Abstract
Recent phylogeographic studies of the endangered Asian elephant (Elephas maximus) reveal two highly divergent mitochondrial DNA (mtDNA) lineages, an elucidation of which is central to understanding the species's evolution. Previous explanations for the divergent clades include introgression of mtDNA haplotypes between ancestral species, allopatric divergence of the clades between Sri Lanka or the Sunda region and the mainland, historical trade of elephants, and retention of divergent lineages due to large population sizes. However, these studies lacked data from India and Myanmar, which host approximately 70 per cent of all extant Asian elephants. In this paper, we analyse mtDNA sequence data from 534 Asian elephants across the species's range to explain the current distribution of the two divergent clades. Based on phylogenetic reconstructions, estimates of times of origin of clades, probable ancestral areas of origin inferred from dispersal–vicariance analyses and the available fossil record, we believe both clades originated from Elephas hysudricus. This probably occurred allopatrically in different glacial refugia, the α clade in the Myanmar region and the β clade possibly in southern India–Sri Lanka, 1.6–2.1 Myr ago. Results from nested clade and dispersal–vicariance analyses indicate a subsequent isolation and independent diversification of the β clade in both Sri Lanka and the Sunda region, followed by northward expansion of the clade. We also find more recent population expansions in both clades based on mismatch distributions. We therefore suggest a contraction–expansion scenario during severe climatic oscillations of the Quaternary, with range expansions from different refugia during warmer interglacials leading to the varying geographical overlaps of the two mtDNA clades. We also demonstrate that trade in Asian elephants has not substantially altered the species's mtDNA population genetic structure.
Keywords: phylogeography, divergent mitochondrial clades, Pleistocene refugia, elephant trade, Elephas fossils
1. Introduction
The Asian elephant (Elephas maximus) is endangered, with a wild population of 41 000–52 000 individuals in 6 per cent of the range occupied 4000 years ago (Sukumar 2003). It is the sole surviving species of the Proboscidea in Asia. Studies of its evolutionary history and phylogeography are recent enough that their results have not been integrated into conservation action, although the flagship role of the elephant for broader conservation in Asia has been recognized (Duckworth & Hedges 1998; Sukumar 2003). Fossils and molecular analyses are valuable in reconstructing evolutionary history, so while fossil data for the Elephantidae are limited in Asia, increasing molecular data and new ways of evaluating them are providing a clearer picture of the species' phylogeography (Fernando et al. 2000, 2003; Fleischer et al. 2001; Vidya et al. 2005).
The largest study of fossil elephantid morphology indicated that the genus Elephas originated in Africa after the differentiation of the genus Loxodonta and was present during the Early Pliocene (Maglio 1973). A recent molecular study by Rohland et al. (2007) has estimated that Loxodonta and the Mammuthus–Elephas lineage diverged 7.6 (95% CI 6.6–8.8) million years ago (Myr ago). The fossil record alone suggests that this split is more recent (ca 5.5 Myr ago) (see electronic supplementary material 1). A derivative of the early Elephas ekorensis–Elephas recki complex colonized Asia and is thought to have given rise to Elephas planifrons and Elephas hysudricus (Maglio 1973). The earliest records of both species were found in the Siwalik Hills in the northern Indian subcontinent, E. planifrons appearing ca 3.6 Myr ago and E. hysudricus ca 2.7 Myr ago (see Nanda 2002). Late in the Early Pleistocene, Elephas namadicus, another derivative of E. recki, colonized Asia and displaced the earlier Elephas species across a considerable part of their ranges (Maglio 1973) before disappearing in the Late Pleistocene. However, E. hysudricus, which was widespread, is considered (based on dental and cranial evidence) to have given rise to E. maximus in southern Asia ca 0.25 Myr ago (Maglio 1973) and to Elephas hysudrindicus, a Javan species, ca 0.8–1.0 Myr ago (Maglio 1973; Van den Bergh et al. 1996).
Molecular phylogeographic analyses are often based on mitochondrial DNA (mtDNA) markers. As mtDNA is maternally inherited, stochastic extinctions of mitochondrial lineages through the absence of female offspring are usually the norm unless sufficiently large populations of females exist. The coexistence of divergent lineages of mtDNA within a species is therefore rare and requires an elucidation of the evolutionary and population processes that led to it (Melnick et al. 1993). The Asian elephant has two such divergent lineages of mtDNA haplotypes or clades, the ‘α’ (Fernando et al. 2000, 2003) or ‘B’ clade (Hartl et al. 1996; Fleischer et al. 2001) and the ‘β’ or ‘A’ clade (we use the α and β terminology here), with a sequence divergence of approximately 3 per cent. Haplotypes from these two clades coexist within populations, sometimes within small geographical areas (Fernando et al. 2000; Fleischer et al. 2001), unlike other mammalian species, in which divergent clades are usually geographically separate (e.g. Taberlet & Bouvet 1994; Jensen-Seaman & Kidd 2001). Therefore, understanding the coexistence and distribution of the two clades is vital to understanding Asian elephant evolution.
Hypotheses to explain the distribution of the two Asian elephant clades have invoked introgression of mitochondrial haplotypes from another species through hybridization, and/or allopatric divergence, in which mutations accumulate and lead to sequence divergence among populations that are geographically separated. More specifically, these hypotheses include: (i) the introgression of mtDNA from E. namadicus or an alternative species of Elephas to E. maximus (Fernando et al. 2000); (ii) allopatric divergence of populations on the mainland giving rise to the α clade and on Sri Lanka giving rise to the β clade, followed by secondary contact and admixture (Fernando et al. 2000); (iii) introgression of mtDNA from E. hysudrindicus (in the Sunda region), which gave rise to the β clade, into E. maximus, which carried the α clade, followed by extensive trade in elephants bringing the β clade to Sri Lanka and southern India (Fleischer et al. 2001); and (iv) incomplete lineage sorting, or the retention of divergent lineages simply owing to large population size (Fleischer et al. 2001). Importantly, these hypotheses were based on only four to six samples from India, which hosts approximately 60 per cent of the entire Asian elephant population (Sukumar 2003), and zero to five samples from Myanmar, which hosts another 10 per cent.
Here, we expand our phylogeographic analysis by examining mtDNA from 534 Asian elephants across the species's range (figure 1), including larger sample sizes from India (n=244) and Myanmar (n=24). The mtDNA segment analysed (599 base pairs (bp), comprising the C-terminal of cyt-b, t-RNAThr, t-RNAPro and the hypervariable left domain of the control region) corresponds to that used by Fernando et al. (2000, 2003) and Fleischer et al. (2001) (minus 76 bp), making it possible to compare the results across studies. We examined mtDNA diversity and population differentiation, and then constructed phylogenetic trees based on maximum parsimony (MP), minimum evolution (ME) and Bayesian methods in order to examine age relationships between haplotypes, and mismatch distributions to detect historical population expansions. Finally, we used a nested clade analysis (NCA) to look for geographical associations of haplotypes and a dispersal–vicariance analysis to identify areas of distribution of ancestral haplotypes. Based on our results, we propose a revised evolutionary hypothesis in which climatic fluctuations during the Pleistocene were an important factor shaping Asian elephant phylogeography.
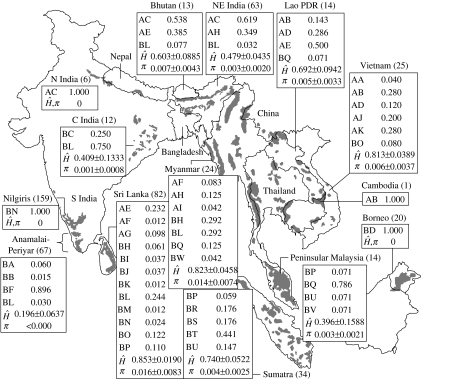
Figure 1.
Present Asian elephant distribution (grey) based on Sukumar (2003) and (for India) Vidya et al. (2005), and the number of individuals sampled (within parentheses), proportions of different haplotypes, and Ĥ and π (expressed as average ±1.96 s.e.) tabled against different populations. Haplotypes beginning with the letter A belong to the α clade and those beginning with the letter B to the β clade. (See electronic supplementary material 2 for more details about figure 1.)
2. Material and methods
(a) Samples and molecular analysis
We sequenced 365 new individuals and added 169 published sequences of Fernando et al. (2000, 2003). Most (332) new samples were non-invasively collected fresh dung, mostly from free-ranging elephants, while the remaining were blood samples from captive elephants with unambiguous capture records. Sequences of the African elephant (Loxodonta africana) were obtained from three zoo animals (see Fernando et al. 2003) and those of the woolly mammoth (Mammuthus primigenius) from GenBank sequences NC007596 (Krause et al. 2006) and DQ316067 (Rogaev et al. 2006). DNA extraction, PCR amplification and squencing were based on Fernando et al. (2000, 2003).
(b) MtDNA diversity and differentiation
We aligned and edited sequences using Sequencher v. 3.1.1 (Gene Codes Corporation 1999). Population structure was assessed based on locus-by-locus analysis of molecular variance (AMOVA; Excoffier et al. 1992) and FST (Weir & Cockerham 1984) values calculated using Arlequin v. 3.1 (Excoffier et al. 2005).
(c) Phylogenetic analyses
Phylogenetic analyses were conducted using MP, ME and Bayesian approaches, with woolly mammoth and the African elephant haplotypes as outgroups. MP and ME trees were constructed using PAUP v. 4 (Swofford 1998), employing a heuristic search with random addition of sequences and TBR branch swapping procedure. The Bayesian analysis was carried out using the HKY+I model, relaxed molecular clock (uncorrelated lognormal) assumption and normal priors for HKY kappa (the corrected transition/transversion ratio), I, and the coalescent population size parameter, in the program BEAST v. 1.4.6 (Drummond & Rambaut 2006). Normal priors were also used for divergence times between the Loxodonta and Mammuthus–Elephas lineages (mean=7.6 Myr ago) and between Mammuthus and Elephas (mean=6.7 Myr ago) based on Rohland et al.'s (2007) findings, and between the α and β clades in the Asian elephant (mean=1.85 Myr ago; see below). These three calibration points allowed dating of other internal nodes (see electronic supplementary material 3 for more details on phylogenetic analyses).
Mismatch distributions (Rogers & Harpending 1992) were constructed and similarity between the observed and simulated mismatches tested using Arlequin v. 3.1. To arrive at the times of expansion, we calculated a mutation rate for our 599 bp segment using HKY-corrected pairwise distances between haplotypes from the software Phylo_Win (Galtier et al. 1996) and a cyt-b-based time calibration. The latter was carried out with cyt-b-based divergences between the Asian and African elephants, and between the two Asian elephant clades as provided by Fleischer et al. (2001), but using divergence times of 6.6–8.8 Myr between the two species (Rohland et al. 2007) instead of the previous 5 Myr estimate. This calculation gave cyt-b-based divergence times between the two Asian elephant clades of 1.6–2.1 Myr, instead of 1.2 Myr calculated by Fleischer et al. (2001).
(d) Analyses of phylogeography
The NCA was carried out using 530 individuals with locational data to test the null hypothesis of no geographical association of haplotypes and, if the null hypothesis was rejected, to discriminate between the alternative hypotheses of restricted gene flow, range fragmentation and range expansion or long-distance colonization (Templeton 1998). A haplotype network based on statistical parsimony was created using TCS v. 1.13 (Clement et al. 2000) and the network nested into a series of nested clades. Geographical associations of clades were tested statistically using GeoDis v. 2.0 (Posada et al. 2000). The inference key of Templeton (2004) was used to distinguish among alternative hypotheses. We used relatively small population units/locations (see electronic supplementary material 4) and land (including past land bridges) distances (1326 distance measurements) between locations.
Dispersal–vicariance analysis to reconstruct ancestral area distributions of the MP and ME phylogenies was carried out using the program DIVA (Ronquist 1996). In the dispersal–vicariance analysis of the MP consensus tree, polytomies were present on individual trees and were not the result of creating consensus trees. Therefore, polytomies were broken down into various combinations and reanalysed.
3. Results
(a) Mitochondrial diversity and differentiation
We discovered a total of 33 substitutions (30 transitions, 3 transversions; 31 sites), which resulted in 32 unique mitochondrial haplotypes, 11 from the α clade and 21 from the β clade, among the 534 individuals sampled (see electronic supplementary material 5). Analysed as a single population, Asian elephant haplotype diversity (Ĥ; Nei 1987) is (average ±1.96 s.e.) 0.871±0.0101, and nucleotide diversity (π; Nei 1987), 0.0157±0.0080. Sri Lanka, Myanmar and Vietnam showed the highest haplotype diversities (figure 1). In addition, Sri Lanka and Myanmar had the highest nucleotide diversities, while Vietnam had haplotypes of only the α clade and, therefore, lower nucleotide diversity (figure 1). Haplotype and nucleotide diversities in the α clade were 0.801±0.0198 and 0.003±0.0020, respectively, and, in the β clade, 0.781±0.0178 and 0.007±0.0040, respectively. The average sequence divergence within clades was 0.37 per cent (range: 0.01–1%) within the α clade and 0.87 per cent (range: 0.01–1.8%) within the β clade, and the average sequence divergence between the α and β clades was 3.0 per cent (range: 2.1–4.0%). Population structure based on AMOVA showed significant differentiation among regions, among populations within regions and within populations (see electronic supplementary material 6). Fifty of the fifty-five pairwise comparisons of FST between populations were significant after Bonferroni corrections were applied (see electronic supplementary material 7).
(b) Phylogenetic analyses
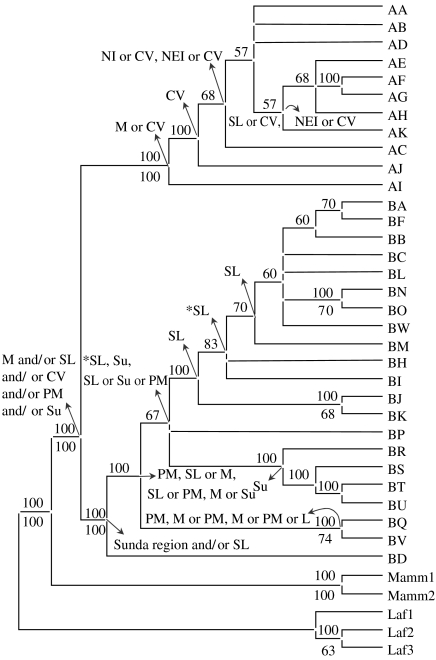
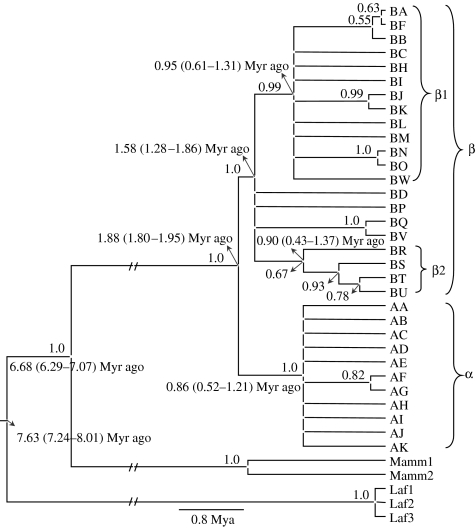
MP analysis yielded 5040 equally parsimonious trees (tree score=125). The majority rule consensus tree is shown in figure 2. The ME tree (not shown) was similar in topology to the MP tree with minor rearrangements of the terminal branches. Log posteriors of the three Bayesian runs (−1914.28, −1914.21 and −1914.25) indicated convergence of results (p>0.05 in pairwise tests based on Bayes factors; see Kass & Raftery 1995). The average log likelihood of the trees obtained from the three runs was −1428.72 (95% CI of the highest posterior densities=−1440.84 to 1416.99). A consensus tree of the 300 003 Bayesian trees (from the three runs) is shown in figure 3. All three types of tree construction revealed the distinction between α and β clades with certainty. A subclade within the β clade, referred to here as the β1 subclade, that consisted of 13 haplotypes—BA, BB, BC, BF, BH–BO (alphabetically) and BW—was present in the Bayesian and ME trees and in the MP consensus tree (figures 2 and 3). Support was poor for this clade in the MP tree (38%), but this clade consistently appeared in all the 5040 most parsimonious trees (figure 2), supporting this structure. This clade was also present in the previous ME analysis of Fernando et al. (2003). Of the eight β clade haplotypes that were not part of the β1 subclade, haplotypes BR, BS, BT and BU formed a monophyletic group, henceforth referred to as the β2 subclade, in the Bayesian tree (figure 3). Bootstrap support for this clade was also lacking in the MP tree (37%), but, again, all the most parsimonious trees showed this node. The β2 subclade is likely to be the Sumatran subclade of Fleischer et al. (2001), although we could not confirm this as their sequences are not publicly available. Haplotype AI was identified as basal to the α clade in the MP and ME trees, while no structure was detected within this clade in the Bayesian tree (figures 2, 3). Dating of nodes in the Bayesian tree indicated that the β clade is older (mean 1.58, 95% CI 1.28–1.86 Myr ago) than the α clade (mean 0.86, 95% CI 0.52–1.21 Myr ago). The β1 and β2 subclades were of roughly the same age as the α clade (figure 3).
Figure 2.
Consensus MP tree of Asian elephant haplotypes, rooted with African elephant (Laf1, Laf2 and Laf3) and woolly mammoth (Mamm1 and Mamm2) haplotypes as outgroups. Consensus values are given above the branch and the bootstrap values (based on 1000 replicates) above 50 per cent are given below the branch adjoining the corresponding node. Ancestral areas inferred from dispersal–vicariance analysis are shown with arrows from the corresponding nodes. Abbreviations used: B, Borneo; CV, Cambodia–Vietnam; L, Laos; M, Myanmar; NEI, northeastern India and Bhutan; NI, northern India; PM, Peninsular Malaysia; SI, southern India; SL, Sri Lanka; Su, Sumatra. Asterisks indicate results based on shown as well as alternative arrangements at those nodes.
Figure 3.
Phylogram from the Bayesian analysis with posterior probabilities of nodes (greater than 0.5) and mean and 95% CI of posterior node heights.
Analysis of mismatch distributions showed that observed data were explained by the fitted models, with the combined data of the two clades indicating demographic stability and the α and β clades indicating population expansion (see electronic supplementary material 8). The HKY-corrected distance between clades was 0.025 and the mutation rate in the mtDNA segment was estimated to be 1.6 and 1.2 per cent per Myr based on the mean divergence times, between the Asian and African elephants, of 6.6 and 8.8 Myr ago, respectively. This was considerably lower than the rate (3.5%) calculated by Fleischer et al. (2001), partly owing to the different time estimate used and partly owing to the smaller HKY-corrected distance between clades in our larger sample. Based on the values of τ (=2μt generations; μ is the mutation rate over the entire sequence analysed and t is the time to expansion of a population with initial effective number of females=N0 to a final size N1) from the mismatch distributions, a generation time of 27 years estimated (as the average age of reproducing females) from field data in southern India (C. Arivazhagan, T. N. C. Vidya & R. Sukumar 2001–2003, unpublished data) and mutation rates of 1.6 per cent (divergence time between clades of 1.6 Myr) and 1.2 per cent (divergence time between clades of 2.1 Myr), respectively the mean times of expansion of the α clade were approximately 128 000 and 170 000 yr ago and the mean times of expansion of the β clade were 383 000 and 511 000 yr ago. N0 was very low in both clades but confidence limits were large and upper limits of N0 were 2190 in the α clade and 4808 in the β clade based on a 1.6 Myr divergence between clades and 2920 in the α clade and 6410 in the β clade based on a 2.1 Myr divergence. The values of N1 (=θ1/2μ; estimation of mutation parameter θ1 thought to be biased upwards; Schneider & Excoffier 1999) were (based on 1.6 and 2.1 Myr divergences, respectively, between clades) 17 488 and 318 in the α clade, and 15 566 and 20 754 in the β clade. N0 and N1 were both approximately 23 600 (based on a 1.6 Myr divergence) or 31 500 (based on a 2.1 Myr divergence) based on the combined data of both clades.
(c) Phylogeographic analyses
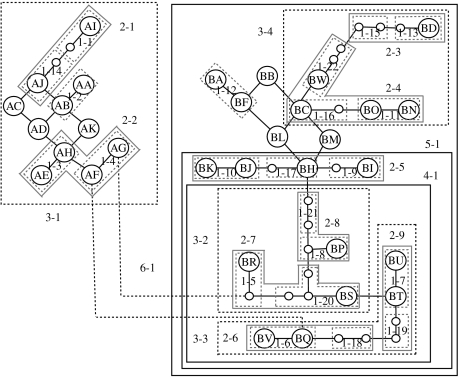
Hierarchical nesting of the cladogram generated by statistical parsimony produced 22 one-step clades, 9 two-step clades, 4 three-step clades, and 1 each of four-, five- and six-step clades (figure 4). Connections between the α and β clades did not adhere to the limits of 95 per cent parsimony, and the two clades were treated as disjointed networks. The distribution of the α and β clades was clinal as suggested by Fleischer et al. (2001), with the β clade showing higher frequencies in the southern areas of the species' range, and the α clade largely distributed towards the northern and eastern areas of the range (figure 1). Nesting level is indicative of the age of haplotypes in an NCA. Based on the levels of nesting, clades 2-6 to 2-9 (nested within 3-2 and 3-3, both in turn nested within 4-1) contained the oldest haplotypes within the β clade (figure 4). These seven haplotypes in clade 4-1 were distributed largely across Peninsular Malaysia and Sumatra, but haplotype BP was also fairly common and widespread in Sri Lanka and BQ was found in Myanmar and Laos (figure 1; electronic supplementary material 4).
Figure 4.
Hierarchical levels of nesting of the haplotype network generated by statistical parsimony. Each clade is denoted by two numbers, the first denoting the level of nesting and the second indicating the number of the clade at that level of nesting. Each branch between two haplotypes denotes a single mutation. Dashed lines show non-parsimonious connections. Empty circles indicate assumed haplotypes.
The NCA uncovered strong geographical associations from 14 out of the 18 clades that could be tested (see electronic supplementary material 9). Within the α clade, contiguous range expansion was observed in clades 1-3 and 2-2, and restricted gene flow and isolation by distance in clade 1-4. Within the β clade, restricted gene flow and isolation by distance was observed in clades 1-11, 1-12, 2-5 and 4-1, allopatric fragmentation in clade 2-4 and contiguous range expansion in clade 3-3. Past fragmentation followed by range expansion was seen in clades 3-4 and 5-1 (see electronic supplementary material 9). Further analysis of the pattern in clade 5-1, by calculating the average pairwise distances between haplotypes and between clades of different levels present in the region (see Templeton 2001), identified Myanmar as a zone of secondary contact of haplotypes, following range expansion. The pairwise distance between the geographical centres of clade 5-1 haplotypes found in Myanmar (1626 km) was higher than those in the Sunda region (Peninsular Malaysia and Sumatra; average=783 km; 95% CI=492–1074) and Sri Lanka (average=657 km; 95% CI=376–938), both of which also harbour this clade. In addition, the average pairwise distances between geographical centres of clades, which is expected to decrease with increasing clade level under the scenario of restricted gene flow (Templeton 2001), remained high instead up to the clade level that marked the fragmentation event (average pairwise distance between haplotypes in Myanmar, 1626 km; between one-step clades, 1645 km; between two-step clades, 1876 km; between clades 2-5 and 3-3, 2047 km; between clades 2-5 and 4-1, 1776 km). Similar analysis could not be carried out for clade 3-4 since almost all haplotypes of this clade were confined to one region each. NCA inference at the level of the entire α or β clade was inconclusive in the absence of interior-tip contrast of clades. While α and β clades were analysed separately thus far in the NCA, when a parsimony network was constructed adding woolly mammoth haplotypes to Asian elephant haplotypes in order to examine the relationship between the two clades, past fragmentation followed by range expansion was inferred (steps 1-2-11-12-13 followed in inference key of Templeton 2004).
Dispersal–vicariance analysis on both MP and ME phylogenies pointed to Sri Lanka as the ancestral area of the β1 subclade and Sumatra as the ancestral area of the β2 subclade. The area occupied by ancestors of the entire β clade (in both MP and ME trees) could not be ascertained unambiguously and was identified as the Sunda region and/or Sri Lanka (figure 2). The ancestral area of the α clade was pointed out to be Myanmar/Cambodia–Vietnam based on the MP tree (figure 2) and Myanmar/northeastern India/Sri Lanka based on the ME tree. The latter is contingent on the α clade not having been introduced to Sri Lanka through trade in elephants (see below).
4. Discussion
(a) Evolutionary history of the α and β clades
The time of divergence, varying complexity and geographical distributions of the α and β clades suggest distinct histories. We examine the hypotheses previously proposed in the context of our results to understand the phylogeography of the Asian elephant.
(i) Introgression of mtDNA from E. namadicus or an alternative species of Elephas to E. maximus
Elephas namadicus would seem a logical candidate for the progenitor of either α or β clade based on its broad distribution in Asia during the Middle and Late Middle Pleistocene (ca 0.70–0.15 Myr ago; see electronic supplementary material 10). However, as pointed out by Fernando et al. (2000), who first examined this hypothesis, and Fleischer et al. (2001), the divergence (including our calculation of 1.6–2.1 Myr) between the α and β clades is not compatible with the proposed phylogenetic relationship between E. namadicus and E. maximus, which are estimated to have diverged over 3.5 Myr ago, based on morphological and stratigraphic data (Maglio 1973). Fernando et al. (2000) suggested that introgression of mtDNA could have alternatively occurred from a different species of Elephas that arose in Asia, but did not specify the species (see below for an examination of other species). Unlike Maglio (1973), who suggested that E. hysudricus gave rise to E. maximus, Aguirre (1969) suggested a common ancestor of E. hysudricus and E. maximus in E. planifrons in the Early Pleistocene. If that were true, it is possible that the two clades originated in E. hysudricus and E. maximus. However, E. maximus is considered by others as a recent species and its fossils date back only to the Late Pleistocene (more recent than 0.25 Myr ago; see electronic supplementary material 10).
(ii) Allopatric divergence of populations on the mainland that gave rise to the α clade and on Sri Lanka that gave rise to the β clade, followed by secondary admixture
The overall geographical distribution of the α clade does suggest a mainland origin and the inferred ancestral areas of the α clade were largely on the mainland. However, within the β clade we identified two subclades, β1 and β2, probably having arisen in Sri Lanka and the Sunda region, respectively. While one of the interior haplotypes BP (figure 4) was found in fairly high frequency in Sri Lanka, others (BP–BV in alphabetical order and BD) were distributed across the Sunda region and other areas. Results from the dispersal–vicariance analysis could not attribute the ancestry of the entire β clade unambiguously to Sri Lanka or the Sunda region alone. The fossil record, however, leads us to believe that the β clade may have arisen in or near Sri Lanka rather than the Sunda region (see below). The hypothesis of allopatric divergence and secondary admixture of the two clades appears correct, with Sri Lanka as an important refugial area during glacial periods. However, the divergence between clades is not likely to have occurred between Sri Lanka and India, with the repeated absence of land bridges between the two causing the differentiation, because southern India and central India show only β clade haplotypes. We elaborate below on a probable mechanism of allopatric divergence between clades.
(iii) Introgression of mtDNA from E. hysudrindicus (in the Sunda region), which gave rise to the β clade, into E. maximus,which already carried the α clade, followed by extensive trade in elephants bringing the β clade to Sri Lanka and southern India
As mentioned above, the origin of the entire β clade is not immediately clear, based on molecular data, but it probably originated in or near Sri Lanka rather than the Sunda region, based on fossil data. Even if the β clade did arise in the Sunda region, the fossil-based time of divergence between E. hysudrindicus and E. maximus of 0.8–1 Myr ago is lower than our newly calculated divergence time of 1.6–2.1 Myr ago between the α and β clades, suggesting that E. hysudrindicus (endemic to Java) did not give rise to the β clade. In addition, being a recent species that appeared only in the Late Pleistocene (more recent than 0.25 Myr ago), E. maximus was presumably still evolving when the β subclades were diversifying independently in Sri Lanka and the Sunda region. Since, based on fossil evidence, E. maximus evolved from E. hysudricus and not E. hysudrindicus, and since fossils of the latter have been recovered only from Java (see electronic supplementary material 10) and not Sri Lanka, it is unlikely that the β clade introgressed from E. hysudrindicus into E. maximus. It is plausible that the α clade originated in E. maximus (E. hysudricus) as we discuss below.
The high frequencies of β clade haplotypes in southern India (100%) and Sri Lanka (66%), and the Sri Lankan origin of the β1 subclade, are not concordant with long-distance trade in elephants, dating back to 2000 years at most, having brought the β clade to these regions.
(iv) Revised hypothesis
Based on our results, fossil data and previous hypotheses, we present the following hypothesis to explain the distribution of the two clades of haplotypes. As with E. namadicus, other Elephas species such as E. planifrons, E. celebensis and E. platycephalus are also precluded from being progenitors of either clade owing to fossil-based divergence times of ca 3.5 Myr between each of them and E. maximus (Maglio 1973). Similarly, the discordance between fossil-based E. hysudrindicus–E. maximus divergence and molecular data-based α–β clade divergence eliminates E. hysudrindicus as a progenitor of either clade of haplotypes. It therefore appears that both clades had their origin in E. hysudricus. Elephas hysudricus fossils have been recorded widely, from the Pinjor horizon of northern India, the upper Irrawaddies of Myanmar, Kiangsu of southern China and the Ratnapura fauna of Sri Lanka (see electronic supplementary material 10). Since E. hysudricus gave rise to E. hysudrindicus, which was found only on Java (Maglio 1973), E. hysudricus was presumably found in the Sunda region also.
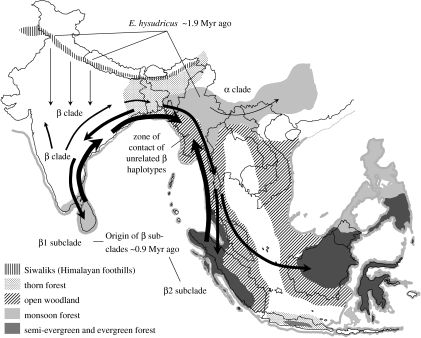
Our new calculation of the divergence between the α and β clades coincides with the beginning of the Pleistocene. The initiation of the Pleistocene in India (as inferred by the magnetostratigraphically dated Olduvai subchron ca 1.9 Myr ago; Krishnamurthy et al. 1986) was associated with a cold period (Krishnamurthy et al. 1986; Singh & Srinivasan 1993) and increasing tectonic activity in and near the Himalayas in northern India (see Sahni & Kotlia 1993; Nanda 2002). For ca 2 Myr ago, E. hysudricus formed part of the ‘Pinjor Fauna’, which were distributed across the Siwalik Hills (foothills of the Himalayas) and the upper Irrawaddies of Myanmar (electronic supplementary material 10). But from ca 1.9 Myr ago, there was a change in lithostratigraphic formation in the Siwaliks and the extinction and southward migration into Peninsular India of the Siwalik Pinjor-associated fauna (Sastry 1997; Nanda 2002, 2008). We think that the Siwalik population of E. hysudricus that migrated southwards probably gave rise to the β clade and the population in Myanmar (the inferred ancestral area of the α clade from the dispersal–vicariance analysis) extending to southern China, to the α clade.
Thus, the origin of divergent clades from E. hysudricus probably took place due to the climatic and/or tectonic changes at the beginning of the Pleistocene, which resulted in allopatric fragmentation, as supported by the results of the NCA. Repeated climatic oscillations in the form of glacial and interglacial periods since the beginning of the Pleistocene, and the ensuing range changes, adaptations and reorganization of populations, are thought to have resulted in large divergences between populations of various species (Hewitt 2000). Based on geography and inferred climatic conditions during the LGM ca 18 000 ya, it is believed that certain relatively warmer areas including Sumatra and southern Borneo, northern and eastern Myanmar, Sri Lanka and the extreme south of India served as glacial refugia during the Pleistocene (figure 5). Given available data on fossils and palaeovegetation, it is difficult to identify the exact region of origin of the β clade. If vegetation during the beginning of the Pleistocene was similar to that reconstructed for the LGM (figure 5), the β clade is likely to have originated in southern India/Sri Lanka. A single fossil of E. hysudricus thought to date back to the beginning of the Pleistocene is known from southern India (Prasad & Daniel 1968). However, if patterns of vegetation were different from that of the LGM during the period, one cannot rule out the rest of Peninsular India as refugia since E. hysudricus fossils of similar ages are known from various sites in central India and Sri Lanka (electronic supplementary material 10), and fossil beds older than 1 Myr are very rare in the former and absent in the latter.
Figure 5.
A simplified schematic of our revised hypothesis to explain the observed distribution of mitochondrial haplotypes. Arrows of increasing thickness indicate more recent time periods. There were at least two southward migrations, the first ca 1.9 Myr ago resulting in allopatric fragmentation and, consequently, the origin of the α and β clades, and the second ca 0.9 Myr ago resulting in the origin of β1 and β2 subclades. A subsequent northward expansion gave rise to the zone of contact of unrelated β clade haplotypes in Myanmar. Palaeovegetation types during the last glacial maximum based on Adams & Faure (1997) and Gathorne-Hardy et al. (2002) are also shown. Monsoon forests and semi-evergreen and evergreen forests are thought to have served as Pleistocene glacial refugia.
Our results from the NCA, showing restricted gene flow through isolation by distance in clades 2-5 (three out of the four haplotypes of which are restricted to Sri Lanka) and 4-1 (distributed mostly across the Sunda region), and from the dispersal–vicariance analysis, showing Sri Lanka and the Sunda region as ancestral areas of the β1 and β2, subclades, respectively, imply independent diversification of β clade haplotypes within each of these regions well after the origin of the β clade. More intriguingly, the secondary contact of unrelated haplotypes in Myanmar suggests that these haplotypes did not arise within Myanmar, but instead resulted from a northward range expansion of β clade haplotypes from both Sri Lanka and the Sunda region followed by subsequent admixture in this region. This may be best explained by the expansion of the β clade from peninsular India/Sri Lanka to the eastern and southeastern regions during warm periods, a second southward migration of the clade such that it became isolated in the southerly areas of Sri Lanka and the Sunda region (where the β1 and β2 subclades originated), and subsequent recolonization of the mainland from both regions when warmer, wetter conditions returned (figure 5).
There were possibly several such north–south movements, as suggested by inferences of range expansion and past fragmentation followed by the range expansion in our NCA. The clinal distribution of the two clades thus appears to be a consequence of possibly inhabiting different refugia, with range expansions during the warmer interglacial periods leading to varying distributional overlaps of the two clades. Two divergent clades of haplotypes, thought to have expanded from glacial refugia in southern India and possibly Peninsular Malaysia or Indochina, have also been reported in the dhole (Cuon alpinus; Iyengar et al. 2005). It was also found that Myanmar had haplotypes of both dhole clades, although the limited sampling precluded testing for a secondary zone of contact. In an earlier study, two divergent clades of haplotypes were also found in the rhesus macaque (Macaca mulatta; Melnick et al. 1993) in Asia, and Pleistocene glaciations were thought to have created the separation between the clades around the Brahmaputra River in northeastern India.
An alternative hypothesis to allopatric fragmentation of E. hysudricus populations giving rise to the two mtDNA clades is that of lineage retention within a single population. We find that the historical effective population size during the last population bottleneck (less than 50 females in each clade with upper limits of a few thousand females in each clade, and approximately 23 600 and 31 500 females, based on 1.6 and 2.1 Myr divergences between clades respectively, if treated as a single population) is smaller than the 29 800 (based on a 1.6 Myr divergence between clades) or 39 800 (based on a 2.1 Myr divergence) females that would be required at a minimum for lineage retention based on applying the formula of Georgiadis et al. (1994). The plausibility of several such bottlenecks in the past makes lineage retention an unlikely explanation for the coexistence of the α and β clades.
Thus, we also invoke extensive movement to explain the distribution of the two Asian elephant mtDNA clades, but unlike Fleischer et al. (2001) we maintain that this movement was mostly ancient, largely as a response to climatic conditions, with details of movement possibly having been additionally influenced by the presence of heterospecific proboscideans competing for similar niches. While we cannot detect human-assisted movement of β-clade haplotypes across Asia, the arrival of the α clade in Sri Lanka due to trade in elephants cannot be ruled out. The fairly high frequency of α-clade haplotypes in Sri Lanka and the presence of a unique haplotype (AG) seem to support a natural colonization of Sri Lanka. However, the conspicuous absence of the α clade in central and southern India, the confinement of the α clade within Sri Lanka to the southern region, the established historical trade in elephants between Sri Lanka and Myanmar (see Sukumar 2003; electronic supplementary material 11) and the presence of two out of the three Sri Lankan α-clade haplotypes (AE and AF; the third haplotype AG differs from AF by a single mutation) in or close to Myanmar suggest a human-assisted transfer of this clade to Sri Lanka. Population genetic modelling of peninsular Indian populations may help in examining whether elephant captures and reduction in population size due to historical habitat loss in these regions could have led to the extinction of the α clade in southern India following a natural colonization of Sri Lanka by this clade.
This study of phylogeography is unique in sampling over 1 per cent of the entire population of a large mammal. We sampled all the major Asian elephant populations with the exception of those in Thailand. However, D-loop sequences were available for elephants from Thailand (GenBank sequences AF317519–AF317535, AF324827–AF324828, AF368903; submitted by J. Fickel, D. Kieckfeldt, T. B. Hildebrandt & C. Pitra 2000–2001, unpublished data) and, although we did not use these sequences as they were not yet published and were 188 bp shorter than our other sequences, based on alignment, we inferred these sequences to be either our haplotypes AB, AC, AD, AE, AH, BH, BO, BP, BQ and BW, or ones closely related to them. The presence of these haplotypes in Thailand is consistent with our proposed phylogeographic explanation: β clade haplotypes are very similar in composition to those in Myanmar, and the increased number of α clade haplotypes geographically close to Myanmar supports the idea of Myanmar being the ancestral area of the α clade.
(b) Potential limitations of the study
As with most phylogenetic studies that calibrate dates based on fossil records, this study also assumes that divergences based on fossil morphology are reflected in the gene tree, which may not always be true. Since divergent clades with coalescence times of over 1 Myr exist in the Asian elephant, the African elephants (Roca et al. 2005) and the mammoth (Barnes et al. 2007; Gilbert et al. 2008), it is possible that extinct species of Elephas also showed similar unusual patterns. That would lead to larger uncertainties in the coalescence times of the two clades and their times of expansions. The molecular dates are calibrated based on the fossil date for elephantid–mastodon divergence (Rohland et al. 2007), and therefore rely on that date being correct (see electronic supplementary material 1).
It has been shown that the NCA produces a large number of false positives when a panmictic population is considered (Panchal & Beaumont 2007). If structured populations show the same results as panmictic populations, it is possible that some of our inferences in the NCA are false positives. However, based on simulations in a panmictic population, the inference of past fragmentation followed by range expansion almost never appears (Panchal & Beaumont 2007). Therefore, our main inference of the two clades having arisen due to the past fragmentation followed by range expansion is not likely to be a false positive. The test for secondary contact of unrelated haplotypes in Myanmar also suggests that the pattern is real. In addition, since the dispersal–vicariance analysis inferred Sri Lanka and Sumatra as the areas of origin of the β1 and β2 subclades, respectively, and Myanmar as an ancestral area of the α clade, and these areas coincide with Pleistocene glacial refugia, our inference of a contraction–expansion scenario would be supported even in the absence of the NCA.
Future work is required to investigate whether the pattern shown by mtDNA is also shown by nuclear DNA so that conservation measures are based on results from multiple genetic markers. A disassociation between mtDNA and nuclear genes has been observed in the African elephants (Roca et al. 2005; Roca 2008). In the Asian elephant, nuclear microsatellite data are available for India and there is a concordance in population genetic structure discerned by mtDNA and nuclear DNA (Vidya et al. 2005).
(c) Population structure
Most of the populations we examined showed distinct frequencies of various haplotypes and were consequently significantly differentiated from one another. It is interesting that while Sri Lanka was significantly differentiated from southern India, it was not significantly differentiated from central India and Myanmar, which are geographically farther away. An analysis of population structure alone would allow this absence of differentiation to be interpreted entirely as a result of past trade in elephants. However, FST is an inadequate measure of whether this movement of matrilines had its origin in ancient colonization patterns or in historically recent trade in elephants, and, as explained above, an analysis of population history supports the former. We therefore emphasize the need for analyses of population history in addition to population structure.
Acknowledgements
Molecular analysis was supported by a USFWS-Asian Elephant Conservation Fund grant to Prithiviraj Fernando and D.J.M., a Center for Environmental Research and Conservation Seed Grant and the Laboratory for Genetic Investigation and Conservation, Columbia University. A visiting scholarship was given to T.N.C.V. by Columbia University. Sequencing from blood-extracted DNA was carried out by the sequencing facility at the Department of Biochemistry, Indian Institute of Science. Field sampling was funded by the Ministry of Environment and Forests, the Government of India. Samples were collected with research permissions from the state forest departments of Uttaranchal, West Bengal, Arunachal Pradesh, Assam, Meghalaya, Orissa, Jharkhand, Tamil Nadu, Karnataka and Kerala, in India; the Ministry of Forestry and the Forest Department, Myanmar; and the Forest Protection Department, Vietnam. We thank S. Varma, C. Arivazhagan, T. R. Shankar Raman, G. Dharmarajan and N. Baskaran from the Indian Institute of Science; G. Polet, WWF Cat Tien National Park Conservation Project, Vietnam; Y. Htut, Forest Department, Myanmar; W. Htun and T. H. Aung, Myanmar Timber Enterprises; S. Tint and M. Thinn, Ministry of Forestry, Myanmar; T. V. Thanh, Forest Protection Department, Vietnam; and N. X. Dang, Institute of Ecology and Biological Resources, Vietnam, for their help in obtaining samples. Field assistance was provided by K. Krishna, R. Mohan and many forest department trackers. We thank Prithiviraj Fernando for his support and comments on the manuscript, and Avinash Nanda, Parth Chauhan and Robin Dennell for their discussion about the fossil records. Prof. W. Hill, three anonymous referees and a Board Member provided comments that helped to improve the manuscript.
Supplementary Material
This contains a write-up on the divergence between the Loxodonta and Elephas-Mammuthus lineages based on the fossil record alone, as opposed to the molecular data-based divergence presented in the main text
Estimated population sizes of the Asian elephant and other details about Figure 1
Additional details about Maximum Parsimony, Minimum Evolution, and Bayesian tree construction
Locations used for the NCA and the haplotypes observed
Polymorphic nucleotide positions in the mtDNA segment analysed (from 534 individuals) and character states at these positions in the observed haplotypes
Percentage of variation, variance components, and Φ-statistics obtained from AMOVAs
Pairwise FSTs between populations
Observed (black bars) and simulated (lines with crosses) mismatch distributions obtained from mtDNA sequences of a) the entire set of individuals sampled, b) only the α clade, and c) only the β clade
Inferences at different clade levels obtained from the NCA
Fossil distribution and time of occurrence of Elephas spp. in south and southeast Asia
Write-up on historical trade in elephants involving Sri Lanka
Impact of the Younger Toba Tuff eruption on Asian elephant mitochondrial DNA based on the present results
A comment on mitochondrial haplotype diversity based on this study
References
- Adams, J. M. & Faure, H. (eds) 1997 Review and atlas of palaeovegetation: preliminary land ecosystem maps of the world since the Last Glacial Maximum Oak Ridge, TN: Oak Ridge National Laboratory. See http://www.esd.ornl.gov/projects/qen/adams1.html
- Aguirre E. Evolutionary history of the elephant. Science. 1969;20:1366–1376. doi: 10.1126/science.164.3886.1366. doi:10.1126/science.164.3886.1366 [DOI] [PubMed] [Google Scholar]
- Barnes I., Shapiro B., Lister A., Kuznetsova T., Sher A. Genetic structure and extinction of the woolly mammoth Mammuthus primigenius. Curr. Biol. 2007;17:1072–1075. doi: 10.1016/j.cub.2007.05.035. doi:10.1016/j.cub.2007.05.035 [DOI] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2006 BEAST v. 1.4.6 See http://beast.bio.ed.ac.uk
- Duckworth J.W., Hedges S. WWF Indochina Programme; Hanoi, Vietnam: 1998. A review of the status of tiger, Asian elephant, gaur, and banteng in Vietnam, Lao, Cambodia and Yunnan Province (China), with recommendations for future conservation action. [Google Scholar]
- Excoffier L., Smouse P., Quattro J. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fernando P., Pfrender M., Encalada S., Lande R. Mitochondrial DNA variation, phylogeography and population structure of the Asian elephant. Heredity. 2000;84:362–372. doi: 10.1046/j.1365-2540.2000.00674.x. doi:10.1046/j.1365-2540.2000.00674.x [DOI] [PubMed] [Google Scholar]
- Fernando P., et al. DNA analysis indicates that Asian elephants are native to Borneo and are therefore a high priority for conservation. PLoS Biol. 2003;1:110–115. doi: 10.1371/journal.pbio.0000006. doi:10.1371/journal.pbio.0000006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer R., Perry E., Muralidharan K., Stevens E., Wemmer C. Phylogeography of the Asian elephant (Elephas maximus) based on mitochondrial DNA. Evolution. 2001;55:1882–1892. doi: 10.1111/j.0014-3820.2001.tb00837.x. [DOI] [PubMed] [Google Scholar]
- Galtier N., Gouy M., Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Cabios. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gathorne-Hardy F.J., Syaukani, Davies R.G., Eggleton P., Jones D.T. Quaternary rainforest refugia in South-East Asia: using termites (Isoptera) as indicators. Biol. J. Linn. Soc. Lond. 2002;75:453–466. doi:10.1046/j.1095-8312.2002.00031.x [Google Scholar]
- Gene Codes Corporation. Gene Codes Corporation; Ann Arbor, MI: 1999. Sequencher: a genetic analysis software. V. 3.1.1. [Google Scholar]
- Georgiadis N., Bischof L., Templeton A., Patton J., Karesh W., Western D. Structure and history of African elephant populations. I. Eastern and southern Africa. J. Hered. 1994;85:100–104. doi: 10.1093/oxfordjournals.jhered.a111405. [DOI] [PubMed] [Google Scholar]
- Gilbert M.T.P., Drautz D.I., Lesk A.M., Ho S.Y.W., Qi J. Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc. Natl Acad. Sci. USA. 2008;105:8327–8332. doi: 10.1073/pnas.0802315105. doi:10.1073/pnas.0802315105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl G., Kurt F., Tiedemann R., Gmeiner C., Nadlinger K., Mar K.U., Rubel A. Population genetics and systematics of Asian elephants (Elephas maximus): a study based on sequence variation at the Cyt b gene of PCR-amplified mitochondrial DNA from hair bulbs. Z. Saugetierkd. 1996;61:285–294. [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. doi:10.1038/35016000 [DOI] [PubMed] [Google Scholar]
- Iyengar A., Babu V.N., Hedges S., Venkataraman A.B., Maclean N., Morin P.A. Phylogeography, genetic structure, and diversity in the dhole (Cuon alpinus) Mol. Ecol. 2005;14:2281–2297. doi: 10.1111/j.1365-294X.2005.02582.x. doi:10.1111/j.1365-294X.2005.02582.x [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman M., Kidd K. Mitochondrial DNA variation and biogeography of eastern gorillas. Mol. Ecol. 2001;10:2241–2247. doi: 10.1046/j.0962-1083.2001.01365.x. doi:10.1046/j.0962-1083.2001.01365.x [DOI] [PubMed] [Google Scholar]
- Kass R.E., Raftery A.E. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. doi:10.2307/2291091 [Google Scholar]
- Krause J., et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. doi:10.1038/nature04432 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy R.V., Bhattacharya S.K., Kusumgar S. Palaeoclimatic changes deduced from 13C/12C and C/N ratios of Karewa lake sediments, India. Nature. 1986;323:150–152. doi:10.1038/323150a0 [Google Scholar]
- Maglio V.J. Origin and evolution of the Elephantidae. Trans. Am. Phil. Soc. 1973;63:1–149. doi:10.2307/1006229 [Google Scholar]
- Melnick D.J., Hoelzer G.A., Absher R., Ashley M.V. mtDNA diversity in rhesus monkeys reveals overestimates of divergence time and paraphyly with neighboring species. Mol. Biol. Evol. 1993;10:282–295. doi: 10.1093/oxfordjournals.molbev.a040004. [DOI] [PubMed] [Google Scholar]
- Nanda A.C. Upper Siwalik mammalian faunas of India and associated events. J. Asian Earth Sci. 2002;21:47–58. doi:10.1016/S1367-9120(02)00013-5 [Google Scholar]
- Nanda A.C. Comments on the Pinjor Mammalian Fauna of the Siwalik Group in relation to the post-Siwalik faunas of Peninsular India and Indo-Gangetic Plain. Quat. Int. 2008;192:6–13. doi:10.1016/j.quaint.2007.06.022 [Google Scholar]
- Nei M. Columbia University Press; New York, NY: 1987. Molecular evolutionary genetics. [Google Scholar]
- Panchal M., Beaumont M.A. The automation and evaluation of nested clade phylogeographic analysis. Evolution. 2007;61:1466–1480. doi: 10.1111/j.1558-5646.2007.00124.x. doi:10.1111/j.1558-5646.2007.00124.x [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A., Templeton A.R. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol. Ecol. 2000;9:487–488. doi: 10.1046/j.1365-294x.2000.00887.x. doi:10.1046/j.1365-294x.2000.00887.x [DOI] [PubMed] [Google Scholar]
- Prasad K.N., Daniel J.A. On the occurrence of Hypselephas hysudricus in the Pleistocene deposits of Tirunelveli valley, Madras State. Curr. Sci. 1968;37:516–517. [Google Scholar]
- Roca A.L., Georgiadis N., O'Brien S.J. Cytonuclear genomic dissociation in African elephant species. Nat. Genet. 2005;37:96–100. doi: 10.1038/ng1485. [DOI] [PubMed] [Google Scholar]
- Roca A.L. The mastodon mitochondrial genome: a mammoth accomplishment. Trends Genet. 2008;24:49–52. doi: 10.1016/j.tig.2007.11.005. doi:10.1016/j.tig.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Rogaev E.I., Moliaka Y.K., Malyarchuk B.A., Kondrashov F.A., Derenko M.V., Chumakov I., Grigorenko A.P. Complete mitochondrial genome and phylogeny of Pleistocene mammoth Mammuthus primigenius. PLoS Biol. 2006;4:403–410. doi: 10.1371/journal.pbio.0040073. doi:10.1371/journal.pbio.0040073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.R., Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rohland N., Malaspinas A., Pollack J.L., Slatkin M., Matheus P., Hofreiter M. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 2007;5:1663–1671. doi: 10.1371/journal.pbio.0050207. doi:10.1371/journal.pbio.0050207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F. 1996 DIVA v. 1.1 Uppsala, Sweden: Uppsala University. See ftp.uu.se or ftp.systbot.uu.se
- Sahni A., Kotlia B.S. Upper Pliocene-Quaternary vertebrate communities of the Karewas and Siwaliks. Curr. Sci. 1993;64:893–898. [Google Scholar]
- Sastry M.V.A. The Pliocene-Pleistocene boundary in the Indian subcontinent. In: Van Couvering J.A., editor. The Pleistocene boundary and the beginning of the Quaternary. Cambridge University Press; Cambridge, UK: 1997. pp. 232–238. [Google Scholar]
- Schneider S., Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.D., Srinivasan M.S. Quaternary climatic changes indicated by planktonic foraminifera of northern Indian Ocean. Curr. Sci. 1993;64:908–915. [Google Scholar]
- Sukumar R. Oxford University Press; New York, NY: 2003. The living elephants: evolutionary ecology, behavior, and conservation. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, UK: 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0.b10. [Google Scholar]
- Taberlet P., Bouvet J. Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of the brown bear Ursus arctos in Europe. Proc. R. Soc. B. 1994;255:195–200. doi: 10.1098/rspb.1994.0028. doi:10.1098/rspb.1994.0028 [DOI] [PubMed] [Google Scholar]
- Templeton A.R. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol. Ecol. 1998;7:381–397. doi: 10.1046/j.1365-294x.1998.00308.x. doi:10.1046/j.1365-294x.1998.00308.x [DOI] [PubMed] [Google Scholar]
- Templeton A.R. Using phylogeographic analyses of gene trees to test species status and processes. Mol. Ecol. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. doi:10.1046/j.1365-294x.2001.01199.x [DOI] [PubMed] [Google Scholar]
- Templeton A.R. Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol. Ecol. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. doi:10.1046/j.1365-294X.2003.02041.x [DOI] [PubMed] [Google Scholar]
- Van den Bergh G.D., Sondaar P.Y., De Vos J., Aziz F. The proboscideans of the South-East Asian islands. In: Shoshani J., Tassy P., editors. The Proboscidea: evolution and palaeoecology of elephants and their relatives. Oxford University Press; Oxford, UK: 1996. pp. 240–248. [Google Scholar]
- Vidya T.N.C., Fernando P., Melnick D.J., Sukumar R. Population genetic structure and conservation of Asian elephants (Elephas maximus) across India. Anim. Conserv. 2005;8:377–388. doi:10.1017/S1367943005002428 [Google Scholar]
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. doi:10.2307/2408641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This contains a write-up on the divergence between the Loxodonta and Elephas-Mammuthus lineages based on the fossil record alone, as opposed to the molecular data-based divergence presented in the main text
Estimated population sizes of the Asian elephant and other details about Figure 1
Additional details about Maximum Parsimony, Minimum Evolution, and Bayesian tree construction
Locations used for the NCA and the haplotypes observed
Polymorphic nucleotide positions in the mtDNA segment analysed (from 534 individuals) and character states at these positions in the observed haplotypes
Percentage of variation, variance components, and Φ-statistics obtained from AMOVAs
Pairwise FSTs between populations
Observed (black bars) and simulated (lines with crosses) mismatch distributions obtained from mtDNA sequences of a) the entire set of individuals sampled, b) only the α clade, and c) only the β clade
Inferences at different clade levels obtained from the NCA
Fossil distribution and time of occurrence of Elephas spp. in south and southeast Asia
Write-up on historical trade in elephants involving Sri Lanka
Impact of the Younger Toba Tuff eruption on Asian elephant mitochondrial DNA based on the present results
A comment on mitochondrial haplotype diversity based on this study