Abstract
The Asellota are a highly variable group of Isopoda with many species in freshwater and marine shallow-water environments. However, in the deep sea, they show their most impressive radiation with a broad range of astonishing morphological adaptations and bizarre body forms. Nevertheless, the evolution and phylogeny of the deep-sea Asellota are poorly known because of difficulties in scoring morphological characters. In this study, the molecular phylogeny of the Asellota is evaluated for 15 marine shallow-water species and 101 deep-sea species, using complete 18S and partial 28S rDNA gene sequences. Our molecular data support the monophyly of most deep-sea families and give evidence for a multiple colonization of the deep sea by at least four major lineages of asellote isopods. According to our molecular data, one of these lineages indicates an impressive radiation in the deep sea. Furthermore, the present study rejects the monophyly of the family Janiridae, a group of plesiomorphic shallow-water Asellota, and several shallow-water and deep-sea genera (Acanthaspidia, Ianthopsis, Haploniscus, Echinozone, Eurycope, Munnopsurus and Syneurycope).
Keywords: 18S rDNA, 28S rDNA, Southern Ocean, molecular phylogeny, multiple colonization
1. Introduction
Covering nearly two-thirds of the Earth's surface, the deep sea is by far the most extensive and enigmatic ecosystem on our planet (Rex 1981; Gage & Tyler 1991). For a long time, it was widely assumed that low temperatures, extreme hydrostatic pressures, the absence of light and nutrient-poor sediments created an environment inhabited by only a limited number of species (see Gage & Tyler 1991). Today, we know that deep-sea regions harbour complex communities with impressively high numbers of species, showing remarkable morphological and physiological adaptations (e.g. Gage & Tyler 1991; Herring 2002). In particular, the deep-sea soft-sediment environment hosts a diverse but highly endemic benthic fauna of, in most cases, unknown origin and age (Hessler & Sanders 1967; Menzies et al. 1973; Brandt et al. 2007).
Owing to the fact that morphological characters in most deep-sea species are highly derived and no fossils exist, the evolutionary history of many elements of the bathyal and abyssal fauna can be inferred with the aid of molecular phylogenetics. However, molecular studies analysing the evolution of deep-sea organisms are still rare, because sampling this remote habitat and preserving of individuals, especially for molecular studies, is very difficult and time-consuming. Hence, the number of specimens available is limited in most groups (Gage & Tyler 1991; Creasey & Rogers 1999). To date, the majority of molecular phylogenetic works on deep-sea organisms focus on phylogeographic patterns or population genetics of single species (e.g. Creasey & Rogers 1999; Etter et al. 2005; Zardus et al. 2006; Raupach et al. 2007), and only a few studies analyse deep-sea colonization patterns and events of more than one species (Shank et al. 1999; Le Goff-Vitry et al. 2004; Raupach et al. 2004; Knudsen et al. 2007).
In most abyssal benthic samples, asellote isopods represent the dominating crustacean taxon and often account for a large fraction of all species present in an area (Sanders et al. 1963; Sanders & Hessler 1969; Brandt et al. 2007). These isopods are mostly very small benthic or epibenthic blind animals with a body size of a few millimetres, which are difficult to identify at the species level. As with all Peracarida, they have no planktonic larvae, which probably limits gene flow and increases the probability of speciation events (Raupach & Wägele 2006; Raupach et al. 2007; Brökeland & Raupach 2008). Deep-sea Asellota show an amazing variety of bizarre and extravagant body forms, including elongated stick-like animals (Ischnomesidae), burrowing dozer-like forms (e.g. Desmosomatidae, Haploniscidae, Janirellidae), spinose species (e.g. Acanthaspidiidae, Mesosignidae) and even some secondarily highly modified swimmers (Eurycope) within the Munnopsidae (Hessler & Strömberg 1989; Hessler 1993; Marshall & Diebel 1995; fig. e3 in the electronic supplementary material). All these bizarre morphological variations among families and the high species diversity of the deep-sea Asellota are probably a result of different evolutionary adaptations (Wägele 1989). Most families and many genera appear to be cosmopolitan in the deep sea, and few species are found in shallow waters under special environmental conditions (e.g. low water temperatures at high latitudes; Hessler & Wilson 1983; Svavarsson et al. 1993). Interestingly, first molecular analyses provided evidence for the existence of complex species flocks within widespread cosmopolitan species (Raupach & Wägele 2006; Raupach et al. 2007), supporting the patchwork theory of closely related but distinct populations in the deep sea (Raupach et al. 2007).
Although deep-sea Asellota are ecologically important in all oceans and are highly interesting from zoogeographic and phylogeographic points of view (Hessler & Wilson 1983; Wägele 1989; Gage & Tyler 1991), many taxonomic problems still exist. Currently, 15 families with more than 950 species of typical deep-sea Asellota are recognized, and some genera are still incertae sedis (Schotte et al. 1995). As a consequence, we have limited knowledge about their colonization of the deep sea. In the absence of a clear fossil record of the deep-sea fauna (except from Foraminifera and Ostracoda), two alternative theories of the origin of the deep-sea Asellota have been suggested: (i) a multiple migration from shallow waters, or (ii) a single origin of all typical deep-sea families, which therefore constitute an ancient deep-sea component. The first theory is based on the hypothesis that the asellote deep-sea fauna was populated mainly through emigration from ‘centres of origin in shallow waters’ (submergence), especially at high latitudes (Wolff 1960; Kussakin 1973; Menzies et al. 1973), where the temperature difference between shallow and deep waters, a potential physiological barrier for many taxa, is small (Gage & Tyler 1991; Childress 1995). For example, most shallow-water species of the family Acanthaspidiidae are widely distributed on the Antarctic shelf, but various related species can be found in the deep sea of the Southern Ocean, a distribution that could have arisen by a southern diversification after multiple polar submergence events (Brandt 1991a; Just 2001). By contrast, other authors (Hessler & Thistle 1975; Hessler et al. 1979; Wägele 1989; Wilson 1999) argue that cold shallow waters should have been colonized from the deep sea (high-latitude emergence) for two main reasons. First, for many species, the centre of generic and species diversity lies in the deep sea. Second, the absence of eyes is a typical feature of deep-sea asellote species, a fact which is also observed for confamilial species living in shallow waters. Since eyes have presumably been lost in the deep sea due to the lack of selective advantage in a dark environment, blind species in shallow water are believed to have emerged from the deep sea.
Phylogenetic studies of the marine Asellota should represent an efficient method to analyse their deep-sea colonization. To date, extensive phylogenetic studies of the deep-sea Asellota are very rare. Various studies (e.g. Kussakin 1973; Hessler et al. 1979; Schultz 1979) were not based on the criteria of Hennig's theory of phylogenetic systematics (Hennig 1950), and most recent morphological works were focused on single selected families or genera (genus Ilyarachna (Munnopsidae): Hessler & Thistle 1975; genus Eurycope (Munnopsidae): Wilson 1982; Munnopsidae: Wilson 1989; Acanthaspidiidae, the munnopsid genera Storthyngura, Acanthocope and Microprotus: Brandt 1991b). Only one work (Wägele 1989) analysed the phylogenetic relationship of all deep-sea taxa of the Asellota, but this study was not able to resolve the details of their phylogeny caused by the high variability of most morphological characters. A first molecular study of the deep-sea Asellota using the complete 18S rRNA gene gave evidence for a multiple colonization of the deep sea (Raupach et al. 2004), but this study was limited to five shallow-water taxa and only 33 deep-sea taxa, and for many families only one species was analysed.
In this study, we provide a comprehensive molecular phylogeny of 116 marine Asellota, with 15 eye-bearing shallow-water species and 101 blind deep-sea species, to infer their colonization of the deep sea. We used the complete 18S rDNA, providing 58 new sequences, and, for the first time for the Asellota, a partial 28S rDNA sequence fragment of 80 different species. Specimens were collected from depths of 0–4900 m and include species from 11 out of 15 globally known families of deep-sea asellote isopods.
2. Material and methods
(a) Specimens, sampling and DNA isolation
Deep-sea Asellota were collected using fishing gear (epibenthic sledge, Agassiz trawl, Rauschert dredge and bottom trawl) employed by RV Polarstern during the expeditions ANT XIX/3 and ANT XIX/4 in 2002 (ANDEEP I and II; ANtarctic benthic DEEP-sea biodiversity: colonization history and recent community patterns), ANT XXI/2 in 2003–2004 and ANT XXII/3 (ANDEEP III) in the Southern Ocean, at depths ranging from 300 to 4900 m (see table eI in the electronic supplementary material). More details and station coordinates are given in the corresponding cruise reports of RV Polarstern (Fütterer et al. 2003; Arntz & Brey 2005; Fahrbach 2006). One species, the shallow-water janirid Iais pubescens, was collected at the shore of Ushuaia, Argentina (kindly provided by Christoph Held, AWI). Since isopods possess highly active nucleases (Dreyer & Wägele 2001, 2002), the samples were stored directly after sampling for at least 36 hours at 0°C in 96 per cent ethanol to prevent DNA digestion. Total genomic DNA was extracted onboard RV Polarstern from dissected legs of specimens using the QIAmp tissue kit (Qiagen GmbH), following the manufacturer's extraction protocol. The specimens are deposited in the collection of the Zoologisches Institut und Museum, Hamburg, Germany. The tissue samples and DNA extracts are deposited in the newly established DNA bank of the Zoologisches Forschungsmuseum Alexander Koenig, Bonn, as part of the DNA bank network (www.dnabank-network.org).
(b) Polymerase chain reaction amplification and DNA sequencing
The complete 18S and partial 28S rDNA sequences (fragments D6–D8) were amplified by polymerase chain reaction (PCR). The primers used in amplification and sequencing for both genes are provided in table eII in the electronic supplementary material. Amplification reactions were carried out on a Progene thermocycler (Techne Ltd) in a 40 μl volume, containing 4 μl 10× Roche PCR buffer, 4 μl dNTPs (2 mmol μl−1), 0.3 μl of each primer (both 100 pmol μl−1), 2–3 μl of DNA template and 0.2 μl Roche Taq (5 U μl−1), and filled up to 40 μl with sterile H2O. The PCR temperature profile for the 28S rDNA fragment consisted of an initial denaturation at 94°C (5 min), followed by 40 cycles at 94°C (denaturation, 1 min), 55°C (annealing, 1 min), 72°C (extension, 90 s) and a final extension at 72°C (6 min). The temperature profile for the complete 18S ribosomal gene amplification follows previous studies (Dreyer & Wägele 2001). Negative and positive controls were included with each round of reactions. Three microlitres of the amplified product were verified for size conformity by electrophoresis in a 1 per cent agarose gel with ethidium bromide using commercial DNA size standards, while the remaining PCR product was purified with the QIAquick PCR purification kit (Qiagen GmbH). Purified PCR products were cycle sequenced and sequenced on a LICOR 4200 or at a contract sequencing facility (Macrogen, Seoul, South Korea) on an ABI3730 XL automatic DNA sequencer. Double-stranded sequences were assembled with the SeqMan II program (DNASTAR, Inc.), and BLAST searches were performed to confirm the identity of all new sequences (Altschul et al. 1997). All new DNA sequences have been deposited in GenBank (see table eI in the electronic supplementary material).
(c) Sequence assembly, alignment and phylogenetic analyses
Datasets were aligned using Muscle v. 3.6 (Edgar 2004) with default settings. Secondary structure information of both rRNAs (Ali et al. 1999; Wuyts et al. 2000) was used to correct the alignment and to identify expansion segments. The two alignments consisted of 2943 bp (18S rDNA) and 2140 bp (28S rDNA). Hypervariable regions in expansion segments within ribosomal RNA genes are a well-known problem in molecular phylogenetics, but sequence differences in these regions are particularly high in Isopoda (Choe et al. 1999; Dreyer & Wägele 2001), with the consequence that most of these sites cannot be aligned unambiguously across all taxa. Therefore, highly variable expansion sections have been excluded from our analyses. Alignment positions excluded in the 18S rDNA are 802–1283 and 2083–2452, and positions excluded in the 28S rDNA are 268–606, 708–769, 1023–1060, 1274–1555 and 1639–1863. The final alignments consisted of 2086 bp (18S rDNA) and 1199 bp (28S rDNA) with, respectively, 543 and 385 parsimony-informative positions.
Previous molecular and morphological data had confirmed the Stenetriidae as a ‘primitive’ marine Asellota (Hessler et al. 1979; Wägele 1989; Raupach et al. 2004). Therefore, the three stenetriid species, Stenetrium sp., Tenupedunculus sp. and Tenupedunculus acutum, were designated as outgroup taxa in our studies. Prior to the phylogenetic analysis, we tested for homogeneity of base frequencies among taxa using the Χ2-test implemented in PAUP* v. 4.0b10 (Swofford 2002) and a tetrahedron model analysis using SeqVis (Ho et al. 2005). A partition homogeneity test was performed using PAUP* v. 4.0b10 with 1000 replicates to test whether the individual maximum-parsimony (MP) trees of the 18S and 28S rDNA datasets contain significantly different phylogenetic signals in comparison with the MP tree of the combined dataset (Farris et al. 1995).
MP trees of the combined datasets were obtained first by using PAUP* v. 4.0b10 to conduct a single long search, and, second, by using the ‘ratchet method’ implemented in PRAP (Müller 2004), which uses PAUP* v. 4.0b10 to perform a ratchet search with 1000 iterations. The results have been verified in a third analysis by computing 100 random addition sequence replicates, for each of which a time-limited heuristic search was carried out such that approximately 8 000 000 000 tree bisection and reconnection branch-swapping rounds were performed for each replicate. The consistency, retention and rescaled consistency indices (CI, RI and RC) were calculated using PAUP* v. 4.0b10 for the most parsimonious topologies to estimate the levels of homoplasy (Kluge & Farris 1969; Farris 1989a,b). Bootstrap support values were obtained by resampling and analysing 1000 replicates. Decay indices (Bremer 1988) were calculated using PAUP* v. 4.0b10 and PRAP with 200 ratchet iterations in each decay search.
Bayesian analyses were conducted using MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003). Appropriate DNA substitution models were determined separately for the 18S rDNA and 28S rDNA datasets using the Akaike information criterion (Akaike 1974) implemented in Modeltest v. 3.7 (Posada & Crandall 1998). In the Bayesian analysis, the combined dataset (18S+28S rDNA) was partitioned (see MrBayes manual for details) to allow unlinked models and model parameters for the two genes. Following the manual of MrBayes, the most appropriate substitution model was specified without fixing the parameter values, allowing them to vary during the annealing process, which leads to more conservative but more realistic posterior probabilities. The Bayesian analysis was conducted by computing 20 000 000 Markov chain Monte Carlo generations in two parallel runs, each with three cold chains and one hot chain. The trees were sampled every 100 generations. The number of burn-in generations was determined by manual inspection of the likelihood score over the 20 000 000 generations.
Using RAxML v. 7.0.0 (Stamatakis 2006), a fast maximum-likelihood (ML) bootstrap analysis with 10 000 replicates was performed (see fig. e1 in the electronic supplementary material). The dataset was partitioned to allow different parameters for each gene under GTRMIX parameter settings (see RAxML v. 7.0.0 manual for details).
Finally, SplitsTree (Huson & Bryant 2005) has been employed to calculate neighbour nets of both data partitions. Neighbour nets visualize the degree of support and conflict in the datasets for the branches in the phylogenetic trees (details are given in the electronic supplementary material).
3. Results
For this study, we newly sequenced the complete 18S rDNA of 58 and partial 28S rDNA of 80 species of marine shallow-water and deep-sea Asellota, which were combined with previously published data (see table eI in the electronic supplementary material). The sequences within each of the two alignments showed no significant differences in the base composition (Χ2-test of the 18S rDNA [28S rDNA], with d.f.=342 [342] and p=1.00 [0.99]). The minor deviations in base frequencies are therefore not expected to influence the phylogenetic reconstructions. All alignments can be obtained from M.J.R. upon request.
For the MP analysis, the two datasets have been concatenated. A partition homogeneity test (Farris et al. 1995) showed that the natural partitioning of the combined dataset into 18S and 28S did not yield a phylogenetic signal significantly different from a random partitioning (p=0.93), indicating a high homogeneity in the combined dataset and thereby justifying the combination of both datasets. Tree reconstruction with MP has been conducted first in a single long search for the best tree and, second, by using the ratchet algorithm implemented in PRAP. Both approaches found multiple best trees, with a tree length of 4409 substitutions. The topology of the final MP tree (figure 1) was obtained by taking the strict consensus of 605 936 equally ‘good’ trees found in the single long search, which was identical to the strict consensus of the best trees found using PRAP. For the most-parsimonious trees, we computed the consistency index (CI 0.423), the retention index (RI 0.743) and the rescaled consistency index (RC 0.315). In the case of the Bayesian analysis, Modeltest indicated that the GTR+I+G model was the appropriate nucleotide substitution models for both datasets with the following parameters (18S rDNA [28S rDNA]): nucleotide frequencies (A: 0.26 [0.24], C: 0.22 [0.22], G: 0.24 [0.28], T: 0.28 [0.26]); substitution rates (RAC: 1.03 [1.23], RAG: 2.15 [2.31], RAT: 1.21 [2.44], RCG: 0.43 [0.50], RCT: 3.18 [4.74], RGT: 1.00 [1.00]); gamma distribution shape=0.45 [0.35]; and a proportion of invariable sites=0.45 [0.46]. For reasons described previously, only the model (not the parameters) has been fixed in the Bayesian analysis. Manual inspection of the likelihood scores of the 2×200 001 tree samples in two parallel runs showed a good convergence after 4000 trees, a value that has therefore been chosen as the burn-in. Posterior probabilities have been computed from a majority-rule consensus tree of the remaining 2×196 001 trees (figure 2). The ML phylogram with bootstrap values (more than 50%) appears very similar to the Bayesian topology (see fig. e1 in the electronic supplementary material).
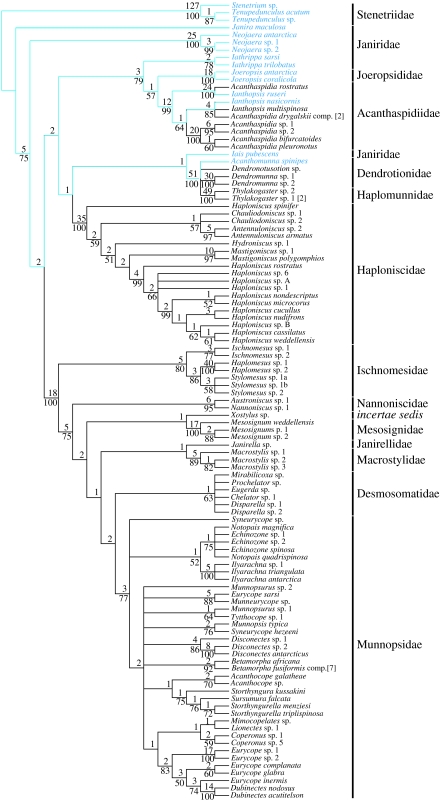
Figure 1.
Strict consensus tree of the most-parsimonious trees, with a tree length of 4409, of the combined dataset (18S and 28S rDNA). The tree is rooted using the Stenetriidae as the outgroup. The numbers above the internal branches are decay indices and those below indicate bootstrap values, which are only given if they are above 50%. Species families are indicated by bold bars. The blue taxon names and branches indicate eye-bearing shallow-water species and their putative ancestral lineages, and the black taxon names and branches represent blind deep-sea species.
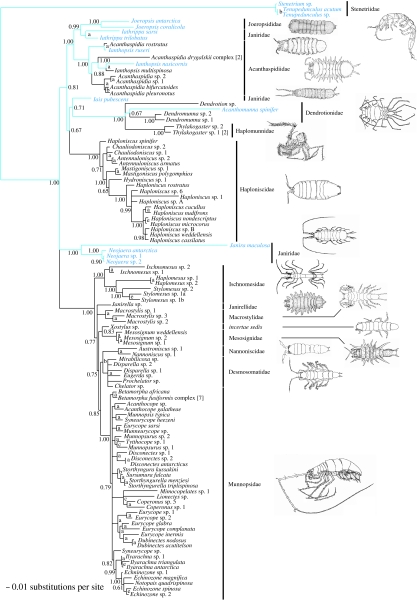
Figure 2.
The 50% majority rule consensus tree from the Bayesian analysis of the combined dataset (18S rDNA and 28S rDNA). The tree is rooted on the Stenetriidae. The numbers represent Bayesian posterior probabilities (a, 1.00–0.95; b, 0.94–0.85; c, 0.84–0.75; d, 0.74–0.65; e, 0.64–0.50). Species families are indicated by bold bars. The blue taxon names and branches indicate eye-bearing shallow-water species and their putative ancestral lineages, and the black taxon names and branches represent blind deep-sea species.
Bayesian inference, ML and MP analyses support the monophyly of most analysed shallow-water and deep-sea families. Posterior probabilities, ML bootstrap values, MP bootstrap values and decay indices support the monophyly of the Joeropsididae (1.00/100/100/18), Acanthaspidiidae (1.00/100/99/12), Haploniscidae (1.00//100/100/35), Ischnomesidae (1.00/100/80/5), Macrostylidae (1.00/91/89/5), Mesosignidae (1.00/100/100/17), Nannoniscidae (1.00/99/95/6), Desmosomatidae (1.00/78/63/1), Munnopsidae (1.00/95/77/3) and a polytomic clade (1.00/100/100/53) including species of the Dendrotionidae and monophyletic Haplomunnidae (1.00/100/100/49). However, all analyses clearly reject the monophyletic nature of the shallow-water Janiridae. Despite the fact that both phylograms show complete correspondence in many clades, some important differences between the MP analyses and Bayesian inference are noteworthy to mention. (i) Most strikingly, in the tree inferred from the MP analyses, the janirid genera Neojaera and Janira represent a paraphyletic sister group to all other analysed Asellota. In contrast to this, Bayesian inference and maximum likelihood support a monophyletic clade (1.00/93), which constitutes one of three major clades in the basal polytomy of the tree. (ii) The topology inferred from the MP analysis gives evidence for a sister group relationship of the Joeropsididae and Acanthaspidiidae (57/1), while the Bayesian approach supports a Joeropsididae–Iathrippa clade with a high posterior probability (0.99). The ML bootstrap analysis shows only moderate support for this clade (77). (iii) Within all analyses, the Ischnomesidae represent the most basal group of the major clade, comprising what we call the munnopsoid radiation consisting of the Desmosomatidae, Janirellidae, Macrostylidae, Mesosignidae, Munnopsidae, Nannoniscidae and the enigmatic genus Xostylus. Within this major clade, maximum parsimony suggests a basal position of the Nannoniscidae (75/5) next to the Ischnomesidae, followed by a branch with Xostylus plus Mesosignidae (−/2), a second branch with the Janirellidae plus Macrostylidae (−/1) and finally a clade with the Desmosomatidae and Munnopsidae (−/2). By contrast, Bayesian inference and maximum likelihood give evidence for a clade containing the Desmosomatidae and Nannoniscidae (0.86/57) as a sister group to the Munnopsidae. As an important result, all analyses agree on splitting various shallow-water and deep-sea genera (Acanthaspidiidae: Acanthaspidia and Ianthopsis; Haploniscidae: Haploniscus; Munnopsidae: Echinozone, Eurycope, Munnopsurus and Syneurycope).
The neighbour net graphs of the 18S and 28S partitions (see figure e2 in the electronic supplementary material) provide insight into which of the two genes is more informative for the different levels of the branching patterns. Both nets show parallel edges for deeper nodes of the asellote phylogeny and most families. In comparison with the 18S partition, many terminal branches of the 28S rDNA partition are shorter, and conflicting signals are stronger. Nevertheless, some deeper nodes and families are clearly resolved. A more detailed discussion of both graphs can be found in the electronic supplementary material.
4. Discussion
The evolutionary origin of the deep-sea fauna has been debated for a long time, and the study of different taxa, mostly based on zoogeographic data, revealed a variety of different colonization patterns. For example, microfossil analyses from both deep-sea and Antarctic shallow-water Foraminifera indicate that elements of both faunas have accumulated by several processes. Whereas most foraminiferal species have evolved in situ, there is evidence that migration has occurred in both directions (with probably more species migrating from the deep sea to shallow-water areas of the Antarctic), giving evidence for a coevolution of both environments (Lipps & Hickmann 1982; Hayward 2001). An analysis of the deep-sea molluscan distribution and taxonomy suggests that this fauna was probably shaped by invasions from adjacent shallow-water regions all over the world (Clarke 1962; Allen 1979). In the case of echinoderm taxa, it has been suggested that bathyal forms are most primitive, while those in the abyssal and hadal zones are more recently evolved (Hansen 1975; Smith 2004).
In this context, our phylogenetic analyses show that (i) the deep sea was initially colonized with asellote isopods by a small number of migration events from the shallow water, and (ii) most typical deep-sea families of the Asellota have evolved and radiated in the deep sea. Our data also document multiple colonization events (polar submergence) within the Acanthaspidiidae. Beside this, the molecular data support the monophyly of most deep-sea asellote families, but clearly reject the monophyly of the less derived shallow-water Janiridae, a ‘collection’ of various taxa sharing several plesiomorphic characters. This result is consistent with previous morphological and molecular studies (Wägele 1989; Wilson 1994; Raupach et al. 2004). All phylogenetic trees divided the blind deep-sea Asellota into four well-supported monophyletic clades, confirming four major and independent migrations from the shallow water into the deep sea: the Haploniscidae (1.00/100/100/35); the Dendrotionidae plus Haplomunnidae (1.00/100/100/51); a multiple colonization within the Acanthaspidiidae (1.00/100/99/12); and a highly derived monophylum that we call the munnopsid radiation (1.00/95/100/18). The munnopsoid radiation includes the Desmosomatidae, Janirellidae, Macrostylidae, Mesosignidae, Munnopsidae, Nannoniscidae and the genus Xostylus, which together represent a large clade with more than 750 known species in total (see Schotte et al. 1995). Furthermore, our study indicates basal polytomies within the Munnopsidae and Desmosomatidae in contrast to other deep-sea asellote families such as the Haploniscidae or Ischnomesidae. The lack of the phylogenetic signal in these polytomies might indicate a rapid radiation of these highly derived families within the deep sea, which is also supported by morphological studies (e.g. Hessler 1970; Wägele 1989).
Even though the MP, ML and Bayesian phylogenetic trees were almost identical, there are two notable differences, namely (i) the position of the shallow-water janirid species Janira and Neojaera, and (ii) the position of the Nannoniscidae. While the MP analyses support a basal, paraphyletic position of Neojaera and Janira, Bayesian inference and maximum likelihood clearly give evidence for a monophyletic clade including both genera (1.00/93). The reason for this difference could be that, in the MP analyses, the long branch of Janira maculosa is attracted by the long branch of the outgroup (see Bergsten 2005; Wägele & Mayer 2007), a well-known weakness of MP analyses. In the case of the Nannoniscidae, maximum parsimony favours a more basal position of this group within the munnopsid radiation when compared with the Bayesian phylogeny. A plausible reason for this apparent disagreement is the lack of clear phylogenetic signal for either of the two alternatives in the dataset. While the MP analysis provides no bootstrap support for the observed difference and merely a weak support based on low decay indices, Bayesian and ML analyses support a clade containing the Desmosomatidae and Nannoniscidae as a sister group to the Munnopsidae with a moderate posterior probability (0.86) and a low bootstrap value (less than 50%). Despite the fact that the interpretation of posterior probabilities is still a subject of discussion (e.g. Suzuki et al. 2002; Wilcox et al. 2002; Alfaro et al. 2003; Cummings et al. 2003), a close relationship of the Nannoniscidae, Desmosomatidae and Munnopsidae is also strongly suggested by morphological studies (Hessler 1970; Wägele 1989; Brandt 1999), making the topology of the Bayesian analysis more plausible.
Our molecular dataset also identified seven non-monophyletic genera (Ianthopsis, Acanthaspidia (both Acanthaspidiidae); Haploniscus (Haploniscidae); Echinozone, Eurycope, Munnopsurus and Syneurycope (all Munnopsidae)). This underpins well-known taxonomic problems, especially within the Acanthaspidiidae and the hyperdiverse Munnopsidae, which are caused by many insufficient species descriptions and illustrations in the past. Nevertheless, the molecular data support recent concepts of taxonomic classifications of the munnopsid genera Storthyngura, Storthyngurella and Sursumura into the subfamily Storthyngurinae, and the classification of Dubinectes into the Eurycopinae (Malyutina 2003; Malyutina & Brandt 2006). The Acanthaspidiidae are a good example where morphological characters failed to provide sufficient phylogenetic information for a reliable tree reconstruction (see Just 2001). The distribution of the Acanthaspidiidae is concentrated in the Southern Hemisphere, specifically in the Southern Ocean, and includes both eye-bearing shallow-water and blind deep-sea genera (Brandt 1991a,b; Just 2001). The taxonomic status of this family was unclear, because no valuable synapomorphies define either the family or the genera (Wägele 1989; Brandt 1991a,b; Just 2001). By contrast, all our analyses clearly support the monophyly of the Acanthaspidiidae (1.00/100/99/12) and indicate a close relationship with other shallow-water Asellota (Joeropsididae, Iathrippa). Furthermore, the molecular data reject the monophyly of the two genera Acanthaspidia and Ianthopsis, indicating an independent and multiple colonization of the deep sea even within the Acanthaspidiidae. Our study also reveals insight into the relationship of the Dendrotionidae and Haplomunnidae. Morphological studies suggest a basal position of the shallow-water genus Acanthomunna within the more derived Dendrotionidae including the genera Dendromunna and Dendrotion (Wilson 1976), based on plesiomorphic morphological traits (e.g. retention of eyes, occurrence in shallow bathyal and deep waters). Molecular phylogenetics support a sister clade relationship between Acanthomunna and the morphologically more derived genus Dendromunna, but exclude the genus Dendrotion. As a consequence, our data support a scenario of a multiple colonization of the deep sea within the Dendrotionidae and the Acanthaspidiidae.
Owing to the lack of fossils and suitable molecular clock models, a dating of the asellote radiation and colonization events is not possible. Drastic climatic changes during the Palaeozoic–Mesozoic boundary (Isozaki 1997; Berner 2002; Benton & Twitchett 2003), the early phylogenetic origin of the Asellota within the Isopoda (Wägele 1989; Brusca & Wilson 1991; Dreyer & Wägele 2002) and the zoogeographic pattern of the fresh-water Asellota (which represent the sister group of the marine Asellota; see Wägele 1989; Raupach et al. 2004) across parts of Laurasia and Africa, support the hypothesis that marine Asellota colonized the deep sea prior to the dysoxia events during the end-Permian extinction event (ca 250 Myr ago; Wägele et al. 1995; Wilson 1998, 1999). Therefore, the Mesozoic deep sea is unlikely to have become completely anoxic, owing to vertical halothermal circulations at low altitudes allowing organisms to escape into deeper waters. As a result, elements of the Palaeozoic fauna may have persisted through the Mesozoic without being present in marine shallow waters (Wägele et al. 1995; Wilson 1998, 1999).
In summary, our findings from the complete 18S and partial 28S rRNA gene sequences are consistent with at least four major colonization events of the deep sea. Beside this, our study gives strong support to the current taxonomy of shallow-water and deep-sea Asellota at the familial level, with one exception: the shallow-water taxon Janiridae. Nevertheless, major problems in the taxonomic classification of diverse asellote genera were found. Therefore, extensive taxonomic revisions are essential for several genera and families. In future, other molecular markers should be tested to determine whether they are appropriate for increasing the basal resolution within the Asellota. A first step may include the analysis of the complete 28S rDNA. While classic mitochondrial gene fragments (CO1, 12S and 16S rDNA) are useful for phylogenetic/phylogeographic studies of closely related species or genera (Wetzer 2001, 2002; Raupach & Wägele 2006; Raupach et al. 2007), recent analyses of complete mitochondrial sequence data and gene arrangement information of isopods (Kilpert & Podsiadlowski 2006; Podsiadlowski & Bartolomaeus 2006) are highly promising and may be useful for a more detailed reconstruction of basal asellote phylogenetic relationships. We conclude that future studies should consider samples from rare taxa currently missing, such as specimens of the families Echinothambematidae, Katianiridae or Mictosomatidae, in order to confirm the colonization of the deep sea by the Asellota in more detail.
Acknowledgments
We would like to thank Angelika Brandt and Brigitte Ebbe for the professional and excellent organization of all three ANDEEP expeditions in the Southern Ocean. We also want to thank Dieter K. Fütterer for managing the ANT XIX/3–4 expeditions (ANDEEP I and II), Wolf E. Arntz for managing the ANT XXI/2 expedition and Eberhard Fahrbach for managing the ANT XXII/3 (ANDEEP III) expedition. Saskia Brix, Wiebke Brökeland, Madhumita Coudhury and Gabi Strieso kindly sorted the EBS samples and/or identified species. Furthermore, we thank Wiebke Brökeland and Katrin Linse for providing the images of the Asellota. We also thank Patrick Kück for his help in analysing our dataset using RaxML. Finally, we are thankful to the crew of ‘Polarstern’ and all other expedition participants for their professional help, support and advice. Michael Rex and two anonymous referees provided their helpful comments that improved our paper. Our research project was supported by grants of the German Science Foundation (DFG) to J.-W.W. (WA 530/26-1, 530/28-1 and 28-2). This is ANDEEP publication 106.
Supplementary Material
Maximum likelihood topology
Neigbor-net analysis of both 18S and 28S data sets
Details to the neighbor-net structures
Accession numbers and taxonomic classifications
PCR-primer informations
A Mesosignum sp. (Mesosignidae), B Acanthaspidia drygalskii (Acanthaspidiidae), C Ischnomesus sp. (Ischnomesidae) D Janirella sp. (Janirellidae), E Haploniscus sp. (Haploniscidae), F Chelator sp. (Desmosomatidae) and G Eurycope sp. (Munnopsidae)
References
- Akaike, H. 1974 Information theory and an extension of the maximum likelihood principle. In Second International Symposium on Information Theory (eds B. N. Petrov & F. Csaki), pp. 267–281. Budapest, Hungary: Akademiai Kiado.
- Alfaro M.E., Zoller S., Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003;20:255–266. doi: 10.1093/molbev/msg028. doi:10.1093/molbev/msg028 [DOI] [PubMed] [Google Scholar]
- Ali A.B., Wuyts J., De Wachter R., Meyer A., van de Peer Y. Construction of a variability map for eukaryotic large subunit ribosomal RNA. Nucl. Acids Res. 1999;27:2825–2831. doi: 10.1093/nar/27.14.2825. doi:10.1093/nar/27.14.2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.A. The adaptations and radiation of deep-sea bivalves. Sarsia. 1979;64:19–27. [Google Scholar]
- Altschul S.F., Madden T.L., Schäffner A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arntz W.E., Brey T. The expedition ANTARKTIS XXI/2 (BENDEX) of RV “Polarstern” in 2003/2004. Rep. Polar Mar. Res. 2005;503:1–149. [Google Scholar]
- Benton M.J., Twitchett R.J. How to kill (almost) all life: the end-Permian extinction event. Trends Ecol. Evol. 2003;18:358–365. doi:10.1016/S0169-5347(03)00093-4 [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. doi:10.1111/j.1096-0031.2005.00059.x [DOI] [PubMed] [Google Scholar]
- Berner R.A. Examination of hypotheses for the Permo–Triassic boundary extinction by carbon cycle modelling. Proc. Natl Acad. Sci. USA. 2002;99:4172–4177. doi: 10.1073/pnas.032095199. doi:10.1073/pnas.032095199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A. A revision of the Acanthaspidiidae Menzies, 1962 (Isopoda: Asellota) Zool. J. Linn. Soc. 1991a;102:203–252. doi:10.1111/j.1096-3642.1991.tb00001.x [Google Scholar]
- Brandt A. Zur Besiedlungsgeschichte des antarktischen Schelfs am Beispiel der Isopoda (Crustacea, Malacostraca) Ber. Polarforsch. 1991b;98:1–240. [Google Scholar]
- Brandt A. Hypotheses on the Southern Ocean peracarid evolution and radiation (Crustacea, Malacostraca) Ant. Sci. 1999;12:269–275. doi:10.1017/S095410200000033X [Google Scholar]
- Brandt A., et al. First insights into the biodiversity and biogeography of the Southern Ocean deep sea. Nature. 2007;447:307–311. doi: 10.1038/nature05827. doi:10.1038/nature05827 [DOI] [PubMed] [Google Scholar]
- Bremer K. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution. 1988;42:795–803. doi: 10.1111/j.1558-5646.1988.tb02497.x. doi:10.2307/2408870 [DOI] [PubMed] [Google Scholar]
- Brökeland W., Raupach M.J. A species complex within the isopod genus Haploniscus (Crustacea: Malacostraca: Peracarida) from the Southern Ocean deep sea: a morphological and molecular approach. Zool. J. Linn. Soc. 2008;152:655–706. doi:10.1111/j.1096-3642.2008.00362.x [Google Scholar]
- Brusca R.C., Wilson G.D.F. A phylogenetic analysis of the Isopoda with some classificatory recommendations. Mem. Qld Mus. 1991;31:143–204. [Google Scholar]
- Childress J.J. Are there physiological and biochemical adaptations of metabolism in deep-sea animals? Trends Ecol. Evol. 1995;10:30–36. doi: 10.1016/s0169-5347(00)88957-0. doi:10.1016/S0169-5347(00)88957-0 [DOI] [PubMed] [Google Scholar]
- Choe C.P., Hancock J.M., Hwang U.W., Kim W. Analysis of the primary sequence and secondary structure of the unusually long ssu rRNA of the soil bug, Armadillidium vulgare. J. Mol. Evol. 1999;49:798–805. doi: 10.1007/pl00006602. doi:10.1007/PL00006602 [DOI] [PubMed] [Google Scholar]
- Clarke A.H. On the composition, zoogeography, origin and age of the deep-sea mollusk fauna. Deep-Sea Res. 1962;9:291–306. [Google Scholar]
- Creasey S.S., Rogers A.D. Population genetics of bathyal and abyssal organisms. Adv. Mar. Biol. 1999;35:1–151. [Google Scholar]
- Cummings M.P., Handley S.A., Myers D.S., Reed D.L., Rokas A., Winka K. Comparing bootstrap and posterior probability values in the four-taxon case. Syst. Biol. 2003;52:477–487. doi: 10.1080/10635150390218213. doi:10.1080/10635150390218213 [DOI] [PubMed] [Google Scholar]
- Dreyer H., Wägele J.-W. Parasites of crustaceans (Isopoda: Bopyridae) evolved from fish parasites: molecular and morphological evidence. Zoology. 2001;103:157–178. [Google Scholar]
- Dreyer H., Wägele J.-W. The Scutocoxifera tax. nov. and the information content of nuclear ssu rDNA sequences for reconstruction of isopod phylogeny (Peracarida: Isopoda) J. Crust. Biol. 2002;22:217–234. doi:10.1651/0278-0372(2002)022[0217:TSTNAT]2.3.CO;2 [Google Scholar]
- Edgar R.C. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. doi:10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter R.J., Rex M.A., Chase M.R., Quattro J.M. Population differentiation decreases with depth in deep-sea bivalves. Evolution. 2005;59:1479–1491. doi:10.1554/04-538 [PubMed] [Google Scholar]
- Fahrbach E. The expedition ANTARKTIS-XXII/3 of research vessel ‘Polarstern’ in 2005. Rep. Polar Mar. Res. 2006;533:1–246. [Google Scholar]
- Farris J.S. The retention index and the rescaled consistency index. Cladistics. 1989a;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. doi:10.1111/j.1096-0031.1989.tb00573.x [DOI] [PubMed] [Google Scholar]
- Farris J.S. The retention index and homoplasy excess. Syst. Zool. 1989b;38:783–791. doi:10.2307/2992406 [Google Scholar]
- Farris J.S., Källersjö M., Kluge A.G., Bult C. Constructing a significance test for incongruence. Syst. Biol. 1995;44:570–572. doi:10.2307/2413663 [Google Scholar]
- Fütterer D.K., Brandt A., Poore G.C.B. The expeditions ANTARKTIS-XIX/3-4 of the Research Vessel POLARSTERN in 2002 (ANDEEP I and II: Antarctic benthic deep-sea biodiversity—colonisation history and recent community patterns) Rep. Polar Mar. Res. 2003;470:1–174. [Google Scholar]
- Gage J.D., Tyler P.D. Cambridge University Press; Cambridge, UK: 1991. Deep-sea biology: a natural history of organisms at the deep-sea floor. [Google Scholar]
- Hansen B. Systematics and biology of the deep-sea holothurians. Galathea Rep. 1975;13:1–262. [Google Scholar]
- Hayward B.W. Global deep-sea extinctions during the Pleistocene ice ages. Geology. 2001;29:599–602. doi:10.1130/0091-7613(2001) [Google Scholar]
- Hennig W. Deutscher Zentralverlag; Berlin, Germany: 1950. Grundzüge einer Theorie der phylogenetischen Systematik. [Google Scholar]
- Herring P. Oxford University Press; Oxford, UK: 2002. The biology of the deep ocean. [Google Scholar]
- Hessler R.R. The Desmosomatidae (Isopoda, Asellota) of the Gay Head-Bermuda transect. Bull. Scripps Inst. Oceanogr. 1970;15:1–189. [Google Scholar]
- Hessler R.R. Swimming morphology in Eurycope cornuta (Isopoda: Asellota) J. Crust. Biol. 1993;13:667–674. doi:10.2307/1549097 [Google Scholar]
- Hessler R.R., Sanders H.L. Faunal diversity in the deep-sea. Deep-Sea Res. 1967;14:65–78. [Google Scholar]
- Hessler R.R., Strömberg J.O. Behaviour of janiroidean isopods (Asellota) with special reference to deep-sea genera. Sarsia. 1989;74:145–159. [Google Scholar]
- Hessler R.R., Thistle D. On the place of origin of deep-sea isopods. Mar. Biol. 1975;32:155–165. doi:10.1007/BF00388508 [Google Scholar]
- Hessler R.R., Wilson G.D.F. The origin and biogeography of malacostracan crustaceans in the deep sea. In: Sims R.W., Price J.H., Whalley P.E.S., editors. Evolution, time and space: the emergence of the biosphere. Academic Press; London, UK: 1983. pp. 227–254. [Google Scholar]
- Hessler R.R., Wilson G.D.F., Thistle D. The deep-sea isopods: a biogeographic and phylogenetic overview. Sarsia. 1979;64:67–75. [Google Scholar]
- Ho J.W.K., et al. SeqVis: visualization of compositional heterogeneity in large alignments of nucleotides. Bioinformatics. 2005;22:2162–2163. doi: 10.1093/bioinformatics/btl283. doi:10.1093/bioinformatics/btl283 [DOI] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005;23:254–267. doi: 10.1093/molbev/msj030. doi:10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- Isozaki Y. Permo–Triassic boundary superanoxia and stratified superocean: records from the lost deep sea. Science. 1997;276:235–238. doi: 10.1126/science.276.5310.235. doi:10.1126/science.276.5310.235 [DOI] [PubMed] [Google Scholar]
- Just J. New species of Mexicope, stat. nov. and Ianthopsis from Australia and a rediagnosis of Acanthaspidiidae (Isopoda: Asellota) Inv. Tax. 2001;15:909–925. doi:10.1071/IT00037 [Google Scholar]
- Kilpert F., Podsiadlowski L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 2006;7:241. doi: 10.1186/1471-2164-7-241. doi:10.1186/1471-2164-7-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge A.G., Farris J.S. Quantitative phyletics and the evolution of anurans. Syst. Zool. 1969;18:1–32. doi:10.2307/2412407 [Google Scholar]
- Knudsen S.W., Møller P.R., Gravlund P. Phylogeny of the snailfishes (Teleostei: Liparidae) based on molecular and morphological data. Mol. Phyl. Evol. 2007;44:649–666. doi: 10.1016/j.ympev.2007.04.005. doi:10.1016/j.ympev.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Kussakin O.G. Peculiarities of the geographical and vertical distribution of marine isopods and the problem of the deep-sea fauna origin. Mar. Biol. 1973;23:19–34. doi:10.1007/BF00394108 [Google Scholar]
- Le Goff-Vitry M.C., Rogers A.D., Baglow D. A deep-sea slant on the molecular phylogeny of the Sceractinia. Mol. Phyl. Evol. 2004;30:167–177. doi: 10.1016/s1055-7903(03)00162-3. doi:10.1016/S1055-7903(03)00162-3 [DOI] [PubMed] [Google Scholar]
- Lipps, J. H. & Hickmann, C. S. 1982 Origin, age and evolution of Antarctic and deep-sea faunas. In The environment of the deep sea, pp. 324–356. Englewood Cliffs, NJ: Prentice Hall.
- Malyutina M.V. Revision of Storthyngura Vanhöffen, 1914 (Crustacea: Isopoda: Munnopsisidae) with descriptions of three new genera and four new species from the deep South Atlantic. Org. Div. Evol. 2003;3:1–101. doi:10.1078/1439-6092-00054 Electronic supplement 13. [Google Scholar]
- Malyutina M.V., Brandt A. A revaluation of the Eurycopinae (Crustacea, Isopoda, Munnopsidae) with a description of Dubinectes gen. nov. from the southern Atlantic deep sea. Zootaxa. 2006;1272:1–44. [Google Scholar]
- Marshall N.J., Diebel C. ‘Deep-sea spiders’ that walk through the water. J. Exp. Biol. 1995;198:1371–1379. doi: 10.1242/jeb.198.6.1371. [DOI] [PubMed] [Google Scholar]
- Menzies R.J., George R.Y., Rowe G.T. Wiley; New York, NY: 1973. Abyssal environment and ecology of the world oceans. [Google Scholar]
- Müller K. PRAP—computation of Bremer support for large data sets. Mol. Phyl. Evol. 2004;31:780–782. doi: 10.1016/j.ympev.2003.12.006. doi:10.1016/j.ympev.2003.12.006 [DOI] [PubMed] [Google Scholar]
- Podsiadlowski L., Bartolomaeus T. Major rearrangements characterize the mitochondrial genome of the isopod Idotea baltica (Crustacea: Peracarida) Mol. Phyl. Evol. 2006;40:893–899. doi: 10.1016/j.ympev.2006.04.008. doi:10.1016/j.ympev.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Raupach M.J., Wägele J.-W. Distinguishing cryptic species in Antarctic Asellota (Crustacea: Isopoda)—a preliminary study of mitochondrial DNA in Acanthaspidia drygalskii. Ant. Sci. 2006;18:191–198. doi:10.1017/S0954102006000228 [Google Scholar]
- Raupach M.J., Held C., Wägele J.-W. Multiple colonization of the deep-sea by the Asellota (Crustacea: Peracarida: Isopoda) Deep-Sea Res. II. 2004;51:1787–1795. doi:10.1016/j.dsr2.2004.06.035 [Google Scholar]
- Raupach M.J., Malyutina M., Brandt A., Wägele J.-W. Molecular data reveal a highly diverse species flock within the munnopsoid deep-sea isopod Betamorpha fusiformis (Barnard, 1920) (Crustacea: Isopoda: Asellota) in the Southern Ocean. Deep-Sea Res. II. 2007;54:1820–1830. doi:10.1016/j.dsr2.2007.07.009 [Google Scholar]
- Rex M.A. Community structure in the deep-sea benthos. Annu. Rev. Ecol. Syst. 1981;12:331–353. doi:10.1146/annurev.es.12.110181.001555 [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sanders H.L., Hessler R.R. Ecology of the deep-sea benthos. Science. 1969;163:1419–1424. doi: 10.1126/science.163.3874.1419. doi:10.1126/science.163.3874.1419 [DOI] [PubMed] [Google Scholar]
- Sanders H.L., Hessler R.R., Hampson G.R. An introduction to the study of deep-sea benthic faunal assemblages along the Gay Head-Bermuda transect. Deep-Sea Res. 1963;12:845–867. [Google Scholar]
- Schotte, M., Kensley, B. F. & Shilling, S. 1995 (onwards) World list of marine, freshwater and terrestrial crustacea isopoda Washington, DC: National Museum of Natural History, Smithsonian Institution. See http://www.nmnh.si.edu/iz/isopod/
- Schultz G.A. Aspects of the evolution and origin of the deep-sea isopod crustaceans. Sarsia. 1979;64:77–83. [Google Scholar]
- Shank T.M., Black M.B., Halanych K.M., Lutz R.A., Vrijenhoek R.C. Miocene radiation of deep-sea vent shrimp (Caridea: Bresilidae): evidence from mitochondrial cytochrome oxidase subunit I. Mol. Phyl. Evol. 1999;13:244–254. doi: 10.1006/mpev.1999.0642. doi:10.1006/mpev.1999.0642 [DOI] [PubMed] [Google Scholar]
- Smith A.B. Phylogeny and systematics of holasteroid echinoids and their migration into the deep-sea. Palaeontology. 2004;47:123–150. doi:10.1111/j.0031-0239.2004.00352.x [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. doi:10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Glazko G.V., Nei M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl Acad. Sci. USA. 2002;99:16 138–16 143. doi: 10.1073/pnas.212646199. doi:10.1073/pnas.212646199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svavarsson J., Stromberg J.O., Brattegard T. The deep-sea asellotes (Isopoda, Crustacea) fauna of the Northern Seas: species composition, distribution patterns and origin. J. Biography. 1993;20:537–555. doi:10.2307/2845725 [Google Scholar]
- Swofford, D. L. 2002 PAUP*: phylogenetic analysis using parsimony (* and other methods), v. 4.0b10. Sunderland, MA: Sinauer Associates.
- Wägele J.-W. Evolution und phylogenetisches System der Isopoda: Stand der Forschung und neue Erkenntnisse. Zoologica. 1989;140:1–262. [Google Scholar]
- Wägele J.-W., Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol. Biol. 2007;7:147. doi: 10.1186/1471-2148-7-147. doi:10.1186/1471-2148-7-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wägele J.-W., Voelz N.J., McArthur J.V. Older than the Atlantic Ocean: discovery of a fresh-water Microcerberus (Isopoda) in North America and erection of Coxicerberus, new genus. J. Crust. Biol. 1995;15:733–745. doi:10.2307/1548822 [Google Scholar]
- Wetzer R. Hierarchical analysis of mtDNA variation and the use of mtDNA for isopod (Crustacea: Peracarida: Isopoda) systematics. Cont. Zool. 2001;70:23–39. [Google Scholar]
- Wetzer R. Mitochondrial genes and isopod phylogeny (Peracarida: Isopoda) J. Crust. Biol. 2002;22:1–14. doi:10.1651/0278-0372(2002)022[0001:MGAIPP]2.0.CO;2 [Google Scholar]
- Wilcox T.P., Zwickl D.J., Heath T.A., Hillis D.M. Phylogenetic relationships of the dwarf boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Mol. Phyl. Evol. 2002;25:361–371. doi: 10.1016/s1055-7903(02)00244-0. doi:10.1016/S1055-7903(02)00244-0 [DOI] [PubMed] [Google Scholar]
- Wilson G.D.F. The systematics and evolution of Haplomunna and its relatives (Isopoda, Haplomunnidae, New family) J. Nat. Hist. 1976;10:569–580. doi:10.1080/00222937600770451 [Google Scholar]
- Wilson G.D.F. Systematics of a species complex on the deep-sea genus Eurycope, with a revision of six previously described species (Crustacea, Isopoda, Eurycopidae) Bull. Scripps Inst. Oceanogr. 1982;25:1–64. [Google Scholar]
- Wilson G.D.F. A systematic revision of the deep-sea subfamily Lipomerinae of the isopod crustacean family Munnopsidae. Bull. Scripps Inst. Oceanogr. 1989;27:1–138. doi:10.2307/1548354 [Google Scholar]
- Wilson G.D.F. A phylogenetic analysis of the isopod family Janiridae (Crustacea) Inv. Tax. 1994;8:749–766. doi:10.1071/T9940749 [Google Scholar]
- Wilson G.D.F. Historical influences on deep-sea isopod diversity in the Atlantic Ocean. Deep-Sea Res. II. 1998;45:279–301. doi:10.1016/S0967-0645(97)00046-5 [Google Scholar]
- Wilson G.D.F. Some of the deep-sea fauna is ancient. Crustaceana. 1999;72:1019–1030. doi:10.1163/156854099503915 [Google Scholar]
- Wolff T. The hadal community: an introduction. Deep-Sea Res. 1960;6:95–124. [Google Scholar]
- Wuyts J., de Rijk P., van de Peer Y., Pison G., Rousseeuw P., de Wachter R. Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of the eukaryotic small subunit ribosomal RNA. Nucl. Acids Res. 2000;28:4698–4708. doi: 10.1093/nar/28.23.4698. doi:10.1093/nar/28.23.4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardus J.D., Etter R.J., Chase M.R., Rex M.A., Boyle E.E. Bathymetric and geographic population structure in the pan-Atlantic deep-sea bivalve Deminucula atacellana (Schenck, 1939) Mol. Ecol. 2006;15:639–651. doi: 10.1111/j.1365-294X.2005.02832.x. doi:10.1111/j.1365-294X.2005.02832.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood topology
Neigbor-net analysis of both 18S and 28S data sets
Details to the neighbor-net structures
Accession numbers and taxonomic classifications
PCR-primer informations
A Mesosignum sp. (Mesosignidae), B Acanthaspidia drygalskii (Acanthaspidiidae), C Ischnomesus sp. (Ischnomesidae) D Janirella sp. (Janirellidae), E Haploniscus sp. (Haploniscidae), F Chelator sp. (Desmosomatidae) and G Eurycope sp. (Munnopsidae)