRCA) of moth-pollinated plants is contemporary with that of Epicephala, a single origin of the mutualism followed by multiple losses would still be a viable hypothesis. Alternatively, evolution of pollinating behaviour post-dating initial host divergence would provide strong support for the multiple-origins hypothesis.
A likelihood ratio test rejected the assumption of a strict molecular clock in both the plant and moth phylogenies (p<0.0001); thus, we used the penalized-likelihood (Sanderson 2002) and Bayesian methods (Thorne & Kishino 2002) to estimate divergence times using the ML phylogenies for each group. For the penalized-likelihood approach, we used the program r8s (Sanderson 2002) with smoothing parameters of 10 and 5 obtained by cross-validation for the plant and moth phylogenies, respectively. Confidence intervals were obtained by means of non-parametric bootstrapping, which consisted of generating 100 bootstrap datasets with SeqBoot (Felsenstein 1993), estimating branch lengths of the ML topology with each of these datasets under the original substitution model and estimating divergence times as described. The Bayesian relaxed-clock approach used was that implemented in multidivtime (Thorne & Kishino 2002), with model parameters obtained using PAML (Yang 1997). The analysis consisted of 100 000 burn-in cycles and 1 million post-burn-in cycles, with sampling at every 100th tree. Prior distributions on parameters for the Bayesian analysis are provided in the electronic supplementary material.
We used the following four non-redundant fossils as minimum age constraints for the Phyllanthaceae phylogeny (table S7 in the electronic supplementary material): Bischofia-type pollen from Bartonian, Middle Eocene (37.2 Myr ago); Actephila-type pollen from Late Eocene (33.9 Myr ago); Phyllanthus-type pollen from Early Eocene (48.6 Myr ago; Gruas-Cavagnetto & Köhler 1992); and Glochidion leaf impressions from Middle Miocene (11.6 Myr ago; Prasad 1994; Antal & Prasad 1996). We initially constrained the root node (i.e. the node splitting Phyllanthaceae and Picrodendraceae) to be no older than 125 Myr ago, a well-established maximum age of the eudicot clade based on the earliest known occurrence of tricolpate pollen (Magallón et al. 1999). However, because Phyllanthaceae is nested well within the phylogeny of eudicots, we also used a more conservative maximum age of 108 Myr ago for our basal node, which is the oldest estimate of the corresponding node in a previous study of Malpighiales radiation (Davis et al. 2005). Because attribution of some of the Phyllanthaceae fossils may still need refinements (Gruas-Cavagnetto & Köhler 1992), caution may be necessary when taking the precise dates resulting from our analysis. For the details of Phyllanthaceae fossils, see the electronic supplementary material.
Because gracillariid moths are extremely scarce in the fossil record (Lopez-Vaamonde et al. 2006), Epicephala divergence times were obtained by estimating relative divergence times for the Epicephala phylogeny and calibrating node ages based on the substitution rate of the COI gene. We used only the COI substitution rate because it is generally conserved across arthropod taxa (Gaunt & Miles 2002), has been widely used for dating in insects (Kandul et al. 2004; Quek et al. 2007; Ueda et al. 2008) and clusters at approximately 1.5% Myr−1 in several arthropod groups (Farrell 2001; Quek et al. 2004; Sota & Hayashi 2007). To obtain relative divergence times for the Epicephala phylogeny, we forced the root node (i.e. the split between Epicephala and C. diospyrosella) to have one arbitrary time unit in both penalized-likelihood and Bayesian analyses. Every node on the obtained ultrametric tree was then individually used as a calibration point to obtain the range of age estimates for the MRCA of Epicephala. COI distances were corrected using the appropriate substitution model (GTR+Γ+I) and model parameters inferred by Modeltest, and the average pairwise COI distance across each node was transformed to absolute age using the 1.5% Myr−1 substitution rate.
In addition to using relative node ages inferred from relaxed-clock methods, we also estimated the age of the Epicephala root node by calculating branch lengths of the ML tree with the COI data under the assumption of a strict clock. Branch lengths were calculated in PAUP* with the above GTR+Γ+I substitution model, and node height of the Epicephala root node on the resulting ultrametric tree was transformed to absolute age using the 1.5% Myr−1 substitution rate. We obtained 95% credibility interval of the estimated age by generating 100 bootstrap datasets with SeqBoot and calculating the root age as described. Although a likelihood ratio test rejected a strict molecular clock in the Epicephala phylogeny (see above), we conducted this analysis as complementary to the relaxed-clock methods because the use of constant COI substitution rate inherently assumes an underlying molecular clock.
3. Results and discussion
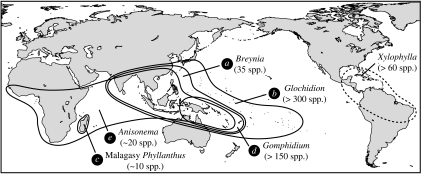
We found two additional Phyllantheae lineages that are obligately pollinated by host-specific Epicephala moths (see the electronic supplementary material), namely Phyllanthus section Anisonema with approximately 20 species throughout the Palaeotropics and an unclassified group of Phyllanthus endemic to Madagascar (approx. 10 species). The remaining species were pollinated by diurnal insects that visited flowers for nectar and pollen, and did not have associations with pollinating Epicephala (tables S1 and S3 in the electronic supplementary material). However, Flueggea suffruticosa and three herbaceous Phyllanthus species were parasitized by seed-parasitic Epicephala species that did not pollinate the flowers. These plants were effectively pollinated by bees, flies, beetles or ants (table S8 in the electronic supplementary material); thus, the associated Epicephala are truly parasitic on their hosts. Overall, species with different pollination syndromes had non-overlapping degrees of style spreading (figure 2); thus, we were able to reliably assign pollination systems to plant species for which sufficient ecological data were not available.
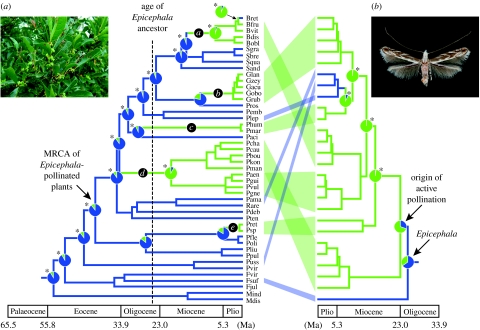
To investigate the origin of the Phyllantheae–Epicephala mutualism, we reconstructed the phylogeny of 46 Phyllantheae species and mapped whether each species is pollinated by Epicephala. Data matrix of the combined chloroplast matK, ndhF, atpB and nuclear PHYC genes consisted of 4059 bp aligned sequences. Maximum-parsimony, likelihood and Bayesian analyses all produced a highly resolved and well-supported phylogeny for Phyllantheae (fig. S1 in the electronic supplementary material). Species pollinated by Epicephala were not monophyletic (approximately unbiased test; p<0.0001), indicating that there have been multiple shifts in pollination systems. To determine the direction of pollinator shifts, we reconstructed ancestral character states for pollination systems using the local maximum likelihood. The results clearly indicated five independent origins of the obligate pollination mutualism in Phyllantheae, with a single reversal to non-Epicephala pollination in Breynia retusa (figure 3).
Figure 3.
Phylogenetic analysis and the origin of the Phyllantheae–Epicephala mutualism. ML chronograms for (a) Phyllantheae plants (Phyllanthaceae root node constrained to be no older than 108 Myr ago) and (b) associated Epicephala moths. Pie charts indicate the probability of Epicephala and non-Epicephala pollination systems (Phyllantheae) or the presence/absence of active pollination behaviour (Epicephala) occurring at ancestral nodes. Asterisks indicate significant differences in likelihoods. The Epicephala tree is calibrated with 25 Myr ago at the root node. Mutualism is represented in green, and associations between major plant and moth clades are indicated. See table 1 for terminal taxon names.
We also reconstructed the phylogeny of 26 Epicephala species associated with the above Phyllantheae species. Analysis of the combined 2933 bp aligned sequences of the mitochondrial COI, nuclear ArgK, EF-1α, Wg and 18S rDNA genes produced a well-resolved phylogeny, although the earliest branching lineage within Epicephala remained ambiguous (fig. S2 in the electronic supplementary material). The pollinator species were non-monophyletic (approximately unbiased test; p<0.0001), and ancestral character state reconstruction indicated a likely single origin of pollination behaviour with a single event of secondary loss (figure 3). Major clades of Epicephala, generally, had specific associations with well-defined taxonomic groups of Phyllantheae (figure 3; fig. S2 in the electronic supplementary material), but relationships at higher levels were not different from random (ParaFit; p=0.39), indicating that host shifts have occurred repeatedly.
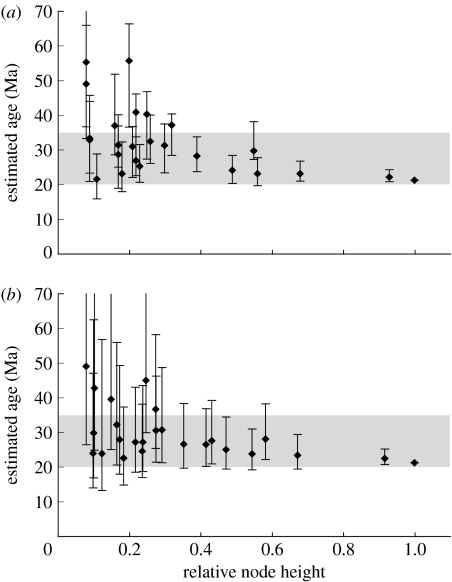
Divergence time estimates using relaxed-clock methods and fossil calibrations indicated that the MRCA of Epicephala-pollinated plants occurred 41.0 Myr ago (penalized likelihood; 95% credibility interval, 39.3–48.3 Myr ago) or 40.4 Myr ago (Bayesian method; 33.4–48.2 Myr ago), when constraining the root node at 108 Myr ago (fig. S3 in the electronic supplementary material). When using the older maximum age for the root node (125 Myr ago), the corresponding estimates were 47.4 Myr ago (penalized likelihood; 45.0–55.2 Myr ago) and 42.2 Myr ago (Bayesian method; 34.2–51.8 Myr ago). By contrast, estimated ages of the Epicephala root node clustered within a time frame between 20 and 35 Myr ago (figure 4; also see the electronic supplementary material and fig. S4 therein) and converged at 20–30 Myr ago as deeper nodes were used to calibrate the root age. Because fixing ages at shallowly placed nodes for deep extrapolations can be prone to large systematic errors (Ho et al. 2005; Sota & Hayashi 2007), the range of 20–30 Myr ago probably provides a robust time frame for the Epicephala root node, which is consistent with the estimates obtained from the COI molecular clock phylogeny (optimal, 22.76 Myr ago; 95% confidence interval, 18.6–29.1 Myr ago). These estimates for the age of Epicephala post-date initial host divergence by roughly 10–20 Myr, which is consistent with delayed radiation of Epicephala and hence multiple origins of the obligate pollination mutualism in Phyllantheae. Although our estimates of the timing of Epicephala divergence depend largely on the accuracy of the COI substitution rate, the assumed 1.5% Myr−1 is among the slowest of known substitution rates for the arthropod COI gene (1.3–2.3% Myr−1; Brower 1994; Quek et al. 2004), and using higher rates would only give younger estimates for the age of the Epicephala root node; thus, our method is conservative with respect to providing young ages.
Figure 4.
Estimated ages for the most recent common ancestor of Epicephala. Optimal ages and 95% credibility intervals are plotted against relative node heights of those used to calibrate the Epicephala chronogram. The majority of estimates fall within a time window of 20–35 Myr ago (shaded). (a) Penalized-likelihood and (b) Bayesian estimation.
Because our taxon sampling was limited to 20 per cent of the global diversity of Phyllantheae at the section level (Kathriarachchi et al. 2006) and less than 5 per cent at the species level, the entire picture of the evolutionary history of Epicephala pollination in Phyllantheae is probably much more complex than as presented here. However, inclusion of other lineages would probably only strengthen our conclusion of repeated independent evolution because these plants have bifid, horizontally spread styles that are characteristic of non-Epicephala-pollinated plants (figure 2). An exception is the New World Phyllanthus subgenus Xylophylla, which consists of approximately 60 species having reduced, columnar styles (Webster 1958) and thus may represent an additional origin of Epicephala moth pollination in Phyllantheae (figure 5).
Figure 5.
Five independent origins of the Phyllantheae–Epicephala obligate pollination mutualism. The New World subgenus Xylophylla shares the reduced, columnar style morphology with other Epicephala-pollinated plants and may represent an additional origin of the mutualism. Letters in circles correspond to clade names in figure 3.
Our finding that the obligate pollination mutualism arose repeatedly in Phyllantheae is in stark contrast with the situations in the fig–fig wasp and yucca–yucca moth mutualisms. Recent coevolutionary analyses in the fig and yucca systems have indicated that these associations arose only once in each partner lineage 40–60 Myr ago (Pellmyr & Leebens-Mack 1999; Rønsted et al. 2005). An exception is Hesperoyucca whipplei, which is phylogenetically distant from the rest of the yuccas and independently established the mutualism with a yucca moth (Bogler et al. 1995; Pellmyr et al. 2007; Smith et al. 2008). In the Phyllantheae–Epicephala system, major lineages of Phyllantheae had already emerged when Epicephala colonized these plants ca 30 Myr ago. Sequential radiation of Epicephala on an already-diverged host lineage has probably provided opportunities for the moth pollinators to establish new mutualistic associations in distant host lineages. Thus, specialization to moth pollination occurred multiple times independently in Phyllantheae as Epicephala evolved to use a broad range of the Phyllantheae lineage.
Our results also indicate that colonization of new host lineages by the pollinators sometimes results in a loss of mutualistic traits. A derived clade of Epicephala has completely lost the pollinating behaviour after colonizing herbaceous species of Phyllanthus. These plants regularly attain full seed set through ant pollination (table S8 in the electronic supplementary material); thus, time and energetic costs required during pollination probably outweighed the benefit of assuring seed set in these moth lineages. At the same time, effective pollination by ants probably swamped the mutualistic effect of pollination by moths; thus, selection did not favour these Phyllanthus to specialize to moth pollination.
Taken together, this study provides two general implications for the coevolutionary dynamics of mutualisms. First, although species associations are phylogenetically conserved in most coevolving interactions (Thompson 2005), rare shifts by a partner possessing the mutualistic trait can give rise to new mutualisms in phylogenetically distant partner lineages. In this sense, the active pollination behaviour in Epicephala has been of critical importance for the establishment and maintenance of the Phyllantheae–Epicephala mutualism and thus represents a key innovation in this association. Second, the outcome of a species interaction can vary greatly depending on the community context in which it occurs (Thompson & Pellmyr 1992; Thompson & Cunningham 2002; Westerbergh 2004); thus, transitions between mutualism and antagonism can occur repeatedly within a single phylogenetic lineage. This parallels recent findings in other mutualisms where derived parasitic taxa are nested within ancestrally mutualistic clades (Pellmyr et al. 1996b; Machado et al. 2001; Als et al. 2004). Of particular relevance to future studies is our finding that the mutualism arose independently in several Phyllantheae lineages, which provides outstanding opportunities for comparative analyses of character evolution, diversification rates and factors affecting mutualism stability. Thus, the Phyllantheae–Epicephala association is a promising new model system for studies of mutualism and the coevolutionary process.
Acknowledgments
We thank Y. Kosaka, S. Takeda, Y. Masuhara, S. Gnophanxay, H. Chanthavong, T. Phimminith, D. Thongphanh, K. Phengchanthamaly, N. Murakami, T. Y. Chiang, T. W. Hsu, M. E. Rahelivololona, M. R. H. Vololona, P. Robert, J. S. Ascher, T. Sota, Y. Takami and A. Takimura for field assistance; T. Okamoto and R. Goto for specimens; Y. Kameda for assistance with data analysis and figure preparation; P. Hoffmann and the late G. L. Webster for communication on Phyllantheae classification; and Y. Okuyama for manuscript comments. This work was supported by grant-in-aid from the Japan Ministry of Education, Culture, Sports, Science and Technology (to M.K.) and by the Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to A.K.).
Supplementary Material
References
- Als T.D., Vila R., Kandul N.P., Nash D.R., Yen S.-H., Hsu Y.-F., Mignault A.A., Boomsma J.J., Pierce N.E. The evolution of alternative parasitic life histories in large blue butterflies. Nature. 2004;432:386–390. doi: 10.1038/nature03020. doi:10.1038/nature03020 [DOI] [PubMed] [Google Scholar]
- Antal J.S., Prasad M. Some more leaf-impressions from the Himalayan foot-hills of Darjeeling District, West Bengal, India. Palaeobotanist. 1996;43:1–9. [Google Scholar]
- Beattie A.J., Turnbull C.L., Knox R.B., Williams E.G. Ant inhibition of pollen function: a possible reason why ant pollination is rare. Am. J. Bot. 1984;71:421–426. doi:10.2307/2443499 [Google Scholar]
- Bogler D.J., Neff J.L., Simpson B.B. Multiple origins of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1995;92:6864–6867. doi: 10.1073/pnas.92.15.6864. doi:10.1073/pnas.92.15.6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. doi:10.1073/pnas.91.14.6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower A.V.Z., DeSalle R. Patterns of mitochondrial versus nuclear DNA sequence divergence among nymphalid butterflies: the utility of wingless as a source of characters for phylogenetic inference. Insect Mol. Biol. 1998;7:73–82. doi: 10.1046/j.1365-2583.1998.71052.x. doi:10.1046/j.1365-2583.1998.71052.x [DOI] [PubMed] [Google Scholar]
- Cook J.M., Rasplus J.Y. Mutualists with attitude: coevolving fig wasps and figs. Trends Ecol. Evol. 2003;18:241–248. doi:10.1016/S0169-5347(03)00062-4 [Google Scholar]
- Davis C.C., Webb C.O., Wurdack K.J., Jaramillo C.A., Donoghue M.J. Explosive radiation of Malpighiales supports a Mid-Cretaceous origin of modern tropical rain forests. Am. Nat. 2005;165:E36–E65. doi: 10.1086/428296. doi:10.1086/428296 [DOI] [PubMed] [Google Scholar]
- Douzery E.J.P., Snell E.A., Bapteste E., Delsuc F., Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Sci. USA. 2004;101:15 386–15 391. doi: 10.1073/pnas.0403984101. doi:10.1073/pnas.0403984101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell B.D. Evolutionary assembly of the milkweed fauna: cytochrome oxidase I and the age of Tetraopes beetles. Mol. Phylogenet. Evol. 2001;18:467–478. doi: 10.1006/mpev.2000.0888. doi:10.1006/mpev.2000.0888 [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1993 Phylip (phylogeny inference package), v. 3.6. Seattle, WA: Department of Genetics, University of Washington.
- Fleming T.H., Holland J.N. The evolution of obligate pollination mutualisms: senita cactus and senita moth. Oecologia. 1998;114:368–375. doi: 10.1007/s004420050459. doi:10.1007/s004420050459 [DOI] [PubMed] [Google Scholar]
- Gaunt M.W., Miles M.A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographical landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Gruas-Cavagnetto C., Köhler E. Pollens fossiles d'Euphorbiacées de l'Eocène français. Grana. 1992;31:291–304. [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Heraty J., Hawks D., Kostecki J.S., Carmichael A. Phylogeny and behaviour of the Gollumiellinae, a new subfamily of the ant-parasitic Eucharitidae (Hymenoptera: Chalcidoidea) Syst. Entomol. 2004;29:544–559. doi:10.1111/j.0307-6970.2004.00267.x [Google Scholar]
- Ho S.Y.W., Phillips M.J., Cooper A., Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. doi:10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Hoffmann P., Kathriarachchi H., Wurdack K.J. A phylogenetic classification of Phyllantheae (Malpighiales; Euphorbiaceae sensu lato) Kew Bull. 2006;61:37–53. [Google Scholar]
- Kandul N.P., Lukhtanov V.A., Dantchenko A.V., Coleman J.W.S., Sekercioglu C.H., Haig D., Pierce N.E. Phylogeny of Agrodiaetus Hüber 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences of COI and COII and nuclear sequences of EF1-α: karyotype diversification and species radiation. Syst. Biol. 2004;53:278–298. doi: 10.1080/10635150490423692. doi:10.1080/10635150490423692 [DOI] [PubMed] [Google Scholar]
- Kathriarachchi H., Hoffmann P., Samuel R., Wurdack K.J., Chase M.W. Molecular phylogenetics of Phyllanthaceae inferred from five genes (plastid atpB, matK, 3′ndhF, rbcL, and nuclear PHYC) Mol. Phylogenet. Evol. 2005;36:112–134. doi: 10.1016/j.ympev.2004.12.002. doi:10.1016/j.ympev.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Kathriarachchi H., Samuel R., Hoffmann P., Mlinarec J., Wurdack K.J., Ralimanana H., Stuessy T.F., Chase M.W. Phylogenetics of the tribe Phyllantheae (Phyllanthaceae; Euphorbiaceae sensu lato) based on nrITS and plastid matK DNA sequence data. Am. J. Bot. 2006;93:637–655. doi: 10.3732/ajb.93.4.637. doi:10.3732/ajb.93.4.637 [DOI] [PubMed] [Google Scholar]
- Kato M., Takimura A., Kawakita A. An obligate pollination mutualism and reciprocal diversification in the tree genus Glochidion (Euphorbiaceae) Proc. Natl Acad. Sci. USA. 2003;100:5264–5267. doi: 10.1073/pnas.0837153100. doi:10.1073/pnas.0837153100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A., Kato M. Evolution of obligate pollination mutualism in New Caledonian Phyllanthus (Euphorbiaceae) Am. J. Bot. 2004a;91:410–415. doi: 10.3732/ajb.91.3.410. doi:10.3732/ajb.91.3.410 [DOI] [PubMed] [Google Scholar]
- Kawakita A., Kato M. Obligate pollination mutualism in Breynia (Phyllanthaceae): further documentation of pollination mutualism involving Epicephala moths (Gracillariidae) Am. J. Bot. 2004b;91:1319–1325. doi: 10.3732/ajb.91.9.1319. doi:10.3732/ajb.91.9.1319 [DOI] [PubMed] [Google Scholar]
- Kawakita A., Kato M. Assessment of the diversity and species specificity of the mutualistic association between Epicephala moths and Glochidion trees. Mol. Ecol. 2006;15:3567–3581. doi: 10.1111/j.1365-294X.2006.03037.x. doi:10.1111/j.1365-294X.2006.03037.x [DOI] [PubMed] [Google Scholar]
- Kawakita A., Takimura A., Terachi T., Sota T., Kato M. Cospeciation analysis of an obligate pollination mutualism: have Glochidion trees (Euphorbiaceae) and pollinating Epicephala moths (Gracillariidae) diversified in parallel? Evolution. 2004;58:2201–2214. doi: 10.1111/j.0014-3820.2004.tb01598.x. doi:10.1554/04-187 [DOI] [PubMed] [Google Scholar]
- Legendre P., Desdevises Y., Bazin E. A statistical test for host–parasite coevolution. Syst. Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. doi:10.1080/10635150252899734 [DOI] [PubMed] [Google Scholar]
- Lopez-Vaamonde C., Wikström N., Labandeira C., Godfray H.C.J., Goodman S.J., Cook J.M. Fossil-calibrated molecular phylogenies reveal that leaf-mining moths radiated millions of years after their host plants. J. Evol. Biol. 2006;19:1314–1326. doi: 10.1111/j.1420-9101.2005.01070.x. doi:10.1111/j.1420-9101.2005.01070.x [DOI] [PubMed] [Google Scholar]
- Machado C.A., Jousselin E., Kjellberg F., Compton S.G., Herr E.A. Phylogenetic relationships, historical biogeography, and character evolution of fig-pollinating wasps. Proc. R. Soc. B. 2001;268:685–694. doi: 10.1098/rspb.2000.1418. doi:10.1098/rspb.2000.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Crane P.R., Herendeen P.S. Phylogenetic pattern, diversity, and diversification of eudicots. Ann. MO Bot. Gard. 1999;86:297–372. doi:10.2307/2666180 [Google Scholar]
- Morrison D.A. Increasing the efficiency of searches for the maximum likelihood tree in a phylogenetic analysis of up to 150 nucleotide sequences. Syst. Biol. 2007;56:988–1010. doi: 10.1080/10635150701779808. doi:10.1080/10635150701779808 [DOI] [PubMed] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 1999;48:612–622. doi:10.1080/106351599260184 [Google Scholar]
- Pagel M., Meade A., Baker D. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. doi:10.1080/10635150490522232 [DOI] [PubMed] [Google Scholar]
- Pellmyr O. The cost of mutualism: interactions between Trollius europaeus and its pollinating parasites. Oecologia. 1989;78:53–59. doi: 10.1007/BF00377197. doi:10.1007/BF00377197 [DOI] [PubMed] [Google Scholar]
- Pellmyr O. Yuccas, yucca moths, and coevolution: a review. Ann. MO Bot. Gard. 2003;90:35–55. doi:10.2307/3298524 [Google Scholar]
- Pellmyr O., Leebens-Mack J. Forty million years of mutualism: evidence for Eocene origin of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. doi:10.1073/pnas.96.16.9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O., Thompson J.N., Brown J.M., Harrison R.G. Evolution of pollination and mutualism in the yucca moth lineage. Am. Nat. 1996a;148:827–847. doi:10.1086/285958 [Google Scholar]
- Pellmyr O., Leebens-Mack J., Huth C.J. Non-mutualistic yucca moths and their evolutionary consequences. Nature. 1996b;380:155–156. doi: 10.1038/380155a0. doi:10.1038/380155a0 [DOI] [PubMed] [Google Scholar]
- Pellmyr O., Segraves K.A., Althoff D.M., Balcázar-Lara M., Leebens-Mack J. The phylogeny of yuccas. Mol. Phylogenet. Evol. 2007;43:493–501. doi: 10.1016/j.ympev.2006.12.015. doi:10.1016/j.ympev.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Prasad M. Siwalik (Middle Miocene) leaf impressions from the foothills of the Himalayas, India. Tertiary Res. 1994;15:53–90. [Google Scholar]
- Quek S.P., Davies S.J., Itino T., Pierce N.E. Codiversification in an ant–plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae) Evolution. 2004;58:554–570. doi:10.1554/03-361 [PubMed] [Google Scholar]
- Quek S.P., Davies S.J., Ashton P.S., Itino T., Pierce N.E. The geography of diversification in mutualistic ants: a gene's-eye view into the Neogene history of Sundaland rain forests. Mol. Ecol. 2007;16:2045–2062. doi: 10.1111/j.1365-294X.2007.03294.x. doi:10.1111/j.1365-294X.2007.03294.x [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rønsted N., Weiblen G.D., Cook J.M., Salamin N., Machado C.A., Savolainen V. 60 million years of co-divergence in the fig–wasp symbiosis. Proc. R. Soc. B. 2005;272:2593–2599. doi: 10.1098/rspb.2005.3249. doi:10.1098/rspb.2005.3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;14:1218–1231. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. doi:10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. Consel: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. doi:10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Smith C.I., Pellmyr O., Althoff D.M., Balcazar-Lara M., Leebens-Mack J.H., Segraves K.A. Pattern and timing of diversification in Yucca (Agavaceae): specialized pollination does not escalate rates of diversification. Proc. R. Soc. B. 2008;275:249–258. doi: 10.1098/rspb.2007.1405. doi:10.1098/rspb.2007.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota T., Hayashi M. Comparative historical biogeography of Plateumaris leaf beetles (Coleoptera: Chrysomelidae) in Japan: interplay between fossil and molecular data. J. Biogeogr. 2007;34:977–993. doi:10.1111/j.1365-2699.2006.01672.x [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. Sunderland, MA: Sinauer Associates.
- Thompson J.N. The University of Chicago Press; Chicago, IL: 2005. The geographic mosaic of coevolution. [Google Scholar]
- Thompson J.N., Cunningham B.M. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. doi: 10.1038/nature00810. doi:10.1038/nature00810 [DOI] [PubMed] [Google Scholar]
- Thompson J.N., Pellmyr O. Mutualism with pollinating seed parasites amid co-pollinators: constraints on specialization. Ecology. 1992;73:1780–1791. doi:10.2307/1940029 [Google Scholar]
- Thorne J.L., Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. doi:10.1080/10635150290102456 [DOI] [PubMed] [Google Scholar]
- Ueda S., Quek S.P., Itioka T., Inamori K., Sato Y., Murase K., Itino T. An ancient tripartite symbiosis of plants, ants and scale insects. Proc. R. Soc. B. 2008;275:2319–2326. doi: 10.1098/rspb.2008.0573. doi:10.1098/rspb.2008.0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster G.L. A monographic study of the West Indian species of Phyllanthus. J. Arnold Arbor. 1958;39:49–100. (See also pp. 111–212.) [Google Scholar]
- Weiblen G.D. How to be a fig wasp. Annu. Rev. Entomol. 2002;47:299–330. doi: 10.1146/annurev.ento.47.091201.145213. doi:10.1146/annurev.ento.47.091201.145213 [DOI] [PubMed] [Google Scholar]
- Westerbergh A. An interaction between a specialized seed predator moth and its dioecious host plant shifting from parasitism to mutualism. Oikos. 2004;105:564–574. doi:10.1111/j.0030-1299.2004.12820.x [Google Scholar]
- Wiens J.J. Missing data and the design of phylogenetic analyses. J. Biomed. Inf. 2006;39:34–42. doi: 10.1016/j.jbi.2005.04.001. doi:10.1016/j.jbi.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. CABIOS. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.