Abstract
The X-chromosome inversion, Xe, distinguishes Drosophila mojavensis and D. arizonae. Earlier work mapped the breakpoints of this inversion to large intervals and provided hypotheses for the locations of the breakpoints within 3000-bp intergenic regions on the D. mojavensis genome sequence assembly. Here, we sequenced these regions directly in the putatively ancestral D. arizonae X-chromosome. We find that the two inversion breakpoints are near an inverted gene duplication and a common repetitive element, respectively, and these features were likely present in the non-inverted ancestral chromosome on the D. mojavensis lineage. Contrary to an earlier hypothesis, the inverted gene duplication appears to predate the inversion. We find no sequence similarity between the breakpoint regions in the D. mojavensis ancestor, excluding an ectopic-exchange model of chromosome rearrangements. We also found no evidence that staggered single-strand breaks caused the inversion. We suggest these features may have contributed to the chromosomal breakages resulting in this inversion.
Keywords: Chromosomal inversion, gene duplication, genome evolution, transposable element
Introduction
For over half a century, chromosomal rearrangements have been known to play critical roles in adaptation within species and the generation of new species. With respect to adaptation, chromosomal rearrangements such as inversions can capture sets of adaptive alleles into nonrecombining units that spread when conditions are favorable (e.g., Dobzhansky 1951; Kirkpatrick and Barton 2006, Lewontin et al. 1981). These rearrangements may also facilitate the persistence of species despite hybridization if they are either directly underdominant (e.g., White 1978) or via their recombinational effects (e.g., Navarro and Barton 2003; Noor et al. 2001; Rieseberg 2001). As such, understanding the genesis of chromosomal rearrangements in general, and inversions in particular, contributes to our understanding of many evolutionary processes.
Multiple hypotheses have been generated regarding the causes of chromosomal inversions (e.g., Coghlan et al. 2005), and these hypotheses have been tested in recent studies of Drosophila species. Analysis of the Drosophila pseudoobscura genome sequence and comparison to that of D. melanogaster provided support for the role of ectopic exchange between repetitive elements giving rise to the majority of the paracentric inversions separating these species (Richards et al. 2005). Richards et al. (2005) found a particular set of sequence motifs that were disproportionately abundant at the junctions of syntenic regions between these two species, but they noted there might be other possible explanations for the coincidence of the breakpoint repeat and inversion breakpoint. More recently, using genome sequence assemblies from multiple species in the D. melanogaster species group, Ranz et al. (2007) identified numerous inverted duplications associated with most inversion breakpoints and suggested that most inversions in this species group were initiated by staggered double-strand breaks.
Previously, our group investigated the breakpoint regions of an X-chromosome inversion distinguishing D. mojavensis and D. arizonae (Cirulli and Noor 2007). D. arizonae bears the ancestral arrangement, while the derived D. mojavensis inversion has been designated Xe in previous studies based on comparisons with D. navajoa and other outgroup species (Wasserman 1962, Ruiz et al. 1990). The breakpoints were localized to intervals of 14 kilobases (kb) (distal breakpoint) and one megabase (proximal breakpoint, breakpoint orientation from Schaeffer et al. 2008), and an analysis of gene order assuming microsynteny with D. melanogaster identified likely breakpoint regions of under 3000 basepairs (bp). In contrast to the findings of Richards et al. (2005), we failed to identify any sequence similarity between the two regions or to known repetitive elements in the genome. However, the predicted gene CG2056 was duplicated and inverted near the distal breakpoint in D. mojavensis, perhaps resulting from the inversion event.
Here, we build on this earlier study by sequencing the breakpoint regions in D. arizonae and comparing it to the available genome sequence assembly of D. mojavensis. We find that, in contrast to both the ectopic exchange and the staggered double-strand break models, the inversion may have resulted from two clean breaks in the chromatin, independent of shared repetitive elements. We also examined the CG2056 gene copies and find that they likely predate the inversion event, as both are present in D. arizonae. The derived copy appears functional in D. mojavensis, but the ancestral one appears to have degenerated. We discuss our findings in the context of the genesis of inversions and the evolution of duplicate genes.
Methods
Fly Stocks
D. arizonae and D. mojavensis flies derived from strains isolated in Sonora, Mexico, were used in this study (Species Resource Center stock numbers 15081-1271.16 and 15081-1352.26.
Primer Design
Primers for amplifying D. mojavensis and D. arizonae sequences were designed based either on D. mojavensis genome sequence assembly (Clark et al. 2007) taken from the DroSpeGe browser (Gilbert 2007), or our own D. arizonae sequence. Primers for amplifying the distal breakpoint of Xe were designed within exons of the genes CG2056_G1 and CG12111 based on the synteny predictions of Cirruli and Noor (2007). Primers were screened by BLASTn comparison to the D. mojavensis (min Expect = 10) genome to check that they were unique, and chosen to have a Tm ~60°C using IDT’s OligoAnalyser 3.0 (Intergrated DNA Technologies). Primer sequences and locations on the D. mojavensis genome assembly are listed in Supplementary Table 1.
DNA sequencing
D. mojavensis and D. arizonae DNA was extracted with the Puregene DNA Purification Kit (Gentra Systems) following their “DNA Purification from a Single Drosophila melanogaster” protocol. PCR was performed using standard protocols. PCR products were visualized on an agarose gel (1–2% depending on size of PCR products) and bright, single bands chosen for sequencing. DNA sequencing was performed using BigDye v. 3.1 on an ABI 3730 xl by the Duke IGSP DNA Sequencing Facility.
RT-PCR
Pure RNA was extracted following the protocol of Bertucci and Noor (2001) from a mixture of D. mojavensis pupae and adults, both male and female. Primers for RT-PCR were designed as above and cDNA was generated in a 20μl reactions using MMLV Reverse Transcriptase and incubated for 10 minutes at 25°C, 15 minutes at 37°C and 5 minutes at 94°C. Purity of cDNA was assessed on a 2% agarose gel and sequencing was performed as described above.
Sequence Analysis
Raw sequences were edited and most DNA alignments generated using Sequencher v4.7 (Gene Codes Corporation). One DNA sequence alignment and all protein sequence alignments were performed using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/.) using default settings. Pairwise, synonymous and non-synonymous substitution rates were calculated using PAML (Yang 1997; Yang 2007). Analyses involving genomic sequences of D. mojavensis or other Drosophila species were performed using tools from the DroSpeGe suite (Gilbert 2007) including BLAST and GBrowse, and their implementations of a suite of gene prediction programs presented in the following GBrowse tracks: Gnomon: NCBI_GNO, GeneMapper: PACH_GMP, Oxford pipeline: OXFD_GPI, GeneWise: EISE_CGW, SNAP: DGIL_SNP, SNAP_hsp: DGIL_SNO, geneid: RGUI_GID, N-SCAN: BREN_NSC, Contrast: BATZ_CNA, GleanR: EISE, Transposon Repeats: ReAS (Clark et al. 2007; Gilbert 2007).
Results
Precise identification of inversion breakpoints
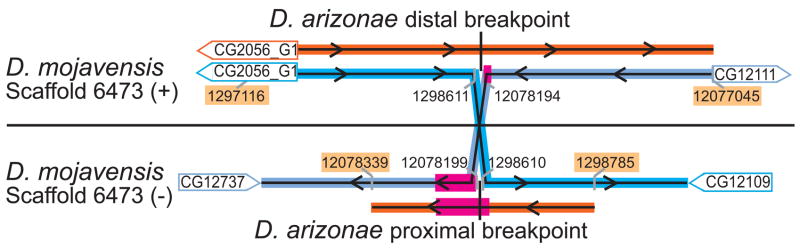
The target regions of the D. mojavensis X-chromosome are illustrated in Fig. 1. Using primers designed within the distal breakpoint region-flanking genes hypothesized by Cirulli and Noor (2007), we generated ~2.5kb of D. arizonae sequence (GenBank accession EU771085) and compared it to the D. mojavensis genome (Clark et al. 2007) using BLAST (Altschul et al. 1990) as implemented by DroSpeGe (Gilbert 2007). Within this single amplicon, regions of high sequence similarity were recovered from two D. mojavensis regions on scaffold 6473 separated by several megabases (With the exclusion of 6 small gaps, of 2531 D. arizonae bases sequenced, 2322/2394 bases (97%) alignable by BLAST are identical in D. mojavensis). These two regions within the D. mojavensis assembly share no sequence similarity with each other, but when concatenated, align to nearly the entire amplified D. arizonae region. Both loci are within the regions on this D. mojavensis scaffold Cirulli and Noor (2007) predicted to contain the Xe breakpoints, and align in opposite orientations. This result was confirmed by generating 279 bp of D. arizonae sequence (GenBank accession EU771089) using primers from the D. mojavensis genomic regions 100–150 bp on either side of the above alignments. The new sequence was highly similar by BLAST to two antiparallel regions of the D. mojavensis genome immediately adjacent to those of the earlier sequence (With the exclusion of 3 small gaps, out of the 279 D. arizonae bases sequenced, 245/263 bases (93%) alignable by BLAST are identical in D. mojavensis). Consensus locations of the proximal and distal breakpoints in D. mojavensis are at bases 12078196 (±4 bp) and 1298611(±6 bp) of scaffold 6473, respectively. The ambiguity is due to slight sequence divergence.
Fig. 1.
Location of sequence homology between D. mojavensis (blue) and D. arizonae (orange) at the breakpoints of the Xe chromosome inversion. Sequence locations are based on the D. mojavensis genome sequence assembly (Clark et al. 2007). The magenta region (50bp) is highly duplicated throughout the D. mojavensis genome (see text). Sequencing primer locations are highlighted in yellow, and genes identified on the D. mojavensis genome assembly adjacent to the breakpoints are labeled (Gilbert 2007).
Features of breakpoint regions
No significant alignment was possible between the breakpoint regions sequenced in D. arizonae using ClustalW. Potential repetitive elements of 99 and 147 bases were identified on the ReAS track of the DroSpeGe D. mojavensis genome browser (Gilbert 2007) at the proximal and distal chromosomal breakpoints, respectively. These sequences were located on opposite strands of he D. mojavensis genome assembly in such a way that they would be joined into one contiguous sequence in the ancestral chromosome, forming a potential 246 bp repetitive element at the proximal breakpoint. This concatenated sequence was compared to the D. mojavensis genome using BLAST. No other copies of this entire sequence were recovered. But, when the D. arizonae sequence from this region was compared to the D. mojavensis genome, two overlapping sequences were found to occur elsewhere in moderate numbers. An 87 bp sequence (generally 71 or 87 bases identical) was repeated at least 18 other times. A smaller 50 bp sequence (generally 43 of 50 bases identical), contained within the larger 87 bp region, was repeated several hundred times. Both of these sequences overlap the proximal breakpoint.
While the DroSpeGe browser’s ReAS track does identify several other repetitive sequences near to each breakpoint in D. mojavensis, none occur at the precise location of the ancestral distal breakpoint, and the two that are within several hundred bases of this location share no sequence similarity with the proximal breakpoint location described above. However, as observed by Cirulli and Noor (2007), two 1 kb regions located close to this breakpoint and separated by 800 bp are nearly-identical oppositely-oriented duplicates of each other. The nearer duplicated region is located ~500 bp from the distal breakpoint. These two regions each compose nearly the entire length of a gene, CG2056, an apparent homologue of a D. melanogaster gene. In D. melanogaster, CG2056 (spirit) is predicted to have serine-type endopeptidase activity based on its sequence-similarity to snk (CG7996). This duplication event appears to be unique to the Drosophila repleta species group, as only one copy of CG2056 is found in the published genome sequencesof every other Drosophila species (Clark et al. 2007). The gene copy nearer to the distal breakpoint (hereafter designated Dmoj_CG2056_G1, FlyBase designation: GI15228) is more likely the derived copy, based on its orientation relative to the neighboring gene CG12065 in other species.
To confirm that the D. mojavensis genome sequence was correctly assembled in this region, we re-sequenced the interior of both duplicated regions, using primers designed to overlap bases that distinguish the two copies (GenBank accessions EU771086 and EU771088). Our new sequence of the region nearer to the distal breakpoint is nearly identical to the published genomic sequence. However, in our sequence of the gene copy more distant from the chromosomal inversion (hereafter Dmoj_CG2056_G2, FlyBase designation: GI15354), a central 12 bp segment replaces a completely different 141 bp sequence in the published genome. Polymorphisms between the two regions outside of this short segment are present in the predicted orientation in our new sequences, suggesting that any further assembly error does not extend beyond this segment.
Description of CG2056 in D. arizonae
We determined that the CG2056 duplication event is present in the ancestral homologous region of D. arizonae. D. arizonae sequences generated using primers designed against the D. mojavensis genome from the distal breakpoint region and the downstream gene CG12065 into CG2056 again identified two distinct copies of CG2056 in this region (GenBank accessions EU771085 and EU771087). This strongly suggests that the duplication event occurred in the Drosophila repleta group ancestor prior to D. arizonae – D. mojavensis speciation, and thus prior to the chromosomal inversion found in D. mojavensis.
Detailed description of CG2056 duplication and inversion in D. mojavensis
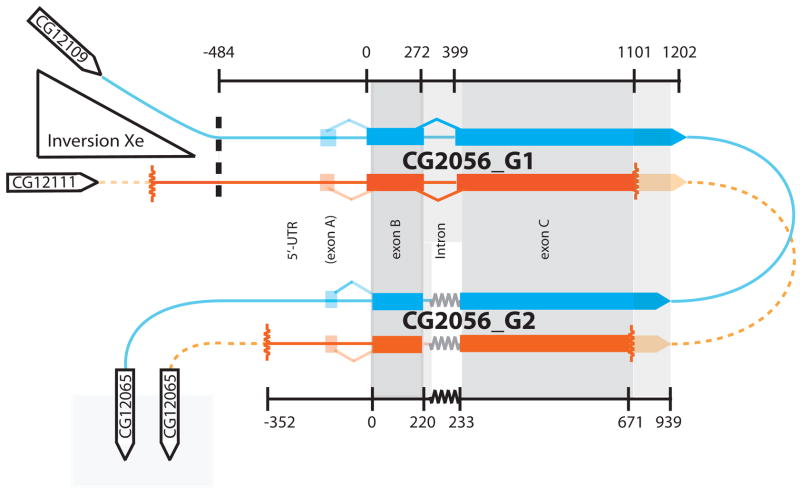
Every gene-prediction program implemented by the DroSpeGe genome browser (Gilbert 2007) identifies both duplicated regions as probable coding sequence. These predicted genes (Dmoj_CG2056_G1 and Dmoj_CG2056_G2, see Fig. 2) contain two or three exons, depending on the prediction algorithm. There is general agreement among these predictions about the existence and intron-exon boundaries of the terminal two exons of each gene. Together with the intervening intron, this region spans only slightly more than the duplication itself. There is much less consistency among predictions of the location of a more 5′ exon of these genes. Since no canonical start codon is contained in the penultimate exon, many programs identify a small earlier region that serves this function.
Fig. 2.
Genomic features near one breakpoint of the X-chromosome inversion in D. mojavensis that distinguishes this species from D. arizonae. D. mojavensis (blue) and D. arizonae (orange) sequences are highly similar to the right of the location of the chromosomal breakpoint (black dashed-line), and duplicated copies of the gene CG2056 are also highly similar in the region highlighted in grey. Dark orange D. arizonae sequence was generated in this study, while light-orange sequence is inferred. Exon predictions are as described in the text. Regions in dark grey were selected as probably exonic sequence and used to assess codon conservation. Horizontal jagged lines indicate gaps in the alignment between CG2056_G1 and CG2056_G2 genes.
We generated cDNA sequence from Dmoj_CG2056_G1 that spanned the terminal two exons of this gene (exons B and C, Fig. 2) and identified the location of their intervening intron. This location corresponded exactly to the predictions given by Gnomon, SNAP, GeneID, N-SCAN, CONTRAST and the consensus generator, Glean-R and further demonstrates expression of this gene copy.
Sequence Variation between paralogs and orthologs of CG2056 in D. mojavensis and D. arizonae
We studied the historical selective constraint on the two copies of CG2056 in both species by comparing the ratio of the rates of synonymous (ds) and non-synonymous (dn) nucleotide changes between paralogous genes within each species and orthologous genes between species (Yang 1997; Yang 2007). For a gene evolving neutrally, the dn/ds ratio should be close to 1.
Since we were unable to experimentally validate each exon boundary, we used two conservative methods to select regions for this analysis. First, based on our sequence analysis described above, we determined that Dmoj_CG2056_G1 is the best annotated of the 4 genes, and that the gene-prediction programs, Gnomon, SNAP, GeneID, N-SCAN, CONTRAST, and GleanR appear to predict the exon structure of this gene better than the others employed by DroSpeGe (Gilbert 2007). We thus compared these exon predictions and selected the regions that were labeled as exonic by all programs. These programs did not all overlap in any part of exon A, so this exon was not included in the further analysis. Secondly, we aligned Dmoj_CG2056_G1 exons B and C against the predicted amino acid sequences of the CG2056 gene prediction in D. virilis, a more distant relative of D. mojavensis and D. arizonae (Supp. Fig. 1, Gnomon prediction is shown. Gilbert 2007). Only regions of each exon that were clearly homologous between these two species were included in the analysis. These restricted regions are shown in Figure 2 and Supplementary Figure 1. After trimming exon C alignments to the ends of our own D. arizonae sequences and concatenating the remaining sequences from exons B and C, we were able to calculate dn/ds values for 759 bases from each gene copy.
The pairwise dn/ds value between CG2056_G1 across the two species was dramatically lower than all comparisons including either species’ CG2056_G2 (Table 1). Dmoj_CG2056_G1 and Dari_CG2056_G1 were, in fact, identical at the amino acid level for exon C, and differed by only 1 amino acid for exon B. Dmoj_CG2056_G2 and Dari_CG2056_G2 differ from each other and from their respective CG2056_G1 by several amino acids. This suggests that CG2056_G2 in both species has been under reduced selective constraint since the duplication of this gene in their ancestor.
Table 1.
| In each cell, the format is: | ||||||
| dn/ds | dn | |||||
| ds | ||||||
| Dari_CG2056_G1 | Dari_CG2056_G2 | Dmoj_CG2056_G1 | ||||
| Dari_CG2056_G2 | 0.711 | 0.0337 | ||||
| 0.0474 | ||||||
| Dmoj_CG2056_G1 | 0.053 | 0.0017 | 0.445 | 0.0351 | ||
| 0.0322 | 0.0788 | |||||
| Dmoj_CG2056_G2 | 0.461 | 0.0366 | 0.333 | 0.0254 | 0.452 | 0.0385 |
| 0.0794 | 0.0762 | 0.0852 | ||||
At a less quantitative level, direct comparison of these sequences yields a similar conclusion. While the exons of all four genes are generally very highly conserved, the sequences around the introns that separate exons B and C differ dramatically. This region is nearly identical between Dmoj_CG2056_G1 and Dari_CG2056_G1, but is largely missing in CG2056_G2 in both species. Additionally, sequence regions extending into each exon from this intron (according to the DroSpeGe gene models of Dmoj_CG2056_G1, but not the regions of homology with the D. virilis CG2056 gene) are barely alignable between paralogs. Finally, the genomic sequences 5’ of exon B and 3’ of exon C are impossible to align in D. mojavensis. Instead, these alignments break down several bases before the end of the predicted exon boundaries, resulting in a premature stop codon in Dmoj_CG2056_G2. If Dmoj_CG2056_G1 is a functional gene, as suggested by our identification of a spliced mRNA (see above), then this evidence further supports the idea that the CG2056_G2 genes have lost or altered functions.
Discussion
We have explored in detail the sequence composition of chromosomal regions that were broken, inverted, and re-combined in the lineage leading to Drosophila mojavensis after its split from D. arizonae. Our results failed to obtain direct support for either a canonical mechanism of transposon- or other repetitive-element-mediated chromosomal rearrangements (e.g., Finnegan 1989) or an alternative model of chromosomal re-modeling proposed by Ranz et al. (2007). We cannot completely exclude these possibilities, though, since, for example, a repetitive element may have been lost from a breakpoint region in the D. arizonae lineage. However, this seems unlikely based on the very low level of sequence divergence between these species. We do find, however, additional sequence features near to the chromosomal breakpoints that may contribute to local chromosomal instability in this region, predisposing it to accelerated breakages and rearrangements.
In the recent model proposed by Ranz et al. (2007), inversions are initiated by two pairs of staggered single-strand breaks in the chromatin. This process does not require common sequence between the two breakpoints in the ancestral chromosome. Rather, nearby nicks in the chromosome backbone cause base-pairing to fail between these breaks and the chromosome regions to separate. Overhanging ends generated by this process can be fixed, either by a loss of 5′overhanging nucleotides, or by filling in the missing strand. Finally, non-homologous end-joining can recombine the chromosomes in a new orientation. Inversions generated in this way may leave clear signatures of the process in the descendant’s genomic sequence. Fixing both 5′ overhangs of the same breakpoint in the same manner results in either inverted duplications of the regions between the nick-points or reciprocal deletions of these regions in the descendant’s chromosome. If the two overhangs of a breakpoint are fixed in opposite ways, though, no DNA is gained or lost, and no evidence of this process will be apparent in the genomic sequence. Hence, although we failed to find evidence for this process, we cannot exclude its operation in this instance.
While our sequence data cannot adequately distinguish between one outcome of the staggered double-strand break model and a model in which clean double-stranded breaks separated out this chromosomal region, the local genome features around the Xe breakpoints suggest possible explanations for why the breakpoints occurred at these locations. Ranz et al. (2007) suggest that certain regions may be predisposed to chromatin breaks and are more likely to be involved in chromosome rearrangements. In the D. mojavensis genome, we observed two key features in the vicinity of the inversion breakpoints on the X-chromosome. The proximal breakpoint is associated with a repeat element that is duplicated in high numbers across the genome. The distal breakpoint is near to a local gene duplication event. Since the sequence homology between the two copies of CG2056 breaks down immediately at the end of the coding sequences (suggesting a fairly ancient duplication), it is impossible to tell if this inversion-duplication event also used this breakpoint. But, its location does seem suggestive that a possible inherent weakness of this genomic region caused both events. Alternatively, base-pairing between the paralogous copies would cause a fairly tight hairpin loop in the chromatin, which may induce local chromosomal instability. Similarly, while the repeat element spanning the proximal breakpoint is not associated with the distal breakpoint, it seems possible that the high-copy number of this region may have increased its susceptibility to chromosome breaks.
Non-repetitive element-associated double-strand breaks in chromatin have been proposed to explain inversions in other Drosophila species. Cirera et al. (1995) suggest such a mechanism for an inversion fixed between D. melanogaster and D. subobscura. They note that alternating purine-pyrimidine sequences (RY repeats) may predispose certain regions towards chromosomal breakages due to topoisomerase II activity. Interestingly, the two Xe breakpoints we identify are centered within short RY repeat sequences (6 and 5 unit repeats for the proximal and distal breakpoint, respectively). This mechanism may be worthy of further study.
Our analysis of the evolution of the duplicated copies of the gene CG2056 in D. mojavensis and D. arizonae provides an interesting aside to the story of the Xe inversion. We have demonstrated that this duplication event predated the inversion in the D. replete group ancestor. Since that point, the function of this gene appears to have been taken over by the derived copy at the expense of the ancestral copy, as evidenced by the expression of CG2056_G1 in D. mojavensis, the amino-acid level conservation of CG2056_G1 in both species, the high level of sequence evolution in both CG2056_G2 genes and the early stop codon in Dmoj_CG2056_G2. Yet, it seems remarkable that Dmoj_CG2056_G1 remains functional despite this inversion event. Much of the 5′ UTR region of the gene was replaced by the rearrangement, which is the region where most regulatory sequences of genes are thought to reside. The nearer breakpoint is located ~500 bases from the start of exon B, but as few as 91 bases from the hypothesized start codons in exon A (Gnomon prediction, Gilbert 2007). It may be that neither exon A nor this 5′UTR are accurately annotated and the entire regulatory region is located within the first 500 bases of exon B, or elsewhere 3′ of this gene, but this result seems worthy of future studies.
The generation of genome-sequences from groups of related organisms is beginning to promote a new level of understanding of the processes underlying genome-evolution. For decades, chromosomal rearrangements have been identified as of great importance in processes of adaptation and speciation, but until recently, models that explain their occurrence have been not been tested against actual genomic data. New results from Drosophila and other taxonomic groups (e.g. Bailey et al. 2004; Ranz et al. 2007) have shown that chromosomes do appear to show pockets of instability, increasing the likelihood of rearrangements. Thus, identifying sequence features that increase this brittleness is of great utility to any studies of genome-evolution and speciation in general. We present here a case study of a recent chromosomal rearrangement that shows two types of sequence features (repetitive elements and adjacent inverted duplicated sequence) that each may be important for predicting chromosomal instability.
Supplementary Material
Amino acid alignment of CG2056 in D. mojavensis, D. arizonae and D. virilis. Amino acids selected for dn/ds-based sequence conservation analysis are highlighted in light orange.
Primers used in this study.
Acknowledgments
The authors would like to thank A. Chang for providing Drosophila mojavensis flies for these analyses, A. Chang, W. Etges, and three anonymous reviewers for helpful comments on the manuscript, and A. Somerville for technical assistance. Funding was provided by NSF grants 0509780 and 0715484 and NIH grant GM076051.
- bp
basepair
- kb
thousand basepairs
- dn
nonsynonymous substitution rate
- ds
synonymous substitution rate
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci LA, Noor MAF. Single fly RNA preparations for RT-PCR. Drosoph Inf Serv. 2001;84:166–168. [Google Scholar]
- Cirera S, Martin-Campos JM, Segarra C, Aguade M. Molecular Characterization of the Breakpoints of an Inversion Fixed Between Drosophila melanogaster and D. subobscura. Genetics. 1995;139:321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Noor MAF. Localization and characterization of X chromosome inversion breakpoints separating Drosophila mojavensis and Drosophila arizonae. J Hered. 2007;98:111–114. doi: 10.1093/jhered/esl065. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dobzhansky TG. Genetics and the origin of species. Columbia University Press; New York: 1951. [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Gilbert DG. DroSpeGe: rapid access database for new Drosophila species genomes. Nucl Acids Res. 2007;35:D480–485. doi: 10.1093/nar/gkl997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, Moore JA, Provine WB, Wallace B. Dobzhansky’s Genetics of Natural Populations. Columbia University Press; New York: 1981. pp. I–XLIII. [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Eanes WF. Sequence variation of alcohol dehydrogenase (Adh) paralogs in cactophilic Drosophila. Genetics. 2003;163:181–194. doi: 10.1093/genetics/163.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence -Accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:1366–1381. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Heed WB, Wasserman M. Evolution of the Mojavensis Cluster of Cactophilic Drosophila with Descriptions of Two New Species. J Hered. 1990;81:30. doi: 10.1093/oxfordjournals.jhered.a110922. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW, Bhutkar A, McAllister BF, Matsuda M, Matzkin LM, et al. Polytene Chromosomal Maps of 11 Drosophila species: The order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman M. Cytological studies of the repleta group of the genus Drosophila. V. The mulleri subgroup. Univ Texas Publ. 1962;6205:85–118. [Google Scholar]
- White MJ. Modes of Speciation. Freeman; San Francisco: 1978. [Google Scholar]
- Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid alignment of CG2056 in D. mojavensis, D. arizonae and D. virilis. Amino acids selected for dn/ds-based sequence conservation analysis are highlighted in light orange.
Primers used in this study.