Summary
The mitochondrial genome (mtGenome) has been very little studied in the turkey (Meleagris gallopavo), for which there is no publicly available whole genome mitochondrial sequence. Here, we used PCR-based methods with 19 pairs of primers designed from the chicken and other species to develop a complete turkey mtGenome sequence. A total length of 16, 717 bp of the whole turkey mtGenome was obtained, with 85% similarity to chicken mtGenome. There were 13 genes and 24 RNA (22 tRNA and 2 rRNA) annotated. The mtGenome-based phylogenetic analysis suggests that the turkey is most closely related to the chicken, Gallus gallus, and quail, Corturnix japonica. Given the importance of the mitochondria genome, the present work adds to the growing genomic resources needed to define the genetic mechanisms that underlie some economic traits in the turkey.
Keywords: turkey, mitochondrial genome, phylogenetics
Introduction
The turkey, Meleagris gallopavo, is native to North America and exists widely as both an important wild bird and the second most widely consumed poultry meat species in developed countries. It is surprising that the turkey, an ecologically and agriculturally important species, lacks a whole mtGenome sequence, which is one of the reasons why the phylogenetic relationship between the turkey and other avian species, especially gallinaceous birds, is also very little understood. The complete turkey mtGenome sequence could provide an opportunity to use non-recombining sequences with diverse rates of evolution for turkey phylogenetics. The few avian phylogenetic studies that involved the turkey include that by Helm-Bychowski and Wilson (1986), which used restriction enzyme-based maps involving 161 restriction sites to describe relationships among the turkey and six other phasianoids that were different from those by traditional and protein-based comparisons. Using allozymes, Randi et al (1991) grouped phasianoids into two main categories, one including the turkey and the other the quails, with the guinea fowl distantly related to both groups. Using DNA-DNA hybridization, Sibley and Ahlquist (1990) reported, in broad terms, that based on DNA-DNA hybridization data and in contrast to some earlier reports involving morphology as well as mtDNA, galliform birds form two clades one of which include chicken, guinea fowl, Old World quail, and turkey. That work subsequently called for more data to permit resolution of the Phasianidae.
Because of the lack of consensus of the relationships among galliform birds, especially among the chicken, quail, and turkey, phylogenetics studies of these poultry species continue. In a recent study, Kaiser et al. (2007) reported that based on insertion events of CR1 retrotransposable elements, the turkey was more closely related to quail and chicken but distant to the guinea fowl. Despite the use of these diverse genetic markers, our understanding of galliform relationships continues to be marginal and the phylogenies continue to be without congruence. The mtGenome sequence of the turkey will provide access to genes with different evolutionary rates than those that have been used to assess galliform phylogenies.
In addition to its utility for phylogeny of galliform birds, the sequence of the turkey mtGenome could be a useful tool for establishing the influence of the mitochondria and its genes on economically important phenotypes. Here, the primary objective was to sequence and annotate the turkey mtGenome. Further, the validity of the sequence and the utility of this genomic resource were evaluated by inter-species phylogenetic analysis.
Materials and methods
Mitochondrial genome sequencing
Two Blue Slate turkeys were used to develop two whole mtGenome sequences from PCR products or amplicons obtained using heterologous primers. The primers included universal oligos previously described by Sorenson et al. (1999) and those developed for the present work (VT Primers, Table 1). All but four primer-pairs used to generate amplicons that were sequenced are new. In addition to the conventional criteria for selecting primers, oligos were designed and chosen by Primer 3 (Rozen and Skaletsky, 1997) for their ability to produce overlapping amplicons of 2 to 4 kb. Primers were optimized at an annealing temperature of 56°C using the FailSafe™ PCR PreMixes kit according to the manufacturer's recommendation (Epicentre Technology, Madison, WI). Following optimization, the successful premix that produced a single amplicon was used to carry out PCR for the specific primer pair. For each primer-pair, at least two independent PCR products were purified and sequenced using both reverse and forward primers as previously described (Lin et al., 2006). Internal primers were also developed to complete the sequencing of some long-range PCR products. The internal primers also ensured that some regions of the turkey mtGenome were sequenced at least three times including in those in the regions were two or more primers produced overlapping amplicons. The sequences were assembled using a combination of bioinformatics tools including of Phred, Phrap and Consed (Gordon et al., 1998).
Table 1.
Sequences of primers used in the polymerase chain reaction
| Primer ID | Primer sequence |
|---|---|
| TL1F* | 5′-AARCCMGAATGRTAYTTYCTWTTYGC-3′ |
| TL1R* | 5′-GTGGCTGGCACARGATTTACC-3′ |
| TL1in1F | 5′-AACCCGCGTACAAGCTCTAA-3′ |
| TL1in1R | 5′-TCTTCAGTGCCATGCTTTTG-3′ |
| TL1in2F | 5′-TCCTACCCCCAACATCCATA-3′ |
| TL1in2R | 5′-GCTTAAGGTTAATTACTGCTGAATACC-3′ |
| TL2F* | 5′-YAAAGCATGRCACTGAA-3′ |
| TL2R* | 5′-TYTCAGGYGTARGCTGARTGCTT-3′ |
| TL3Fnew | 5′-GCCCTTGGAAGGAGGATTTA-3′ |
| TL3Rnew | 5′-CAGTTCTGCACGGATTAGCA-3′ |
| TL3in1F* | 5′-CAACCGTACCGTAAGGGAAA-3′ |
| TL3in1R | 5′-CGTCTGGTTTGCACTCAGAA-3′ |
| TL3in2F | 5′-AGCCCCCTCGAAAAAGAATA-3′ |
| TL3in2R | 5′-AGGCCGGCTAGAGATAGGAG-3′ |
| TL3in3F | 5′-GTGTTCTCGTGCAAAAACGA-3′ |
| TL3in3R | 5′-GGTGGTGGGATTTTGAGATG-3′ |
| TL3in4F | 5′-CTCGGCAAATGCAAAAGACT-3′ |
| TL3in4R | 5′-TGGGAGGTTCAGGAAACTTG-3′ |
| TL4F* | 5′-CCYCTGTAAAAAGGWCTACAGCC-3′ |
| TL4in2F | 5′-CATAAAACCCCCAGCACTGT-3′ |
| TL4Rnew2 | 5′-TAATTTGCTGGGTCGAAACC-3′ |
| TL4in3F | 5′-TGGAGGTCTTACGGGAATTG-3′ |
| TL4in3R | 5′-GGGTTGTTTGAGCGAGAAGA-3′ |
| TL4in4F | 5′-GAAGGAATCGAACCCTCACA-3′ |
| TL4in4R | 5′-CTGCTTTCGGTTTCCTTCTG-3′ |
| TL4in5F | 5′-GCCTGATCCTCCCTCCTATC-3′ |
| TL4in5R | 5′-ATGTCCGGCTGTAAGGTTTG-3′ |
| TL5Fnew | 5′-CAAACAACCCCAGACACAGA-3′ |
| TL5Rnew | 5′-GGCTGAGTAGGAAGGCAGTTT-3′ |
| TL5in1F | 5′-AAAACCAAACCCCATCCTTC-3′ |
| TL5in1R | 5′-GGGTTGTAGGCCTCGTGTAA-3′ |
| TL6F* | 5′-ATCCRTTGGTCTTAGGARCCA-3′ |
| TL6R* | 5′-CTTCANTYTTTGGYTTACAAGRCC-3′ |
| TL6in1F | 5′-ACAAGCAATCCAACCAGACC-3′ |
| TL6in1R | 5′-GTTTGGGATTGAGCGTAGGA-3′ |
| TL6in2F | 5′-TCCGCATGACACTGCTAGTC-3′ |
| TL6in2R | 5′-GATGAAGAAGAATGAGGCGC-3′ |
Universal primers described by (Sorenson et al., 1999) were also used.
Sequence validation and annotation
The whole genome sequence was validated at two levels: multiple sequencing of each region and sequence comparison with GenBank mtGenome sequences from other birds. An additional validation of the turkey mitochondrial DNA sequences was based on sequence similarity as revealed by a Clustal-X (Thompson et al., 1997) based multi-alignment using mitochondrial DNA sequences publicly available for Coturnix japonica (NC_003408), Numida meleagris (NC_006308), and Gallus gallus gallus (NC_007236). To annotate the sequence, BLAST 2 (Tatusova & Madden, 1999) and GeneDoc (Nicholas et al., 1997) were used to compare the assembled sequence to the database of mtGenome sequences. Additionally, ORF-Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and vsfold4 (http://www.rna.itchiba.ac.jp/∼vsfold/vsfold4/), a DNA sequence viewer and annotation tool and an RNA secondary structure prediction program, respectively, were also used to further annotate and/or validate the BLAST 2 and GeneDoc annotation of the turkey mitochondrial DNA sequence.
Phylogenetic analysis
Using the rationale that different segments of the mtGenome undergo varying rates of evolution, the two phylogenetic analyses carried out to evaluate the turkey's relationship with other birds were based on the 16S rRNA and the coding region that included 12 protein-coding genes but not ND6 which is encoded on a different strand, two rRNA genes and 19 tRNA genes. A total of 57 species, including the turkey, chicken, quail, and guineafowl were used (supplementary Table S1). In the 16S rRNA-based phylogenetic analysis, the American alligator and human sequences were used as outgroups for rooting. Based on the results of 16S rRNA phylogenetic analysis, Anseriformes were selected as outgroup for the coding region-based phylogenetic analysis. The outgroups were selected based on conventional criterion and as advanced by Caspers et al. (1997) and van Tuinen et al. (2000). Following Clustal-X (Thompson et al., 1997) based multiple sequence alignment, phylogenetic analysis, tree construction, model selection and statistical tests were carried out as described above. One thousand bootstrap replicates were used to assess the confidence in the grouping in minimum evolution, neighbor-joining, and maximum parsimony methods (Felsenstein & Kishino, 1993). Again, using the Akaike information criterion as the basis for selecting models for the 16S rRNA and coding region phylogenetic analyses, the General Time Reversible + Gamma + Proportion Invariant (GTR+G+I) model of evolution were selected with gamma distribution shape parameter of 0.6002 and 0.9201, respectively.
Results
Full mtDNA sequence of the turkey
A total sequence length of 16,717 bp, representing the BS turkey mtGenome was sequenced, validated and annotated. The sequence has been submitted to GenBank and assigned accession number EF153719. The sequence showed 85, 84 and 83% average similarity with the chicken, Japanese quail and guinea fowl mtGenome sequences in GenBank, respectively. The length of turkey mtGenome was similar as that of the chicken, Japanese quail and guinea fowl mtGenomes that are 16,785, 16,697 and 16,726 bp, respectively. The variable region or the D-loop was 1,164 bp, which was homologous to the only turkey mtDNA sequences available in GenBank from previous investigations including those by Drovetski (2002), Lucchini et al. (2001), and Mock et al. (2001).
Within the turkey mtGenome sequence, the most frequent nucleotide in the H-strand was A, followed by C, T and G. In the chicken, the most frequent nucleotide is a C, followed by A, T, and G. The GC content of 43.5% observed in the turkey sequence is also consistent with that reported in other birds ranging from Apteryx hastii to Aythya Americana with 42 and 48%, respectively. Though the GC content of the turkey mtGenome appears to be consistent with the average in vertebrates, it is lower than the chicken (46%) and goose (47%).
As expected, 13 protein coding genes and 24 RNA (22 tRNA and 2 rRNA) were identified within the turkey mtGenome sequence (Table 2). While 12 of the protein coding genes were located on the H strand, the gene for NADH dehydrogenase subunit 6 (ND6) was on the L-strand. On the other hand, while the sequences for the two rRNAs (12S and 16S) were on the H-strand, those for the 9 tRNAs were located on the L-strand. These results are consistent with the mtGenome organization in some vertebrate species (Pereira, 2000). A characteristic that was found previously in only 46 other avian mtGenomes (Mindell et al., 1998a) was an extra base at position 174 of the gene for NADH, subunit 3 (ND3). It was suggested by Harlid et al. (1997) that the additional nucleotide causes a reading frame change which results in multiple stop codons in the ND3 gene sequence. However, since the frame shifting does not affect ND3 function, Mindell et al. (1998a) hypothesized that birds might have, as yet unknown, a mechanism such as translational frame shifting or RNA editing to correct this anomaly.
Table 2.
Sequence annotation of the mitochondrial genome of the turkey, Meleagris gallopavo.
| Genes | Location | Size (bp) | Initial Codon | Terminal Codon |
|---|---|---|---|---|
| tRNA-Phe | 1-68 | 68 | ||
| 12S ribosomal RNA (12S rRNA) | 69-1040 | 971 | ||
| tRNA-Val | 1041-1113 | 73 | ||
| 16S ribosomal RNA (16S rRNA) | 1114-2731 | 1618 | ||
| tRNA-Leu | 2732-2805 | 74 | ||
| NADH dehydrogenase subunit 1 (ND1) | 2821-3795 | 975 | ATG | TAA |
| tRNA-Ile | 3796-3867 | 72 | ||
| tRNA-Gln | 3875-3945* | 71 | ||
| tRNA-Met | 3945-4013 | 69 | ||
| NADH dehydrogenase subunit 2 (ND2) | 4014-5054 | 1041 | ATG | TAG |
| tRNA-Trp | 5053-5131 | 79 | ||
| tRNA-Ala | 5138-5206* | 69 | ||
| tRNA-Asn | 5209-5280* | 72 | ||
| tRNA-Cys | 5283-5347* | 65 | ||
| tRNA-Tyr | 5347-5417* | 71 | ||
| Cytochrome oxidase subunit 1 (COX1) | 5419-6969 | 1551 | GTG | AGG |
| tRNA-Ser | 6961-7035* | 75 | ||
| tRNA-Asp | 7038-7106 | 69 | ||
| Cytochrome oxidase subunit 2 (COX2) | 7108-7791 | 684 | ATG | TAA |
| tRNA-Lys | 7793-7861 | 69 | ||
| ATPase subunit 8 (ATPase8) | 7863-8027 | 165 | ATG | TAA |
| ATPase subunit 6 (ATPase6) | 8018-8701 | 684 | ATG | TAA |
| Cytochrome oxidase subunit 3 (COX3) | 8701-9487 | 787 | ATG | TGC |
| tRNA-Gly | 9485-9553 | 69 | ||
| NADH dehydrogenase subunit 3 (ND3) | 9554-9905 | 352 | ATG | TAA |
| tRNA-Arg | 9907-9974 | 68 | ||
| NADH dehydrogenase subunit 4 light-chain (ND4L) | 9975-10271 | 297 | ATG | TAA |
| NADH dehydrogenase subunit 4 (ND4) | 10265-11645 | 1381 | ATG | TGC |
| tRNA-His | 11643-11711 | 69 | ||
| tRNA-Ser | 11713-11777 | 65 | ||
| tRNA-Leu | 11779-11849 | 71 | ||
| NADH dehydrogenase subunit 5 (ND5) | 11850-13667 | 1818 | ATG | TAA |
| Cytochrome b (Cytb) | 13671-14813 | 1143 | ATG | TAA |
| tRNA-Thr | 14816-14884 | 69 | ||
| tRNA-Pro | 14887-14956* | 70 | ||
| NADH dehydrogenase subunit 6 (ND6) | 14964-15484* | 521 | ATG | TAG |
| tRNA-Glu | 15486-15553* | 68 |
Coded on the complementary (L) strand.
Phylogenetic analyses
The minimum evolution, maximum likelihood, maximum parsimony, and neighbor-joining trees, constructed from both 16S rRNA- and the combined sequences of 12 protein-coding genes, two rRNA genes and 19 tRNA genes were congruent and showed a closer relationship between the turkey, chicken, and quail but relatively more distant to the guinea fowl, also a gallinaceous bird (supplementary Figures 1 and Figure 2). Except for the relationship between the turkey and the guinea fowl, those defined here among the galliformes are consistent with previous reports, thus providing additional support for the vailidity of the sequence described here (Kimball et al., 1999; Sibley and Ahlquist, 1990).
Discussion
The animal mtGenome is generally considered to be under selection for both small size and a conserved gene order (Rand & Harrison, 1986; Quinn & Wilson, 1993; Boore, 1999). Animal mitochondrial genomes rarely contain either introns or intergenic spacers (Quinn and Wilson 1993, McKnight and Shaffer 1997). The turkey, an important agricultural and model avian species remains one of many birds for which there is no publicly available whole mtGenome DNA sequence. Here, we have described the turkey mtGenome sequence and showed that it is similar in length and nucleotide composition to that of most other birds. Compared to mammalian species, only a limited number of birds, which generally exceed mammals in the total number of species, have had their whole mtGenome sequenced. While the mtGenome gene content and gene order is remarkably stable across vertebrate species, the avian species are an exception to this stability. For example, the avian species have several unusual features in their mitochondrial DNA, such as the lack of the traditional origin of replication for the light strand and the as-yet unidentified splicing function to repair the one-base insertion found in the ND3 gene in most avian mtGenomes. Several gene order rearrangements have occurred in avian mtGenomes, primarily affecting the area around the ND6 gene. For instance, the genus Amazona has two duplicated control regions which are found between tRNAGlu and tRNAPhe (Eberhard et al. 2001). Other birds including the forest falcon, kestrel, and the woodpecker, have a mitochondrial genome with a different gene order that involves two control regions: one between tRNAThr and tRNAPro, and a second region between tRNAGlu and tRNAPhe (Mindell et al. 1998b).
The new sequence was also used to evaluate the phylogenetic relationships between the turkey and other birds. To date, the most extensive comparisons of Galliformes have involved partial sequences from mitochondrial (Dimcheff et al., 2002) and nuclear genes (Smith et al., 2005; Kaiser et al., 2007) as well as the hybridization results of Sibley and Ahlquist (1990). The relationships between the turkey and other Galliformes were in general agreement with Sibley and Ahlquist (1990), Smith et al (2005) and Dimcheff et al. (2000, 2002). Briefly, these studies showed the turkey as a sister to clades containing Gallus and Coturnix species. Further, and as observed here, Numida was basal to the Coturnix and Gallus clades but more distant to the turkey. The high bootstrap values provide strong support for the relationships defined here between the turkey and other Galliformes.
Though the phylogenetic analysis done here was to provide examples of the potential of the turkey mtGenome sequence and validation of the sequence quality, it should be noted that the placement of G. varius using GenBank sequences described by others appears to be inconsistent with some previous studies. Helm-Bychowski and Wilson (1986) and Fumihito et al (1996) showed that G. varius should not be more closely related to G. gallus than the Ceylon or Gray Junglefowl. It is possible that the G. varius (GenBank accession number: NC_007238) sequence was from a sample contaminated with domestic chicken DNA or the amplified samples were mixed up and assembled incorrectly.
In summary, the turkey mtGenome sequence was developed and used to evaluate the genetic relatedness between the turkey and other birds. Though our analyses of relationships among birds were limited in the extent of statistical parameters included, the mtGenome sequence provides a resource for extensive phylogenetic analyses. Since the evolutionary relationships among Galliformes continues to be without a general consensus, the whole mtGenome sequence described here will provide an additional tool for generating more data needed to understand the turkey's relationship with other gallinaceous birds. The sequence will also facilitate assignment of function to the mtGenome, especially the role of mitochondrial genes in variation in economically important phenotypes in the turkey. Furthermore, it will provide a foundation to begin to more widely evaluate the role of the genome of this important organelle in the turkey.
Supplementary Material
Table S1: Scientific and common names of birds, for which whole mitochondrial genome sequences are available in GenBank, used to evaluate the newly developed sequence for phylogentic analyses of the turkey
Figure 1.
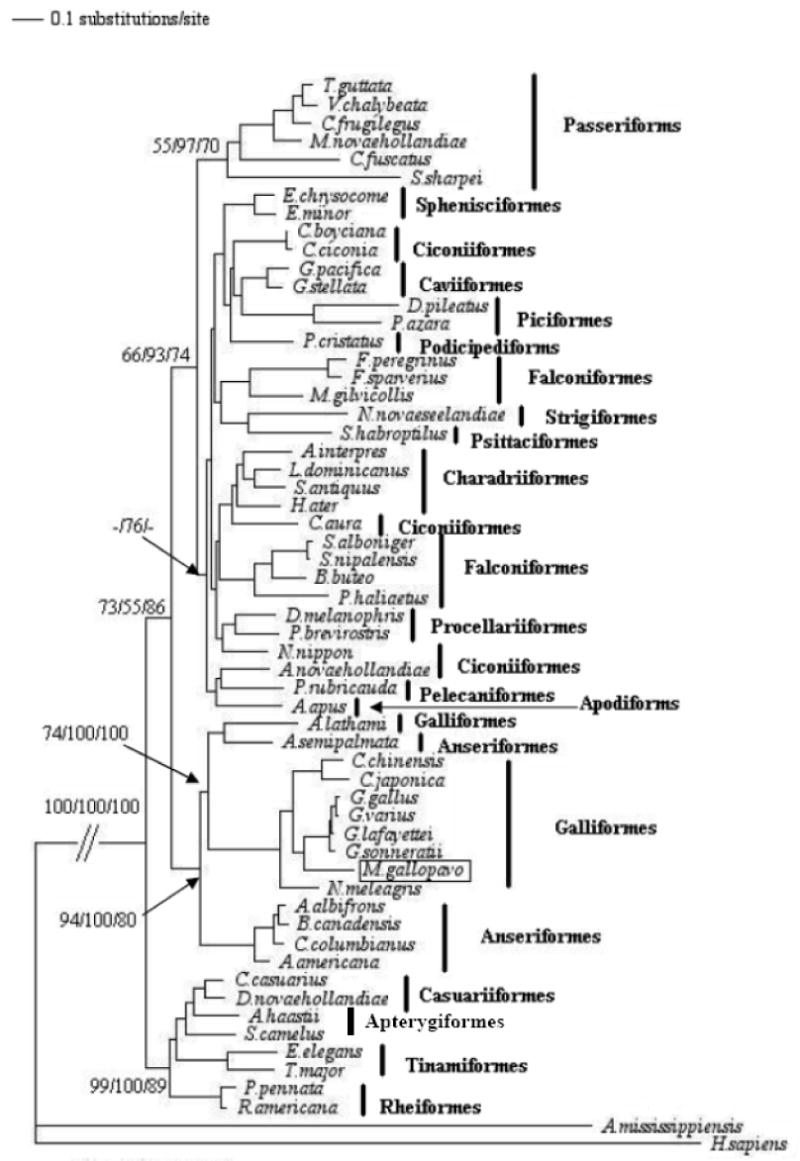
A maximum likelihood-based phylogenetic tree constructed using 16S rRNA from 56 avian species described in Supplementary Table S3. The tree was rooted using A. mississippiensis (American alligator) and H. sapiens (human). The tree was congruent with those from neighbor joining, minimum evolution, and maximum parsimony methods. Confidence of the groupings was estimated using 1000 bootstrap replications. The Arabic numeral at the base of a node is the bootstrap value. The Arabic numerals at the base of a node are the bootstrap values derived from the maximum parsimony, neighbor-joining, and minimum-evolution analysis, respectively. Bootstrap values lower than 50% are not shown.
Figure 2.
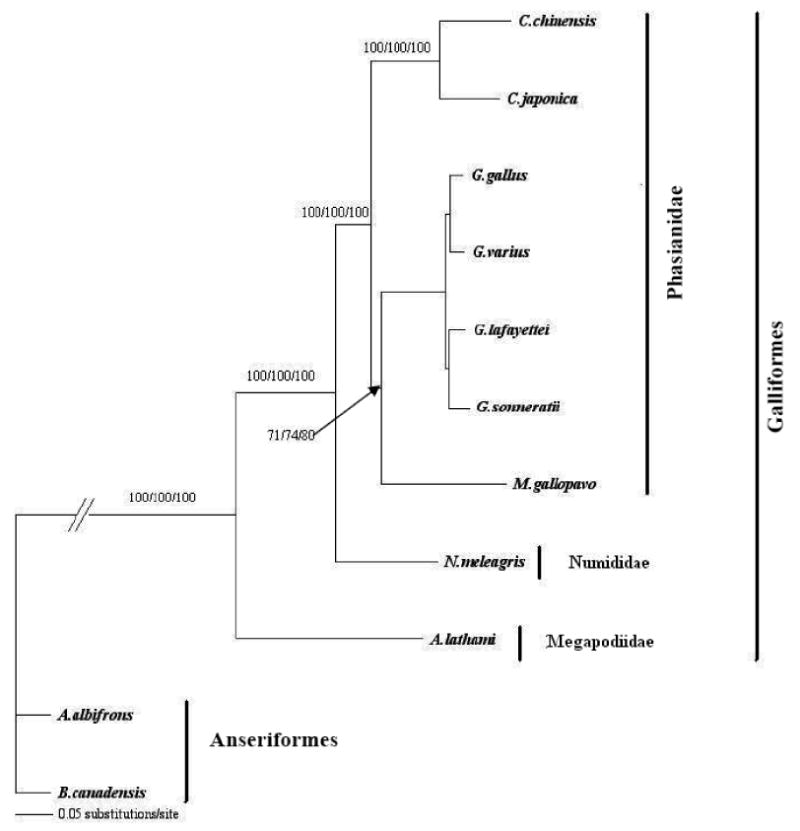
A maximum likelihood phylogenetic tree based on the mitochondrial coding region that included 12 protein-coding genes, two rRNA genes, and 19 tRNA genes from Galliformes. Anserigormes were used as outgroup for rooting. The tree was congruent with those from neighbor joining, minimum evolution, and maximum parsimony methods. Confidence of the groupings was estimated using 1000 bootstrap replications. The Arabic numerals at the base of a node are the bootstrap values derived from the maximum parsimony, neighbor-joining, and minimum-evolution analysis, respectively. Bootstrap values lower than 50% are not shown. Though the tree shows a G. varius relationship that is consistent with the amino acid tree of Nishibori et al. (2005), it is not congruent with those of Helm-Bychowski and Wilson (1986) and Fumihito et al. (1996).
Acknowledgments
We are grateful to technicians at the Virginia Tech Poultry Farm for helping with sample collections, and to Dr. Gliss Smith for editorial help. Funding for this work was provided in part by the National Institutes of Aging, NIH, and the Virginia Ag. Council.
References
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Research. 1999;27:1767–80. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers GJ, Weerd DU, Wattel J, Jong WW. a-Crystallin Sequences Support a Galliform/Anseriform Clade. Molecular Phylogenetics and Evolution. 1997;7:185–88. doi: 10.1006/mpev.1996.0384. [DOI] [PubMed] [Google Scholar]
- Dimcheff DE, Drovetski SV, Krishnan M, Mindell DP. Cospeciation and horizontal transmission of Avian Sarcoma and Leukosis Virus gag genes in galliform birds. Journal of Virology. 2000;74:3984–95. doi: 10.1128/jvi.74.9.3984-3995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimcheff DE, Drovetski SV, Mindell DP. Molecular evolution and systematics of tetraoninae and other Galliformes using mitochondrial 12S and ND2 genes. Molecular Phylogenetics and Evolution. 2002;24:203–15. doi: 10.1016/s1055-7903(02)00230-0. [DOI] [PubMed] [Google Scholar]
- Drovetski SV. Molecular phylogeny of grouse: individual and combined performance of W-linked, autosomal, and mitochondrial loci. Systematic Biology. 2002;51:930–45. doi: 10.1080/10635150290102500. [DOI] [PubMed] [Google Scholar]
- Eberhard JR, Wright TF, Bermingham E. Duplication and Concerted Evolution of the Mitochondrial Control Region in the Parrot Genus Amazona. Molecular Biology and Evolution. 2001;18:1330–42. doi: 10.1093/oxfordjournals.molbev.a003917. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary tree from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–76. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J, Kishino H. Is there something wrong with the bootstrap on phylogenies? A reply to Hillis and Bull. Systematic Biology. 1993;42:193–200. [Google Scholar]
- Fumihito A, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, Kondo N, Ohno S. Monophyletic origin and unique dispersal patterns of domestic fowls. Proceedings of National Academy of Sciences of the United States of America. 1996;93:6792–95. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Research. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Harlid A, Janke A, Arnason U. The mtDNA sequence of the ostrich and the divergence between paleognathous and neognathous birds. Molecular Biology and Evolution. 1997;14:754–61. doi: 10.1093/oxfordjournals.molbev.a025815. [DOI] [PubMed] [Google Scholar]
- Helm-Bychowski KM, Wilson AC. Rates of nuclear DNA evolution in pheasant-like birds: evidence from restriction maps. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:688–92. doi: 10.1073/pnas.83.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, van Tuinen M, Ellegren H. Insertion events of CR1 retrotransposable elements elucidate the phylogenetic branching order in galliform birds. Molecular Biology and Evolution. 2007;24:338–47. doi: 10.1093/molbev/msl164. [DOI] [PubMed] [Google Scholar]
- Kimball RT, Braun EL, Zwartjes PW, Crowe TM, Ligon JD. A molecular phylogeny of the pheasants and partridges suggests that these lineages are not monophyletic. Molecular Phylogenetics and Evolution. 1999;11:38–54. doi: 10.1006/mpev.1998.0562. [DOI] [PubMed] [Google Scholar]
- Lin KC, Xu J, Gyenai KB, Pyle R, Smith EJ. Candidate gene expression analysis of toxin-induced dilated cardiomyopathy in the turkey, Meleagris gallopavo. Poultry Science. 2006;85:2216–21. doi: 10.1093/ps/85.12.2216. [DOI] [PubMed] [Google Scholar]
- Lucchini V, Hoglund J, Klaus S, Swenson J, Randi E. Historical biogeography and a mitochondrial DNA phylogeny of grouse and ptarmigan. Molecular Phylogenetics and Evolution. 2001;20:149–62. doi: 10.1006/mpev.2001.0943. [DOI] [PubMed] [Google Scholar]
- McKnight ML, Shaffer HB. Large, rapidly evolving intergenic spacers in the mitochondrial DNA of the salamander family Ambystomatidae (Amphibia: Caudata) Molecular Biology and Evolution. 1997;14:1167–76. doi: 10.1093/oxfordjournals.molbev.a025726. [DOI] [PubMed] [Google Scholar]
- Mindell DP, Sorenson MD, Dimcheff DE. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Molecular Biology and Evolution. 1998a;15:1568–71. doi: 10.1093/oxfordjournals.molbev.a025884. [DOI] [PubMed] [Google Scholar]
- Mindell DP, Sorenson MD, Dimcheff DE. Multiple independent origins of mitochondrial gene order in birds. Proceedings of the National Academy of Sciences USA. 1998b;95:10693–97. doi: 10.1073/pnas.95.18.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock KE, Theimer TC, Wakeling BF, Rhodes OE, Jr, Greenberg DL, Keim P. Verifying the origins of a reintroduced population of Gould's wild turkey. The Journal of Wildlife Management. 2001;65:871–79. [Google Scholar]
- Nicholas KB, Nicholas HB, Deerfield DW. GeneDoc: analysis and visualization of genetic variation. EMBnet News 1997 [Google Scholar]
- Nishibori M, Shimogiri T, Hayashi T, Yasue H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Animal Genetics. 2005;36:367–375. doi: 10.1111/j.1365-2052.2005.01318.x. [DOI] [PubMed] [Google Scholar]
- Niu D, Fu Y, Luo J, Ruan H, Yu X, Chen G, Zhang Y. The Origin and Genetic Diversity of Chinese Native Chicken Breeds. Biochemical Genetics. 2002;40:163–74. doi: 10.1023/a:1015832108669. [DOI] [PubMed] [Google Scholar]
- Pereira SL. Mitochondrial genome organization and vertebrate phylogenetics. Genetics and Molecular Biology. 2000;23:4. [Google Scholar]
- Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Research. 2006;34:W700–3. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TW, Wilson AC. Sequence evolution in and around the mitochondrial control region in birds. Journal of molecular evolution. 1993;37:417–25. doi: 10.1007/BF00178871. [DOI] [PubMed] [Google Scholar]
- Rand DM, Harrison RG. Mitochondrial DNA transmission genetics in crickets. Genetics. 1986;114:955–70. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E, Fusco G, Lorenzini R, Crowe TM. Phylogenetic relationships and rates of allozyme evolution within the Phasianidae. Biochemical Systematics and Ecology. 1991;19:213–21. [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3. 1997 doi: 10.1385/1-59259-192-2:365. Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.html. [DOI] [PubMed]
- Sibley CG, Ahlquist JE. The phylogeny and classification of birds: a study in molecular evolution. New Haven: Yale University Press; 1990. [Google Scholar]
- Smith EJ, Shi L, Tu Z. Gallus gallus aggrecan gene-based phylogenetic analysis of selected avian taxonomic groups. Genetica. 2005;124:23–32. doi: 10.1007/s10709-004-5184-4. [DOI] [PubMed] [Google Scholar]
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. Primers for a PCR based approach to mitochondrial genome sequencing in birds and other vertebrates. Molecular Phylogenetics and Evolution. 1999;12:105–14. doi: 10.1006/mpev.1998.0602. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0. Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences - a new tool for comparing protein and nucleotide sequences. FEMS microbiology letters. 1999;174:247–50. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuinen M, Sibley CG, Hedges SB. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Molecular Biology and Evolution. 2000;17:451–57. doi: 10.1093/oxfordjournals.molbev.a026324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Scientific and common names of birds, for which whole mitochondrial genome sequences are available in GenBank, used to evaluate the newly developed sequence for phylogentic analyses of the turkey


