Abstract
Cell proliferation studies are an important experimental tool. The most commonly used thymidine analogues, tritiated thymidine and BrdU label cells during S-phase. Both methods have significant drawbacks; low sensitivity in the case of tritiated thymidine and a denaturation step during BrdU detection that destroys most cellular epitopes, requiring careful optimization. The antibody against BrdU is also large and tissue penetration can be difficult. EdU is a closely chemically related to BrdU, with detection achieved by a copper catalyzed reaction requiring a small fluorescently conjugated azide. Cell cultures, flow cytometry and high throughput studies using EdU labeled cells is exceptionally fast and does not require denaturation or antibodies. We have developed a tissue labeling technique in chick embryos using EdU. Following EdU chemistry to detect proliferating cells the tissue can undergo immunolabeling. We demonstrate fluorescent EdU chemistry followed by Tuj1 antibody staining resulting in multiplex fluorescent tissues.
Keywords: BrdU, chick, development, EdU, neural tube, otocyst, proliferation, Tuj1
Introduction
Labeling of proliferating cells is an important technique used by many researchers. Two methods of directly labeling DNA in proliferating cells have been extensively reported: BrdU (5’-bromo-2’-deoxyuridine), a thymine analogue that is incorporated into the proliferating DNA during S-phase of cell division (Gratzner, 1982) and the classical method of [3H]thymidine labeling, which is significantly less sensitive than BrdU. Detection of PCNA (a DNA polymerase cofactor) or phospho-histone H3 (late G2 and M phase) proteins present in mitotic cells using antibody staining are also often utilized as indirect methods of cell proliferation detection. Detection of BrdU incorporated into live cells in culture or whole mount tissues of embryos involves fixation and denaturation of the DNA, immunological processing and colometric/fluorescent visualization. This results in an assay that, in principle, can be done in less than twenty-four hours to measure proliferation of cells. However, BrdU detection is a technical challenge, reliant on a balance between the amount of DNA denaturation, overall epitope destruction and successful antibody binding. Furthermore, BrdU has dose dependent toxic effects on cells, for example, Tuj1 positive neurons (Caldwell et al., 2005) and the protocol has to be carefully optimized to reduce toxic effects, while still enabling detection of the incorporated BrdU.
The Click-iT™ EdU assay (Invitrogen) uses a unique copper catalyzed cycloaddition reaction. A chemical reaction occurs between the terminal alkyne group of EdU (5’-ethynyl-2’-deoxyuridine) and a small fluorophore-conjugated azide molecule. This reaction is used to detect the incorporated thymidine analog in intact double stranded DNA. The copper catalyzed click reaction was first described independently by two groups (Rostovtsev et al., 2002; Tornoe et al., 2002) and has since proven extremely useful in cell culture, flow cytometry and high throughput assays. The use of a large antibody molecule such as anti-BrdU and requisite DNA denaturation step during detection is completely circumvented by the click reaction. Recent reports highlight the use of EdU proliferation studies in cell culture (Buck et al., 2008; Cappella et al., 2008) and intraperitoneal injection into mice (Salic and Mitchison, 2008). EdU detection in cultured cells is easier to perform than immunochemistry and may take as little as 30−90 minutes. We have developed a pulse labeling whole mount and histological protocol using chick embryos in ovo and determined a detection protocol that takes less than 4 hours to complete. This fast, reliable, reproducible and cost effective protocol is a viable alternative to other labeling methods for researchers using embryonic tissues. Moreover, we show that, at least in the case of Tuj1, that antibody immunostaining following EdU chemistry is viable. This is a major benefit of this technique over BrdU, where the denaturation generally precludes double immunochemistry.
Results and Discussion
EdU labeling of proliferating cells in whole mount and sectioned embryos
First, we determined the concentration of EdU needed to label whole mount chick embryos. The manufacturer's protocol recommends 10 μM EdU concentration for cell cultures. Eggs were incubated to 72 hours (E3), windowed and had 400 μl of EdU solution added onto the embryo. Eggs were resealed and incubated (pulsed) for a further 4 hours before harvesting and processing.
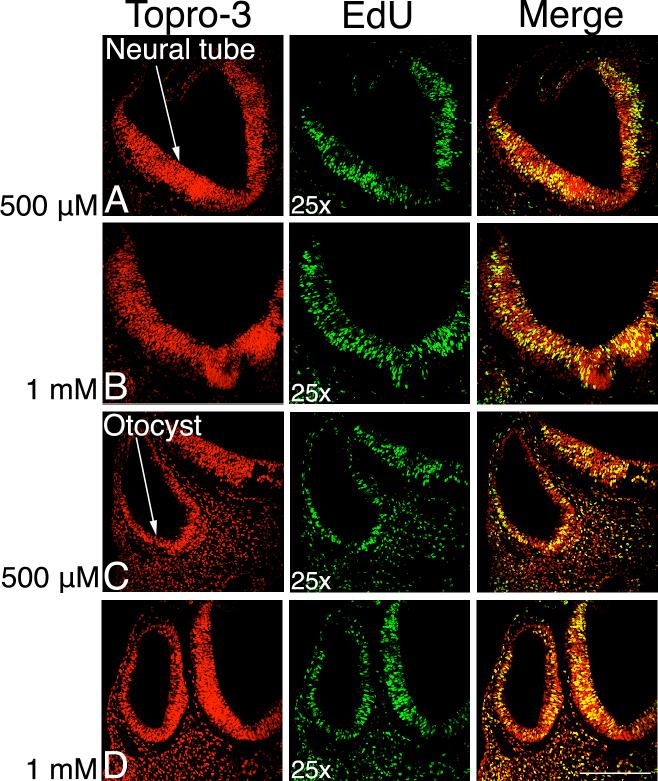
Neither 10 μM (n=0/10) nor 50 μM (n=0/3) labeled any dividing cells (not shown). We attribute this to the different requirements for labeling whole mount embryos with their various surrounding membranes and surface epithelium versus more accessible individual cells in culture. We therefore tested an increased concentration of EdU to compensate for whole mount labeling. We added 400 μl of either a 500 μM, 1 mM or 2 mM solution of EdU to embryos during a 4-hour pulse. In each case, all paraffin labeled sections showed labeling of proliferating cells: 500 μM (n=8/8, Fig. 1A, C), 1 mM (n=8/8, Fig. 1B, D) or 2 mM (n=6/6, not shown). We compared the neural tube and otocyst of E3 embryos and found that at each of the three doses that proliferating cells were labeled.
Figure 1. EdU labels proliferating cells in day 3 chick embryonic tissue.
Dorsal to the top, 10 μm paraffin sections labeled with TO-PRO-3, a DNA marker labeling every cell, in red. EdU is in green, with the merged images in the right hand column. The yellow color in the merged image shows the proliferating cells. Rows A and C labeled with 500 μM EdU, B and D with 1 mM. The neural tube (nt) and otocyst (o) are labeled. Scale bar: 200 μm.
The fluorescence from EdU labeled cells is extremely bright and easy to detect using a confocal or fluorescent microscope. Salic and Mitchison shown that BrdU fluorescence requires five times longer exposure to achieve fluorescence comparable to EdU (Salic and Mitchison, 2008). In our hands, BrdU produces either no staining or inconsistent results (data not shown), in addition to taking two days to complete the immunostaining process, whereas the EdU protocol gives consistent results in under 4 hours.
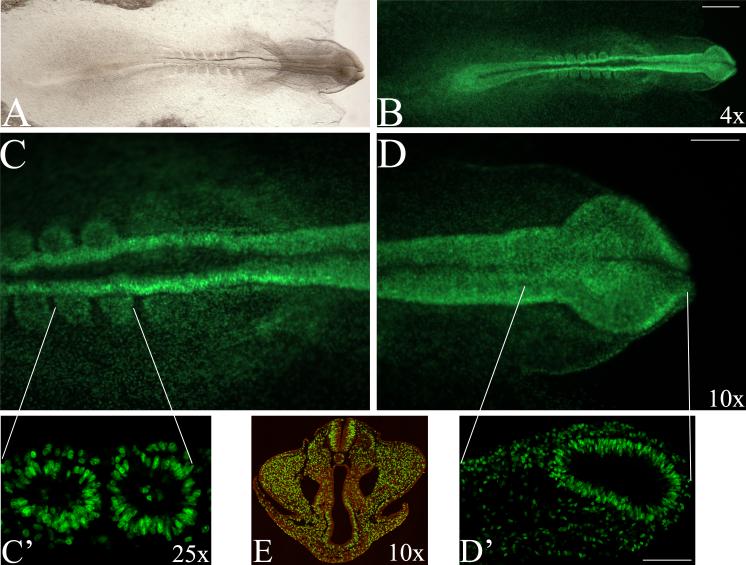
In addition to the labeling of histological sections, we determined that whole mount chick embryos up to stage 18 could be directly processed and visualized (Fig. 2A-D). Embryos were then either cryosectioned (Fig. 2C’, D’) or embedded in paraffin and sectioned (Fig. 2E). In both cases, the fluorescence survived the processing and sections were visualized on a fluorescence stereomicroscope or confocal microscope. In embryos older than 72 hours, EdU did not always fully penetrate the tissue in whole mount embryos (not shown), thus processing for EdU labeling in sectioned tissue is indicated in this case.
Figure 2. Whole mount labeling of chick embryos.
Head to the right, dorsal view. (A, B) Whole mount stage 9 embryo, light and fluorescence images. All green cells are EdU positive. (C, D) Higher magnification view of hindbrain/somites and head using stereomicroscope with respective confocal images of the same regions of the cryosectioned embryo (C’, D’). (C’) Two somites in embryo sagittally sectioned, (D’) forebrain region. The fluorescence survives cryosectioning or paraffin embedding and sectioning. Level of section shown indicated by white lines. (E). Stage 18 embryo labeled as a whole mount with EdU and TO-PRO-3, embedded in paraffin, sectioned and imaged at 25× on a confocal microscope. Scale bars: (A, B) 500 μm, (C-E) 150 μm, (C’, D’) 150 μm.
Increasing doses of EdU labels similar numbers of cells
We analyzed the number of labeled cells in chick neural tubes from 500 μM, 1 and 2 mM EdU doses in E3, 4-hour pulsed, embryos (compare Fig. 1A and B). Our results suggest that increasing EdU dose does not alter numbers of labeled cells in a statistically significant manner (Table 1), suggesting that 500 μM is a saturating dose. EdU used in three days long pulse experiments in cell culture showed mildly increased anti-proliferative activity compared to BrdU (Cappella et al., 2008). However, in a pulse and chase paradigm (a one-hour pulse, followed by three-day chase), anti-proliferative activity was negligible compared to BrdU (Cappella et al., 2008). Previous reports also indicated that BrdU is selectively toxic to Tuj1 positive neurons in culture in line with the administered dose (Caldwell et al., 2005). However, in their in vivo study in adult mice, Cappella and coworkers detected no signs of EdU toxicity (Cappella et al., 2008).
Table 1.
Analysis of EdU labeled cell counts
| Dose | Number of embryos | Number of sections | Cells/unit area | Standard error | Statistically significant |
|---|---|---|---|---|---|
| 500 μM | 3 | 11 | 22.48 | 6.65 | No |
| 1 mM | 5 | 12 | 28.48 | 6.14 | No |
| 2 mM | 2 | 11 | 14.39 | 7.66 | No |
EdU specifically labels proliferating cells
We compared EdU labeling specificity in E3 otocysts with that reported by Lang and coworkers and find that EdU labeling was comparable to that reported for BrdU (Lang et al., 2000). This comparison suggests that EdU is at least as good at labeling proliferating cells as BrdU. This finding is strengthened by Buck and coworkers showing that EdU and BrdU exhibit comparable labeling sensitivity using a flow cytometry assay (Buck et al., 2008).
EdU should be resuspended in aqueous solution
We then tested if reconstituting EdU in DMSO, PBS or Tyrode's solution had any effect on the embryos during reincubation. EdU was first dissolved in DMSO to produce a 2 ml stock at recommended 10 mM. A working dilution was then made with PBS or Tyrode's solution. The solution containing DMSO resulted in a low survival rate after 4 hours (n=5/20). EdU diluted directly in PBS or Tyrode's solution resulted in 100% survival irrespective of the EdU dose (n=16/16) (not shown). Due to the higher EdU concentration required when labeling whole mount embryos DMSO toxicity should be taken into consideration when resuspending EdU. As aqueous media appeared to resuspend EdU equally well, DMSO is probably best avoided. We used freshly made EdU or single freeze aliquots to prevent repeated freeze thawing that might be detrimental to the terminal alkyne group.
Immunostaining of EdU labeled tissues
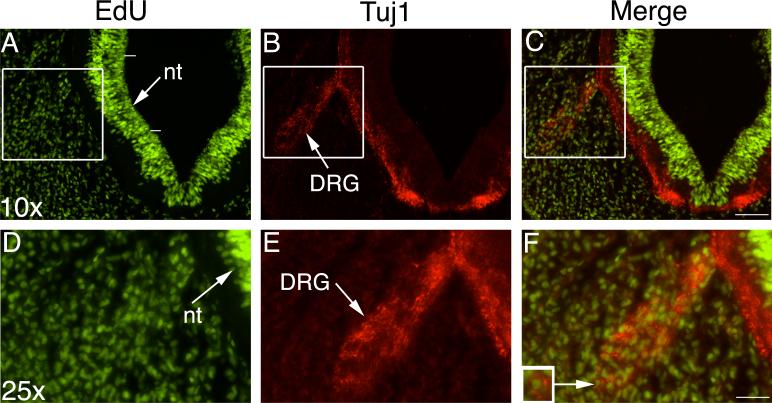
One of the drawbacks of BrdU labeling is that the denaturation step destroys epitopes making double labeling with a second antibody extremely difficult and often impossible (Tang et al., 2007). EdU chemistry avoids the denaturation step and uses reagents 1/500th the size of anti-BrdU antibody enabling easy penetration into tissues. We tested the potential for tissues that had undergone EdU chemistry to be double immunolabeled by further processing using Tuj1 antibody. Tuj1 is an antibody against class III beta-tubulin, a marker of neuronal precursors within one hour of exiting the cell cycle at the start of neuronal differentiation (Hammerle and Tejedor, 2002). Tuj1 does not label glia or neurons that remain in the proliferative zone (Menezes and Luskin, 1994; Memberg and Hall, 1995; Moody et al., 1989). Immunostaining following EdU chemistry was successful (Fig. 3) and completed in the same day as EdU labeling (see Experimental Procedures). It may be possible to perform these steps in reverse order, although we did not specifically test this. In Fig. 3 (A, D) EdU labels proliferating cells in the neural tube and surrounding mesenchyme. Tuj1 labels the cytoplasm and processes of differentiated neurons (Fig. 3B, E). The merged images graphically illustrate double EdU/Tuj1 labeling (Fig. 3C, F). Note the inset showing an EdU labeled cell nucleus surrounded by Tuj1 cytoplasm (Fig. 3F inset).
Figure 3. EdU and Tuj1 double immunolabeling.
Top row shows 10× magnification of an embryonic day 3 section, 10 μm paraffin. Boxed area from top row magnified in second row at 25×. EdU shown in green, Tuj1 in red and merged images are in the right hand column. The neural tube (nt) and dorsal root ganglion (DRG) are arrowed. (F) The inset in the high magnification merged image shows a labeled cell in the DRG surrounded by Tuj1. The white lines adjacent to the neural tube in (A) show the level at which labeled cells were counted for statistical analysis.
As an alternative processing method, EdU visualization could be facilitated by an HRP-conjugated azide, rather than a fluorophore, enabling visualization by light rather than confocal/fluorescence microscopy.
Summary
In summary, utilizing EdU chemistry rather than BrdU immunostaining protocol comes down to a just a few considerations: Is EdU just as reliable as/or better than labeling proliferating cells with BrdU and is the protocol time efficient? Given the faster processing time, extremely bright fluorescence and ability to do double staining we propose that EdU is a viable alternative for direct DNA labeling of proliferating cells. BrdU labeling requires careful calibration and is more than temperamental in our hands. We find that EdU labeling is quick, reproducible and reliable and together with further antibody processing can potentially be used to determine the day of birth of neurons and other cell types and characterize the proliferation prolife of various tissues in a highly efficient and cost effective manner.
Experimental Procedures
Live whole embryo manipulation
Chicken eggs were incubated with blunt end up at 38.5 °C in a humidified chamber until the desired stage was reached. A hole was punched into the blunt end of the egg and a square of clear tape was smoothed over the hole. Curved scissors were inserted through the tape into the hole and a circular opening was cut into the top of the egg. The tape prevents fracturing of the shell and loose pieces falling onto the embryo. If necessary, the shell membranes were opened using fine forceps to reveal the embryo beneath. 400 μl of EdU solution was then pipetted directly on top of the embryo. The egg was resealed with tape and incubated (pulsed) for a further 4 hours, then harvested and the head fixed in 4% PFA/PBS for 1 hr at room temperature, or overnight at 4 °C.
EdU (Click-iT EdU kit, Cat. #C10083 Invitrogen) was diluted in DMSO, 1X PBS or Tyrode's solution at 5 mg in 2 ml giving a stock concentration of 10 mM. Working dilutions were made in either Tyrode's solution or 1X PBS at 10 μM, 50 μM, 500 μM, 1 mM or 2 mM depending on the experiment. A small amount of Fast Green was added to color the solution for visualizing during application.
Paraffin embedding and sectioning
Heads were embedded by washing twice in 1X PBS washes, followed by a graded series of ethanol to absolute alcohol, then twice for 30 min using NeoClear Xylene substitute (EMD, #65351), followed by NeoClear/Paraplast at 60 °C for 30 min and then several changes of Paraplast before a final Paraplast embedding step under vacuum overnight. Whole heads were embedded in peelable embedding molds and the tissue was then sectioned at 10−12 μm using a microtome. Sectioned tissue was dried at 37 °C on a slide warmer and then stored at room temperature until further processing at which time the slides were baked at 50 °C for 1 hour to attach the tissue permanently to the Superfrost slide. Slides were dewaxed by submerging in 2−3 × 10 min each in NeoClear, followed by a series of graded alcohols and finally in running water for 1 min. EdU chemistry was performed immediately.
EdU chemistry on sections
Permeabilization
15 min 4% PFA/PBS, 300 μl was added to each slide, all incubations at room temperature. Slides were washed 2 × 3 min 3% BSA/PBS, 1 × 20 min 0.5% Triton X-100/PBS, 2 × 3 min of 3% BSA/PBS.
Hybridization
The reaction cocktail (reaction buffer, CuSO4, Alexa Fluor 488 azide and buffer additive as per manufacturer's protocol) was added for 30 minutes. Slides were kept in the dark and washed for 3 min 3% BSA/PBS, 3 min 1X PBS, then 1:200 TO-PRO-3 (Invitrogen, #T3605), in 1X PBS for 1 min.
Visualization
Following two rinses with 1X PBS, equilibration buffer was added for 5 min, and then 3 drops of Slowfade (Invitrogen, #S2828) as the embedding medium for cover slipping. Slides were placed at 4 °C in the dark until visualizing the fluorescence on a Zeiss 510 LSM confocal microscope or Nikon SMZ1500 fluorescence steromicroscope.
EdU chemistry on whole mount embryos
Embryos were harvested in saline and fixed in multiwell dishes using 4% PFA/PBS overnight at 4 °C.
Permeabilization
Embryos were transferred to 7 ml glass vials, washed 3 × 3 min 1X PBS, 3 min H2O, 2 × 3 min 3% BSA/PBS, 0.5% Triton X-100/PBS for 20 minutes and finally 2 × 3 min 3% BSA/PBS all at room temperature.
Hybridization
The reaction cocktail (reaction buffer, CuSO4, Alexa Fluor 488 azide and buffer additive as per manufacturer's protocol) was added for 30 minutes, followed by 3 min 3% BSA/PBS, 3 min 1X PBS, then 1:200 TO-PRO-3 in 1X PBS for 1 min all in the dark.
Visualization
Following two rinses with PBS, equilibration buffer was added for 5 min, the embryos mounted onto cavity slides, then 3 drops of Slowfade was used to protect the fluorescence. The cavity slide was cover slipped and embryos imaged. Whole mount embryos were then either paraffin sectioned or cryosectioned to test if the fluorescence survives this processing.
Cryosectioning of EdU labeled whole mount embryos
Following whole mount EdU labeling and imaging on a fluorescence stereomicroscope embryos were washed twice in 1X PBS, followed by 5% and 15% Sucrose (until the embryos sank), then overnight in 15% sucrose/7.5% gelatin at 40 °C in a water bath. Using a bath of methyl butane and dry ice the embryos were mounted in cryomolds, set at − 20 °C and then sectioned on a Leica cryostat at 20 μm. The tissue was air-dried and the sucrose/gelatin removed by washing in 1X PBS at 40 °C for 30 minutes, twice in 1X PBS for 5 min at room temperature, then mounted with Slowfade and imaged on a Zeiss 510 LSM confocal microscope.
Immunohistochemistry
Following the EdU hybridization steps described above, slides were double stained using Tuj1 (Abcam, #ab14545) antibody. In brief, following the reaction cocktail, the slides were washed for 3 min in 3% BSA/PBS and blocked for 30 min using PBT (0.1% Triton-X, 5% goat serum and 0.2% BSA). A 1:400 dilution of Tuj1 antibody was then added for 1 hour at room temperature or overnight at 4 °C in the same solution. Slides were washed several times in 1X PBT, and secondary antibody (goat anti-mouse IgG Alexa Fluor 594, Invitrogen) was added at 1:200 dilution for 1 hour at room temperature. Slides were washed several times in PBT before mounting in Slowfade and imaged.
Statistical Analysis
ImageJ was used to count the number of EdU labeled cells in a defined area of neural tube from several different embryos at each of three concentrations. In each case a cell count from the middle region of one half of the neural tube was selected using the marquee tool. White lines have been placed on Fig. 3A, indicating the level at which cells were counted. The selected area was cropped to include only neural tube, adjusted to a black and white image, and then the ImageJ threshold tool was applied to select cells labeled with EdU and TO-PRO-3 (these appear light gray in the image and are easily distinguished from the dark TO-PRO-3 labeled cells). ImageJ was used to count the cells and the area was measured. The cell count was normalized to give an average number of cells per unit of area. 11, 12 and 11 individual sections, from 3, 5 and 2 individual embryos, for 500 μM, 1 mM and 2 mM concentrations were counted, respectively (see Table 1). The Mixed Procedure applied uses an analysis of variance to compare EdU across the 3 dose levels. Terms in the model include the fixed effect of dose and the random effect of embryos within each dose.
Acknowledgements
We thank Leiyin Zhu for technical assistance, Prof. P. Gerard for statistical analysis and the Jordan Hall Imaging Facility for use of the confocal microscope. This study was supported by The Clemson University, SCLIFE HHMI Project support to K.P. and the NIH grant number DC009236 to S.C.C.
Grant Sponsor: NIH Grant number: DC009236
References
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A. Detection of S-phase cell cycle progression using 5-ethynyl-2'-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2'-deoxyuridine antibodies. Biotechniques. 2008;44:927–929. doi: 10.2144/000112812. [DOI] [PubMed] [Google Scholar]
- Caldwell MA, He X, Svendsen CN. 5-Bromo-2'-deoxyuridine is selectively toxic to neuronal precursors in vitro. Eur J Neurosci. 2005;22:2965–2970. doi: 10.1111/j.1460-9568.2005.04504.x. [DOI] [PubMed] [Google Scholar]
- Cappella P, Gasparri F, Pulici M, Moll J. A novel method based on click chemistry, which overcomes limitations of cell cycle analysis by classical determination of BrdU incorporation, allowing multiplex antibody staining. Cytometry A. 2008;73:626–636. doi: 10.1002/cyto.a.20582. [DOI] [PubMed] [Google Scholar]
- Gratzner HG. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science. 1982;218:474–475. doi: 10.1126/science.7123245. [DOI] [PubMed] [Google Scholar]
- Hammerle B, Tejedor FJ. A method for pulse and chase BrdU-labeling of early chick embryos. J Neurosci Methods. 2002;122:59–64. doi: 10.1016/s0165-0270(02)00278-9. [DOI] [PubMed] [Google Scholar]
- Lang H, Bever MM, Fekete DM. Cell proliferation and cell death in the developing chick inner ear: spatial and temporal patterns. J Comp Neurol. 2000;417:205–220. doi: 10.1002/(sici)1096-9861(20000207)417:2<205::aid-cne6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Falls DL, Li X, Lane T, Luskin MB. Antigen-retrieval procedure for bromodeoxyuridine immunolabeling with concurrent labeling of nuclear DNA and antigens damaged by HCl pretreatment. J Neurosci. 2007;27:5837–5844. doi: 10.1523/JNEUROSCI.5048-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(i)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]