Abstract
The relative importance of ancient demography and climate in determining worldwide patterns of human within-population phenotypic diversity is still open to debate. Several morphometric traits have been argued to be under selection by climatic factors, but it is unclear whether climate affects the global decline in morphological diversity with increasing geographical distance from sub-Saharan Africa. Using a large database of male and female skull measurements, we apply an explicit framework to quantify the relative role of climate and distance from Africa. We show that distance from sub-Saharan Africa is the sole determinant of human within-population phenotypic diversity, while climate plays no role. By selecting the most informative set of traits, it was possible to explain over half of the worldwide variation in phenotypic diversity. These results mirror those previously obtained for genetic markers and show that ‘bones and molecules’ are in perfect agreement for humans.
Keywords: morphology, phenotypic diversity, cranial traits, human evolution, climate, ancient demography
1. Introduction
Recent studies on worldwide human populations have highlighted a decrease in within-population genetic diversity with increasing distance from sub-Saharan Africa (Handley et al. 2007). The pattern has been interpreted as a signature of the recent African origin of anatomically modern humans. The decline in within-population diversity has been ascribed to a sequence of small founder events due to pioneering breakaways experienced during the fast expansion wave that led to the colonization of the world (Prugnolle et al. 2005a; Ramachandran et al. 2005; Liu et al. 2006; Li et al. 2008; Romero et al. in press). A similar decrease in within-population diversity with distance from Africa has been detected on human dental (Hanihara 2008) and morphometric traits (Manica et al. 2007; von Cramon-Taubadel & Lycett 2008), giving further support to the hypothesis of a single origin of modern humans followed by worldwide expansion (Manica et al. 2007; Hanihara 2008; von Cramon-Taubadel & Lycett 2008). In spite of the similar pattern, neutral genetic markers gave a much higher proportion of explained variance than cranial morphometric traits, suggesting that low heritability and/or selective pressures acting on the cranial shape reduce the amount of demographic information that can be extracted from morphometric characters (Relethford 2004a,b; Roseman 2004; Manica et al. 2007).
Climate is recognized to be the most important selective force on cranial traits, but it remains unclear how to disentangle its effect from that of ancient demography. Previous attempts have either ignored climate (Hanihara 2008), tried to minimize its effect by standardizing measurements by size (von Cramon-Taubadel & Lycett 2008) or assumed a priori an effect of climate and modelled its effect directly (Manica et al. 2007). However, climate has been shown repeatedly to affect the size of certain traits (i.e. Beals et al. 1984; Roseman 2004; Harvati & Weaver 2006), and it could be expected to reduce within-population variability in regions under strong selection. In this paper, we develop an explicit framework to quantify the relative role of climate and distance from the putative origin in determining within-population phenotypic variability in humans. Finally, having found the best model to describe the global patterns of pooled phenotypic diversity, we investigate which traits are most powerful at recovering past demographic signatures.
2. Material and methods
(a) Data
We used a large morphometric database of 37 cranial measurements (table S1 in the electronic supplementary material) from 6245 skulls (4666 male and 1579 females), collected from 105 and 39 worldwide distributed human populations for males and females, respectively (Manica et al. 2007). Only skulls younger than 2000 years were included in the analyses, and a minimum sample size of 15 individuals per population was enforced. Polynesian and Micronesian populations were excluded from the dataset because of the possibility of multiple colonization waves, including relatively recent ones (e.g. Melton et al. 1995). Individual trait measures were standardized by transforming into z-scores. The within-population diversity across multiple traits was calculated by averaging the individual diversities for individual cranial traits (the trace of the variance–covariance matrix of the standardized cranial measurements, divided by the number of traits, as defined by Relethford & Blangero (1990)). This measure is proportional to heterozygosity (Relethford & Blangero 1990), making it useful for comparisons with genetic diversity. Three key climatic variables (minimum temperature, maximum temperature and average annual precipitation) were obtained from WORLDCLIM (Hijmans et al. 2005), as set of global climatic GIS layers interpolating data from approximately 15 000 weather stations distributed worldwide.
(b) Evaluating the relative explanatory power of distance from a putative origin and climate
We investigated the predictive power of distance from the putative source and the three climatic variables for a grid of potential origins distributed at intervals of 5° latitude/longitude across the whole of Africa, Eurasia, Australia and America. Distances from the origins were computed using an application of graph theory as detailed in Manica et al. (2005), which allows to model movement over land while avoiding major mountain ranges (more than 2000 m altitude). We started with full models that included the cubic polynomial of distance (linear, quadratic and cubic) and all possible interactions of rainfall, minimum and maximum temperatures. We then selected the best minimal model for each origin by backward stepwise elimination based on the Bayesian information criterion (BIC). Origins within four BIC units of the best origin were deemed to have ‘considerable support’ (Anderson et al. 1998) and were chosen as to give a confidence envelope of possible origins.
The analyses were repeated excluding from the male dataset all Eskimo/Inuit populations and the other populations living in areas with a minimum annual temperature lower than −20°C, in order to test the robustness of the model to extreme environmental conditions. An equivalent test could not be implemented with the female dataset due to its small size.
(c) Defining the most informative set of traits
As the individual cranial traits are determined by genetic and environmental factors in different proportion, we looked for the combination of traits that best represent the genetic signature of the OOA expansion. A forward stepwise addition of traits was carried on, starting with the single cranial trait that showed the best correlation with distance from Africa (using the best origin inferred in previous analysis). At each step, the trait whose addition led to the greatest improvement in the relationship between within-population diversity and distance from Africa was included in the set of most informative traits. The best combination of traits was then defined as the set for which no additional trait could increase the correlation. We also used a backward stepwise elimination approach, starting from the whole set of cranial traits and progressively dropping traits, which did not contribute to the global correlation. This approach led to the same best set of informative traits, which was obtained with the forward stepwise approach.
3. Results
(a) Evaluating the relative explanatory power of distance from a putative origin and climate
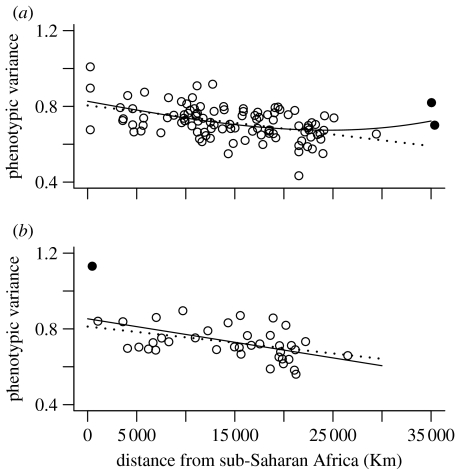
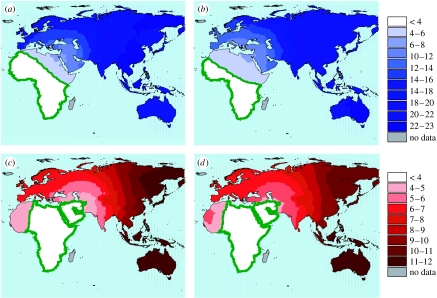
The most likely origin for the male dataset was placed somewhere in sub-Saharan Africa (figures 1a and 2a; table S2 in the electronic supplementary material; R2=0.20, F2,102=12.84, p<0.001), in accordance with previous analyses (Manica et al. 2007; von Cramon-Taubadel & Lycett 2008). The model included only the linear and cubic distances from the origin, while climatic variables were not part of the best model. As the curvature defined by the cubic relationship seemed to be mostly affected by two populations from Patagonia, we repeated the whole analysis without these outliers. The area encompassing the most likely origins did not change markedly (figure 1b), but the best model this time included only the linear term of the distance polynomial (figure 2b; R2=0.21, F1,101=27.22, p<0.001). As for the full dataset, no climatic variable was included in the best model. Repeating the analyses on the smaller dataset of female crania gave similar results. While the area of possible origins is broader than that for males (not surprisingly, given the small sample size), the best model included only the linear geographical distance (figure 1b and 2c; table S3 in the electronic supplementary material; R2=0.28, F1,37=14.44, p<0.001) (figure 1b and 2c). A population from Kenya with extreme phenotypic diversity did not fit the general pattern. However, the removal of this population did not affect the goodness of fit of the relationship (figure 1b; R2=0.28) nor the likely origin (figure 2d). Removing from the male dataset, the populations living in extremely cold regions did not affect the results, returning a similar R2 with a minimal model comprising only linear and cubic distance from the origin (R2=0.20, F2,90=11.46 p<0.001).
Figure 1.
Plot of within-population phenotypic variance versus geographical distance from the best centre of origin for (a) males and (b) females. The solid line refers to the best fit for the complete dataset while the dotted line refers to the dataset without outlier populations. Populations are represented by circles, with outliers highlighted by a filled circle.
Figure 2.
Maps showing the likely area of origin of the out-of-Africa expansion for (a) the full male dataset, (b) the male dataset excluding the two outlier populations from Patagonia, (c) the full female dataset and (d) the female dataset excluding the outlier population from Kenya. Lighter colours represent better fits of the models to the data, and the area containing the most likely centre of origin is highlighted by a green line. The Americas (not displayed) did not contain any likely origin. The areas not investigated as possible origins are shown in grey.
(b) The most informative set of traits
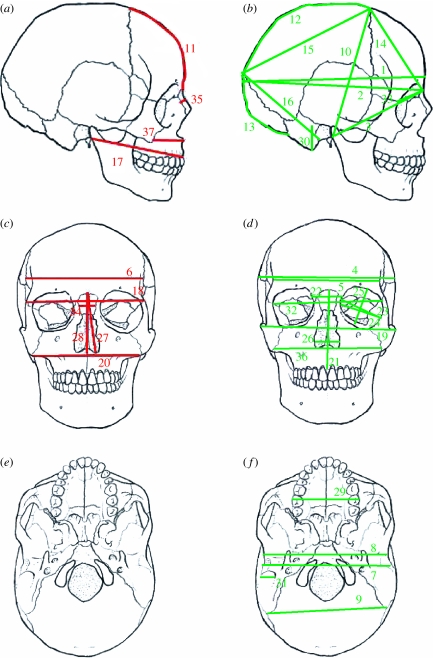
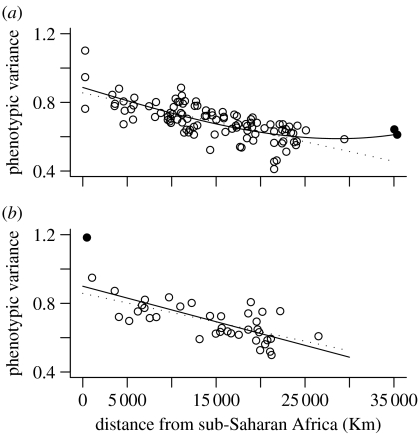
Forward stepwise addition of single cranial traits and backward stepwise elimination from the whole set of traits returned the same combination of 10 cranial variables (figure 3; table S4 in the electronic supplementary material), which provide the best relationship between phenotypic diversity and geographical distance (R2=0.50, F2,102=50.86, p<0.001). This set of traits, when used for the female dataset, explained a very similar proportion of variance (R2=0.53, F1,37=41.76, p<0.001), suggesting that the same traits are informative in the two sexes (figure 4). We also repeated this analysis (table S5 in the electronic supplementary material) on a subset of 19 traits for which heritability is known (Carson 2006). Heritability was a good predictor of the ability of individual traits to recover a clear geographical pattern (heritability was negatively correlated to the order in which traits were selected by the forward stepwise procedure: Kendall's τ=−0.368, p=0.0286; figure S1 in the electronic supplementary material).
Figure 3.
Location of (a,c,e) highly informative traits in red and (b,d,f) less informative traits in green.
Figure 4.
Plot of within-population phenotypic variance versus geographical distance from the best centre of origin, for (a) males and (b) females. The phenotypic variance is calculated over the best combination of traits defined in males. The solid line refers to the best fit for the complete dataset while the dotted line refers to the dataset without outlier populations, which are highlighted by filled circles.
4. Discussion
Our direct test of what determines worldwide human within-population phenotypic variation clearly indicates that distance from Africa, and not climate, plays a role. These results might at first seem at odds with evidence that several traits have been influenced by climate. However, it is important to realize two things. First, selection can change the mean size of traits without affecting their variances. Second, while it is possible to find links between climate and individual traits (Manica et al. 2007), a single multivariate measure of phenotypic diversity should show little relation to climate unless the same climatic variables were affecting a large number of traits in a similar way.
While we used the best available climatic data, these are covering only the last 50 years (Hijmans et al. 2005). Climate has not been constant over the last 50k years, modulating the strength of temperature-mediated natural selection through time. Ideally, one may wish to integrate climatic variation over long time periods when analysing how temperature shaped morphometric traits. However, this is currently impossible. High-quality data on geographical variation in local climate are still unavailable. Moreover, it is unclear how one should integrate such variation over time when modelling morphometric responses. However, this is possibly not as big an issue as it may seem; current climate has repeatedly been shown to capture a large extent of the environmental pressures that affected humans over time as illustrated by the amount of between-population differentiation explained by climate on morphometric (Beals et al. 1984; Roseman 2004; Harvati & Weaver 2006) and genetic (Young et al. 2005; Hancock et al. 2008) traits.
Despite the inherent noise in morphometric data, only two Patagonian populations were outliers to the smooth linear pattern of decreasing phenotypic diversity from sub-Saharan Africa. The peopling of Patagonia is still a debated issue. Some authors have suggested that the Americas have been colonized by two waves, with the second wave having replaced the first, with the possible exception of some populations in South America (including Patagonia), which might be the remnants of the first wave or the result of admixture between the two waves (e.g. Lahr 1995; Neves et al. 1999; Powell & Neves 1999; Gonzalez Jose et al. 2001). The latter scenario would explain the higher than expected diversity observed in our Patagonian samples. Conversely, excluding the 12 populations from very cold regions does not affect the outcome, confirming the robustness of the results to extreme environmental conditions. A strong influence of extremely cold climate on cranial traits has been suggested by various authors (i.e. Hennessy & Stringer 2002; Relethford 2004a; Roseman 2004; Harvati & Weaver 2006; von Cramon-Taubadel & Lycett 2008). von Cramon-Taubadel & Lycett (2008) reported an increase in variance explained by distance Africa after excluding the single population in their dataset that came from an extremely cold environment. However, while suggestive of an effect of cold climate, the removal of a single outlier from a relatively small dataset does not allow reaching any strong conclusion.
The proportion of variance explained by the nonlinear model of distance from Africa represented 21 and 28 per cent, for males and females, respectively, which is in line with the 26 per cent obtained on a much smaller sample size by von Cramon-Taubadel & Lycett (2008) and comparable with the estimates obtained for selected genes (Prugnolle et al. 2005b). Moreover, it is possible to increase the proportion of explained variance to over 50 per cent by selecting a subset of 10 traits. Thus, the whole set of cranial measurements can be far less informative than a subset of traits, in accordance with the conclusions of Harvati & Weaver (2006). This suggests that different traits bear a distinct proportion of genetic information and can be subject to different selective pressures. All the most informative traits were distributed in the anterior region of the cranium (yet not all anterior traits were highly informative). This came as a surprise as the anterior regions of the cranium are generally assumed to be more affected by natural selection (i.e. facial area and cranial breadth), while the temporal bone and some traits of the neurocranium are considered to reflect population history (Beals et al. 1984; Franciscus & Long 1991; Roseman 2004; Harvati & Weaver 2006). Another feature of informative traits was their relatively high heritability. This makes sense if we consider heritability as a measure of genetic information over environmental noise, but contradicts the speculation by Roseman & Weaver (2007) that heritability should not affect the relationship between distance from Africa and phenotypic diversity.
The story told by phenotypic traits bears a striking resemblance to the one based on genetic information. The highest diversity for both types of traits is found in sub-Saharan Africa and declines smoothly as we move away from this hypothetical cradle of humanity. While the relationship for genetic markers is even stronger, by choosing the most informative phenotypic traits we could explain an impressive 50 per cent of within-population diversity without any contribution from climate. It is now time to bury the old adage of ‘bones versus molecules’ and recognize how pervasive the signal from ancient demography can be.
Acknowledgements
L.B. was supported by a travelling studentship from the University of Milan. F.B. acknowledges grant support from the BBSRC.
Supplementary Material
Morphometric traits, phenotypic diversity and R squared explained by individual traits
References
- Anderson D.R., Burnham K.P., White G.C. Comparison of Akaike information criterion and consistent Akaike information criterion for model selection and statistical inference from capture–recapture studies. J. Appl. Stat. 1998;25:263–282. doi:10.1080/02664769823250 [Google Scholar]
- Beals K.L., Smith C.L., Dodd S.M. Brain size, cranial morphology, climate, and time machines. Curr. Anthropol. 1984;25:301–330. doi:10.1086/203138 [Google Scholar]
- Carson E.A. Maximum likelihood estimation of human craniometric heritabilities. Am. J. Phys. Anthropol. 2006;131:169–180. doi: 10.1002/ajpa.20424. doi:10.1002/ajpa.20424 [DOI] [PubMed] [Google Scholar]
- Franciscus R.G., Long J.C. Variation in human nasal height and breadth. Am. J. Phys. Anthropol. 1991;85:419–427. doi: 10.1002/ajpa.1330850406. doi:10.1002/ajpa.1330850406 [DOI] [PubMed] [Google Scholar]
- Gonzalez Jose R., Dahinten S., Hernandez M. The settlement of patagonia: a matrix correlation study. Hum. Biol. 2001;73:233–248. doi:10.1353/hub.2001.0019 [PubMed] [Google Scholar]
- Hancock A.M., Witonsky D.B., Gordon A.S., Eshel G., Pritchard J.K., Coop G., Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. doi: 10.1371/journal.pgen.0040032. doi:10.1371/journal.pgen.0040032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley L.J.L., Manica A., Goudet J., Balloux F. Going the distance: human population genetics in a clinal world. Trends Genet. 2007;23:432–439. doi: 10.1016/j.tig.2007.07.002. doi:10.1016/j.tig.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Hanihara T. Morphological variation of major human populations based on nonmetric dental traits. Am. J. Phys. Anthropol. 2008;136:169–182. doi: 10.1002/ajpa.20792. doi:10.1002/ajpa.20792 [DOI] [PubMed] [Google Scholar]
- Harvati K., Weaver T.D. Human cranial anatomy and the differential preservation of population history and climate signatures. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006;288A:1225–1233. doi: 10.1002/ar.a.20395. doi:10.1002/ar.a.20395 [DOI] [PubMed] [Google Scholar]
- Hennessy R.J., Stringer C.B. Geometric morphometric study of the regional variation of modern human craniofacial form. Am. J. Phys. Anthropol. 2002;117:37–48. doi: 10.1002/ajpa.10005. doi:10.1002/ajpa.10005 [DOI] [PubMed] [Google Scholar]
- Hijmans R.J., Cameron S.E., Parra J.L., Jones P.G., Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. doi:10.1002/joc.1276 [Google Scholar]
- Lahr M.M. Patterns of modern human diversification: implications for Amerindian origins. Yearb. Phys. Anthropol. 1995;38:163–198. doi:10.1002/ajpa.1330380609 [Google Scholar]
- Li J.Z., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. doi:10.1126/science.1153717 [DOI] [PubMed] [Google Scholar]
- Liu H., Prugnolle F., Manica A., Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. doi:10.1086/505436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A., Prugnolle F., Balloux F. Geography is a better determinant of human genetic differentiation than ethnicity. Hum. Genet. 2005;118:366–371. doi: 10.1007/s00439-005-0039-3. doi:10.1007/s00439-005-0039-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A., Amos W., Balloux F., Hanihara T. The effect of ancient population bottlenecks on human phenotypic variation. Nature. 2007;448:346–348. doi: 10.1038/nature05951. doi:10.1038/nature05951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton T., Peterson R., Redd A.J., Saha N., Sofro A.S.M., Martinson J., Stoneking M. Polynesian genetics affinities with southeast Asian populations as identified by mtDNA analysis. Am. J. Hum. Genet. 1995;57:403–414. [PMC free article] [PubMed] [Google Scholar]
- Neves W.A., Powell J.F., Ozolins E.G. Modem human origins as seen from the peripheries. J. Hum. Evol. 1999;37:129–133. doi: 10.1006/jhev.1999.0315. doi:10.1006/jhev.1999.0315 [DOI] [PubMed] [Google Scholar]
- Powell J.F., Neves W.A. Craniofacial morphology of the first Americans: pattern and process in the peopling of the new world. Yearb. Phys. Anthropol. 1999;42:153–188. doi: 10.1002/(sici)1096-8644(1999)110:29+<153::aid-ajpa6>3.3.co;2-c. doi:10.1002/(SICI)1096-8644(1999)110:29+<153::AID-AJPA6>3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Prugnolle F., Manica A., Balloux F. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 2005a;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. doi:10.1016/j.cub.2005.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guegan J.F., Guernier V., Balloux F. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 2005b;15:1022–1027. doi: 10.1016/j.cub.2005.04.050. doi:10.1016/j.cub.2005.04.050 [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA. 2005;102:15 942–15 947. doi: 10.1073/pnas.0507611102. doi:10.1073/pnas.0507611102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford J.H. Boas and beyond: migration and craniometric variation. Am. J. Hum. Biol. 2004a;16:379–386. doi: 10.1002/ajhb.20045. doi:10.1002/ajhb.20045 [DOI] [PubMed] [Google Scholar]
- Relethford J.H. Global patterns of isolation by distance based on genetic and morphological data. Hum. Biol. 2004b;76:499–513. doi: 10.1353/hub.2004.0060. doi:10.1353/hub.2004.0060 [DOI] [PubMed] [Google Scholar]
- Relethford J.H., Blangero J. Detection of differential gene flow from patterns of quantitative variation. Hum. Biol. 1990;62:5–25. [PubMed] [Google Scholar]
- Romero, I. G., Manica, A., Goudet, J., Handley, L. L. & Balloux, F. In press. How accurate is the current picture of human genetic variation? Hered. Adv. (doi:10.1038/hdy.2008.89). [DOI] [PubMed]
- Roseman C.C. Detecting interregionally diversifying natural selection on modern human cranial form by using matched molecular and morphometric data. Proc. Natl Acad. Sci. USA. 2004;101:12 824–12 829. doi: 10.1073/pnas.0402637101. doi:10.1073/pnas.0402637101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman C.C., Weaver T.D. Molecules versus morphology? Not for the human cranium. Bioessays. 2007;29:1185–1188. doi: 10.1002/bies.20678. doi:10.1002/bies.20678 [DOI] [PubMed] [Google Scholar]
- von Cramon-Taubadel N., Lycett S.J. Brief communication: human cranial variation fits iterative founder effect model with African origin. Am. J. Phys. Anthropol. 2008;136:108–113. doi: 10.1002/ajpa.20775. doi:10.1002/ajpa.20775 [DOI] [PubMed] [Google Scholar]
- Young J.H., Chang Y.-P.C., Kim J.D.-O., Chretien J.-P., Klag M.J., Levine M.A., Ruff C.B., Wang N.-Y., Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. doi:10.1371/journal.pgen.0010082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphometric traits, phenotypic diversity and R squared explained by individual traits