Abstract
Mutations in the progranulin gene (GRN) are an important cause of frontotemporal lobar degeneration (FTLD) with ubiquitin and TAR DNA-binding protein 43 (TDP43)-positive pathology. The clinical presentation associated with GRN mutations is heterogeneous and may include clinical probable Alzheimer's disease. All GRN mutations identified thus far cause disease through a uniform disease mechanism, i.e. the loss of functional GRN or haploinsufficiency. To determine if expression of GRN in plasma could predict GRN mutation status and could be used as a biological marker, we optimized a GRN ELISA and studied plasma samples of a consecutive clinical FTLD series of 219 patients, 70 control individuals, 72 early-onset probable Alzheimer's disease patients and nine symptomatic and 18 asymptomatic relatives of GRN mutation families. All FTLD patients with GRN loss-of-function mutations showed significantly reduced levels of GRN in plasma to about one third of the levels observed in non-GRN carriers and control individuals (P < 0.001). No overlap in distributions of GRN levels was observed between the eight GRN loss-of-function mutation carriers (range: 53–94 ng/ml) and 191 non-GRN mutation carriers (range: 115–386 ng/ml). Similar low levels of GRN were identified in asymptomatic GRN mutation carriers. Importantly, ELISA analyses also identified one probable Alzheimer's disease patient (1.4%) carrying a loss-of-function mutation in GRN. Biochemical analyses further showed that the GRN ELISA only detects full-length GRN, no intermediate granulin fragments. This study demonstrates that using a GRN ELISA in plasma, pathogenic GRN mutations can be accurately detected in symptomatic and asymptomatic carriers. The ∼75% reduction in full-length GRN, suggests an unbalanced GRN metabolism in loss-of-function mutation carriers whereby more GRN is processed into granulins. We propose that plasma GRN levels could be used as a reliable and inexpensive tool to identify all GRN mutation carriers in early-onset dementia populations and asymptomatic at-risk individuals.
Keywords: Progranulin, ELISA, frontotemporal lobar degeneration, Alzheimer's disease
Introduction
Frontotemporal lobar degeneration (FTLD) is a genetically complex neurodegenerative disorder (Rademakers and Hutton, 2007). It is the second most common form of early-onset dementia after Alzheimer's disease, accounting for 5–10% of all dementia patients and 10–20% of patients with an onset of dementia before 65 years (Graff-Radford and Woodruff, 2007). Up to 50% of patients with FTLD report a family history of dementia, suggesting a strong genetic component to the disease. The clinical symptoms associated with FTLD are diverse including behaviour and personality changes, language disorders of expression and comprehension, cognitive impairment and sometimes motor neuron disease (McKhann et al., 2001). Three clinical FTLD subtypes have been defined: behavioural variant frontotemporal dementia (bvFTD), progressive non-fluent aphasia (PNFA) and semantic dementia (SD). The most common neuropathology associated with this type of dementia is FTLD with ubiquitin- and TAR DNA-binding protein 43 (TDP-43) positive inclusions (FTLD-U) (Josephs et al., 2004; Lipton et al., 2004; Mackenzie et al., 2006; Neumann et al., 2006).
In 2006, we identified loss-of-function mutations in the progranulin gene (GRN) as a major cause of familial FTLD-U, explaining 25% of FTLD-U patients worldwide (Baker et al., 2006; Cruts et al., 2006; Gass et al., 2006). To date, 63 different pathogenic GRN loss-of-function mutations have been reported in 169 genealogically unrelated FTLD families (Gijselinck et al., 2008a). GRN mutations are found scattered over the gene and include a variety of genetic alterations, such as nonsense and splice-site mutations as well as small insertions and deletions leading to a shift in the normal reading frame. Recently, the first complete genomic GRN deletion and a near complete deletion of GRN exons 1–11 were also reported (Gijselinck et al., 2008b; Rovelet-Lecrux et al., 2008). Together these studies established the notion that all GRN mutations share the same pathogenic mechanism, i.e. the loss of 50% functional GRN, suggesting a haploinsufficiency disease mechanism.
The spectrum of clinical presentations associated with mutations in GRN is highly heterogeneous (van Swieten and Heutink, 2008). Behaviour changes are the most common symptoms, although prominent language impairment is a frequent occurrence during the course of the disease (Gass et al., 2006; Le Ber et al., 2007; Rademakers et al., 2007; Beck et al., 2008). Importantly, episodic memory deficits occur in the initial stage in 10–30% of GRN mutation carriers, occasionally leading to patients being diagnosed with Alzheimer's disease or an amnestic variant of mild cognitive impairment (Benussi et al., 2008; Brouwers et al., 2007; Josephs et al., 2007; Kelley et al., 2007; Rademakers et al., 2007).
GRN encodes a secreted precursor protein composed of a signal peptide sequence and 7.5 tandem repeats of a rare 12 cysteinyl motif (Bateman and Bennett, 1998). While GRN has growth factor properties and a weak anti-inflammatory effect, it can be proteolytically cleaved by extracellular proteases, such as elastase, to form a family of peptides, ranging in size from 6 to 25 kDa, called granulins, which are strongly pro-inflammatory (Zhu et al., 2002). Both GRN and the granulins are widely expressed and have been implicated in a range of biological processes including development, wound repair, inflammation and tumorigenesis (He and Bateman, 2003; He et al., 2003). The exact function of GRN in the brain still remains to be determined; however, a recent in vitro study suggested that GRN may function as a neurotrophic factor (Van Damme et al., 2008).
Based on the uniform haploinsufficiency disease mechanism associated with GRN mutations, we hypothesized that the analyses of GRN levels in plasma using an ELISA could be used to predict GRN mutation status in patients with FTLD and their asymptomatic family members. In addition, we sought to test the usefulness of a GRN ELISA in the identification of GRN mutation carriers in patient populations with other forms of early-onset dementia, such as probable Alzheimer's disease (McKhann et al., 1984).
Methods
Participants
Our FTLD study cohort consisted of 219 patients from a consecutive clinical case series seen at Mayo Jacksonville by the Behavioural Neurology section. All patients agreed to be in the study and biological samples were obtained after informed consent. DNA and plasma samples were collected and stored using standard procedures. Plasma was available for 94.5% (207/219) of the patients. Each patient underwent a full neurological evaluation and all who were testable had a neuropsychological evaluation. Structural neuroimaging was performed in all patients, the majority having MRI but a few have CT. Functional imaging using either single photon emission tomography (SPECT) or PET was also performed in most patients. The majority of patients were followed over time clinically. The diagnoses of the bvFTD, SD and PNFA were made using the Neary criteria (Neary et al., 1998). Patients who had Primary Progressive Aphasia and were fluent (FA) met Mesulam criteria (Mesulam, 2003). Retrospectively, many but not all of them would meet criteria for logopenic or mixed aphasia (Gorno-Tempini et al., 2008). Those with the corticobasal syndrome (CBS) met the criteria outlined by Boeve (2007). Out of 219 patients with FTLD (50.7% women), 74 patients were diagnosed with bvFTD (including one with FTD-ALS), 33 with PNFA, 44 with SD, 42 with FA and 26 with CBS. The mean age at disease onset was 63.9 years (SD 8.3 years, range: 44–85 years). In 39.7% (87/219) of the patients a positive family history, defined as having at least one first degree relative with a history of dementia or disease in the FTLD spectrum including ALS, progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD), was observed. Twenty-one patients underwent autopsy: 16 patients showed pathological diagnoses within the FTLD spectrum, while five patients showed Alzheimer's disease at autopsy.
In addition to the patients included in the consecutive FTLD series, DNA and plasma samples of eight affected and 18 unaffected at-risk individuals from three previously published GRN mutation families ascertained at Mayo Jacksonville and Mayo Rochester were available (Baker et al., 2006; Gass et al., 2006; Kelley et al., 2007; Rademakers et al., 2007). DNA and plasma samples of 72 early-onset probable Alzheimer's disease patients (mean age at onset ± SD: 57.9 ± 6.4 years; range: 37–65 years; 61% women) and 70 unrelated control individuals (mean age ± SD: 66.3 ± 11.0 years; range: 46–85 years; 50% women) were also ascertained at Mayo Jacksonville. The clinical diagnoses of Alzheimer's disease were made using the NINCDS/ADRDA criteria (McKhann et al., 1984). In collaboration with Dr Deramecourt we further obtained plasma samples from the French patient carrying a deletion of GRN exons 1–11 and two unaffected unrelated control individuals from France (Rovelet-Lecrux et al., 2008).
Procedures
For sequencing analyses, all 12 coding exons and the non-coding exon 0 of GRN and exon 1 and exons 9–13 of MAPT were amplified by PCR using our previously published primers and protocols (Hutton et al., 1998; Baker et al., 2006). PCR products were purified using AMPure (Agencourt Biosciences, Beverly, MA) then sequenced in both directions using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA). Sequencing reactions were purified using CleanSEQ (Agencourt Biosciences) and analysed on an ABI3730 Genetic Analyser (Applied Biosystems). GRN sequencing was previously performed in 64 patients from this cohort as part of the Mayo Clinic FTLD series, while MAPT sequencing was not previously performed in this cohort (Gass et al., 2006).
To investigate whether patients who carry the same GRN mutation are descendants from a common founder, we did a haplotype sharing study with eight short tandem repeat (STR) markers spanning a region of 6.9 Mb flanking GRN at chromosome 17q21. The STR markers D17S1299, D17S951, D17S1860, D17S934, D17S950, D17S806, TAUPROM [an STR marker located in intron 0 of MAPT (Gass et al., 2006)] and the newly developed GRN_GT15 were PCR amplified with one fluorescently labeled primer and analysed on an automated ABI3730 DNA analyser. PCR primers used for GRN_GT15 were F: TCCCATTTCTCCCTTCTAGTTG and R: AAGTTGAGGCTGCAGGGTG. Alleles were scored using the GENOTYPER software (Applied Biosystems).
Genotyping of control individuals for GRN variant c.415T>C (p.C139R) was performed with a TaqMan chemistry-based allelic discrimination assay, with Assay by Design probes (Applied Biosystems) and an Applied Biosystems 7900 PCR system, followed by analysis with Sequence Detection System 2.2.1 software (Applied Biosystems).
To determine GRN expression levels in human plasma samples, we used the human Progranulin ELISA kit (Adipogen Inc., Seoul, Korea) using a 1:100 dilution of the plasma samples in 1× diluent following manufacturer's instructions. The wash solution was aspirated after each third wash to ensure that all residual wash solution was removed. To increase accuracy all samples were analysed twice in two independent experiments using six interplate control samples for normalization. Based on our FTLD population (n = 207), the median coefficient of variation (CV) was 3.2%. Recombinant human GRN provided with the ELISA kit was used as a standard.
For Western blot analyses, albumin depleted plasma samples (SwellGel Blue Albumin Removal Kit, Pierce, Rockford, IL) and protein samples extracted from cerebellar brain homogenates of FTLD patients with and without GRN mutations, were separated by SDS–PAGE using pre-cast 4–20% Tris–Glycine gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes. GRN levels were assessed using the GRN capture antibody and the GRN detection antibody used in the ELISA (Adipogen Inc.). GRN cleavage products were obtained by elastase treatment of recombinant GRN for 30 min at 25°C (Athens Research & Technology, Athens, GA).
Statistical analyses
GRN expression levels were compared between two groups using Mann–Whitney tests. The Kruskal–Wallis test was used to compare GRN expression across clinical FTLD subtypes. Results were considered statistically significant for P ≤ 0.05; no adjustments were made for multiple testing; however, results were interpreted cautiously in the light of the number of tests conducted in these exploratory analyses. Since there was no overlap in distributions of GRN expression levels in the GRN loss-of-function mutation carriers compared to the non-GRN carriers, we explored sensitivity and specificity for various cut-off points on a continuous scale by fitting normal distributions to the square root of the GRN levels. Ten thousand bootstrap samples were then obtained to construct 95% confidence intervals for sensitivity and specificity at a range of potential cut-off values.
Results
We performed systematic mutation analyses of GRN and MAPT through direct sequencing in all 219 patients diagnosed with FTLD from a consecutive clinical case series ascertained at Mayo Jacksonville. In MAPT, we identified the known pathogenic mutation, IVS10 + 16C>T, in intron 10 in a single patient (NGR180), who presented with personality changes at the age of 55 and was diagnosed with bvFTD. His father, paternal uncle and aunt also had early-onset dementia. Imaging analyses in this patient showed bilateral temporal atrophy on MRI as well as bilateral temporal hypometabolism on PET.
In GRN, we identified five different pathogenic loss-of-function mutations in a total of eight patients with FTLD and a previously published partial deletion of exon 11, IVS10-15_Ex11 + 178del, in a single patient with FTLD (Table 1). We further identified four missense mutations, two silent mutations and one regulatory variant, 5′UTR-22C>T, in eight patients with FTLD (Table 1). With the exception of the silent mutation, p.P578, these GRN variants were previously published, however, their pathogenic nature largely remains unknown. Samples were not available from other family members to test for segregation of these GRN variants. The presence of missense mutation p.C139R (c.415C>T), which had not been identified in control individuals in previous studies, was further excluded in 248 control individuals ascertained at Mayo Jacksonville. A summary of the clinical and pathological characteristics of the GRN mutation carriers is provided in Table 1.
Table 1.
Clinical and pathological characteristics of GRN mutation carriers
| Patient (alias) | Predicted cDNA mutation Predicted protein mutation | Sex | Age at onset | Family history | Clinical Dx | Initial clinical | Structural neuroimaging | Functional neuroimaging | Pathology | Reference to patient |
|---|---|---|---|---|---|---|---|---|---|---|
| (death) | symptom | |||||||||
| GRN loss-of-function mutations | ||||||||||
| NGR019 | c.26C>A | F | 56 | Y | PNFA | Aphasia | MRI: perisylvian | Not done | (Gass et al., 2006; | |
| (11696) | p.A9D | (L>R) atrophy | Kelley et al., 2007) | |||||||
| NGR001 | c.388_391delCAGT | M | 65 (75) | N | PNFA | Aphasia | MRI: temporal and | Not done | FTLD-U (NII) + | This study |
| p.Q130SfsX125 | perisylvian (L) atrophy | HpScl | ||||||||
| NGR077 | c.388_391delCAGT | M | 50 | Y | bvFTD | Personality | MRI: generalized | FDG-PET: frontal | This study | |
| p.Q130SfsX125 | atrophy | (L>R) hypometabolism | ||||||||
| NGR228 | c.911_912insTG | M | 63 | N | bvFTD | Aphasia/ | MRI: frontal (L>R) | FDG-PET: frontal | This study | |
| p.W304LfsX58 | Personality | atrophy | (L>R) hypometabolism | |||||||
| NGR192 | c.911_912insTG | F | 49 | Y | FA | Aphasia | MRI: parietal, | FDG-PET: parietal and | This study | |
| p.W304LfsX58 | perisylvian and temporal (L) atrophy | temporal (L) hypometabolism | ||||||||
| NGR003 | c.998delG | M | 65 (73) | Y | PNFA | Aphasia | MRI: perisylvian | Not done | FTLD-U (NII) | (Gass et al., 2006; |
| (PPA-1D) | p.G333VfsX28 | (L) atrophy | + HpScl | Mesulam et al., 2007) | ||||||
| NGR043 | c.1477C>T | F | 64 (69) | Y | bvFTD | Personality | MRI: moderate | Not done | (Rademakers et al., 2007) | |
| (2619) | p.R493X | frontal and temporal (L+R) atrophy | ||||||||
| NGR068 | c.1477C>T | M | 48 (50) | N | FA | Aphasia | MRI: mild | FDG-PET: temporal | (Rademakers et al., 2007) | |
| (7756) | p.R493X | ventricular enlargement in frontal horn | (L+R), frontal and parietal (L) hypometabolism | |||||||
| NGR247 | c.1414-15_1590del p.A472_Q548del | F | 79 (86) | N | SD | Anomia | CT: temporal atrophy (L) | Not done | This study | |
| GRN mutations with unknown pathogenic nature | ||||||||||
| NGR022 | c.55C>T | F | 61 | N | bvFTD | Personality | MRI: frontal and | Not done | (Gass et al., 2006) | |
| p.R19W | temporal (L+R) atrophy | |||||||||
| NGR139a | c.415T>C | M | 76 | Y | FA | Aphasia | MRI: mild | FDG-PET: parietal and | This study | |
| p.C139R | generalized atrophy | temporal (L) hypometabolism | ||||||||
| NGR221 | c.970G>A | M | 64 (70) | N | PNFA | Apraxia of | MRI: temporal and | Not done | CBD | This study |
| p.A324T | speech | parietal (L>R) atrophy | ||||||||
| NGR039 | c.1297C>T | M | 66 | N | PNFA | Aphasia | MR: sylvian and | Not done | (Gass et al., 2006) | |
| p.R433W | temporal (L) atrophy | |||||||||
| NGR105 | c.1297C>T | F | 61 | Y | SD | Anomia | MRI: temporal | FDG-PET: left | This study | |
| p.R433W | (L>R) atrophy | hemisphere hypometabolism | ||||||||
| NGR167 | c.1297C>T | M | 68 | Y | FA | Aphasia | MRI: slight | FDG-PET: temporal | This study | |
| p.R433W | enlargement of the sylvian fissure (L) | and perisylvian (L) hypometabolism | ||||||||
| NGR084 | c.1341C>T | F | 64 | Y | bvFTD | Personality | MRI: frontal (R>L) | FDG-PET: frontal | This study | |
| p.H447 | atrophy | (R>L) hypometabolism | ||||||||
| NGR225 | c.1734G>A | F | 55 | N | PNFA | Aphasia | MRI: temporal | Not done | This study | |
| p.P578 | (L>R) atrophy | |||||||||
aPatient NGR139 also carries the rare 5′UTR-22C>T promoter variant. PNFA = Progressive non-fluent aphasia. bvFTD = behavioural variant frontotemporal dementia. FA = Fluent aphasia. SD = Semantic dementia. CBD = Corticobasal degeneration. R = right. L = Left. MRI = Magnetic resonance imaging. CT = computerized tomography. FDG-PET = 18F-fluorodeoxyglucose positron emission tomography. FTLD-U = frontotemporal lobar degeneration with ubiquitin inclusions. NII = intranuclear inclusions. HpScl = Hippocampal sclerosis.
Haplotype analyses using eight STR markers in a 6.9 Mb genomic region flanking GRN showed shared alleles for seven consecutive markers for patients NGR001 and NGR077 carrying p.Q130SfsX125 and shared alleles for all eight consecutive markers for patients NGR192 and NGR228 carrying p.W304LfsX58, suggesting a common genetic origin for each of these GRN mutations. A common founder for patients NGR043 and NGR068 carrying p.R493X had previously been reported (Gass et al., 2006).
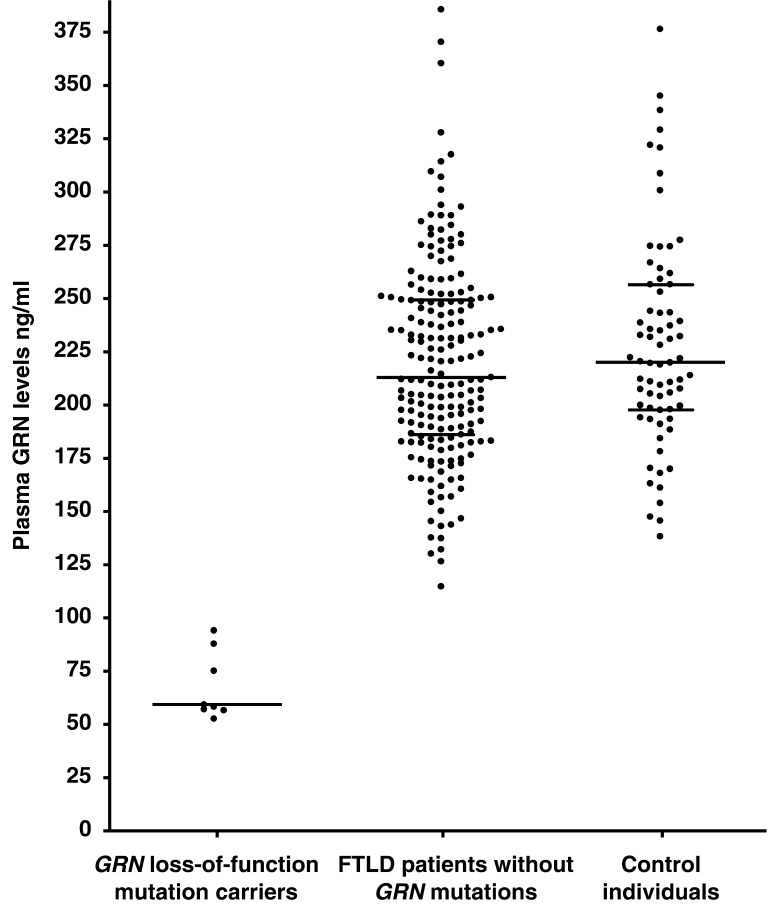
We then determined whether the levels of GRN in plasma could be used to distinguish GRN loss-of-function mutation carriers from non-GRN carriers in our FTLD series. Using a GRN ELISA we analysed a subset of 207 patients with FTLD from our consecutive cohort with plasma availability, including seven loss-of-function GRN mutation carriers and patient NGR247 carrying the partial GRN deletion, p.A472_Q548del. A significant reduction in GRN levels was observed in all patients carrying GRN loss-of-function mutations to about one third of the level observed in non-GRN carriers (Fig. 1; Table 2). GRN levels ranged from 53 to 94 ng/ml (mean value ± SD: 68 ± 16 ng/ml) in mutation carriers, while non-GRN carriers showed levels from 115 to 386 ng/ml (mean value ± SD: 220 ± 47 ng/ml). Interestingly, GRN levels were variable among GRN mutation carriers with slightly higher expression in NGR019 (88 ng/ml) carrying the p.A9D mutation in the signal peptide sequence and NGR247 (94 ng/ml) carrying p.A472_Q548del, compared to the typical frameshift and nonsense mutation carriers (Fig. 2). Based on our data alone, any cut-off value between 94 and 115 ng/ml is associated with crude estimates of both sensitivity and specificity of 100%. By assuming normality of GRN levels after a square root transformation, we were able to explore continuous estimates of sensitivity and specificity with different cut-off values (Table 3). A cut-off value of 112 ng/ml maximized the average of the estimates of sensitivity and specificity. GRN levels in 70 control individuals ranged from 138 to 376 ng/ml (mean value ± SD: 228 ± 50 ng/ml) comparable to patients with FTLD without GRN mutations (Fig. 1). As expected, normal GRN expression (259 ng/ml) was observed in our MAPT mutation carrier.
Figure 1.
Plasma GRN levels in FTLD patients and control individuals. Plasma GRN protein levels (ng/ml) in patients with FTLD carrying loss-of-function GRN mutations (n = 8), FTLD patients without GRN mutations (n = 191) and healthy control individuals (n = 70). Each data point represents an individual. Plasma levels are significantly decreased in loss-of-function GRN mutation carriers compared to non-GRN mutation carriers (P < 0.001; Mann–Whitney test). For each group the median plasma level is indicated with a wide horizontal line. For larger groups (FTLD patients without GRN mutation carriers and healthy control individuals) 25% and 75% quantiles are shown with short horizontal lines.
Table 2.
Summary of plasma GRN expression levels in patients with FTLD
| Plasma GRN levels (ng/ml) |
||||||
|---|---|---|---|---|---|---|
| Variable | N | Median | IQR | Range | P-value | |
| Loss-of-function GRN mutationa | ||||||
| Yes | 8 | 59 | 57–81 | 53–94 | <0.001 | |
| No | 191 | 213 | 186–249 | 115–386 | ||
| Genderb | ||||||
| Male | 95 | 203 | 183–233 | 130–370 | 0.0011 | |
| Female | 96 | 233 | 196–258 | 115–386 | ||
| Family historyb | ||||||
| Yes | 74 | 219 | 185–249 | 115–386 | 0.80 | |
| No | 117 | 211 | 186–250 | 130–307 | ||
| Clinical FTLD subtypesb | ||||||
| bvFTD | 62 | 218 | 189–258 | 143–370 | 0.61 | |
| PNFA | 27 | 216 | 187–260 | 115–386 | ||
| SD | 40 | 212 | 187–235 | 137–278 | ||
| FA | 37 | 205 | 180–248 | 127–310 | ||
| CBS | 25 | 230 | 188–249 | 130–360 | ||
aPatients carrying GRN mutations with unknown pathogenic nature were excluded.
bPatients carrying GRN mutations were excluded.
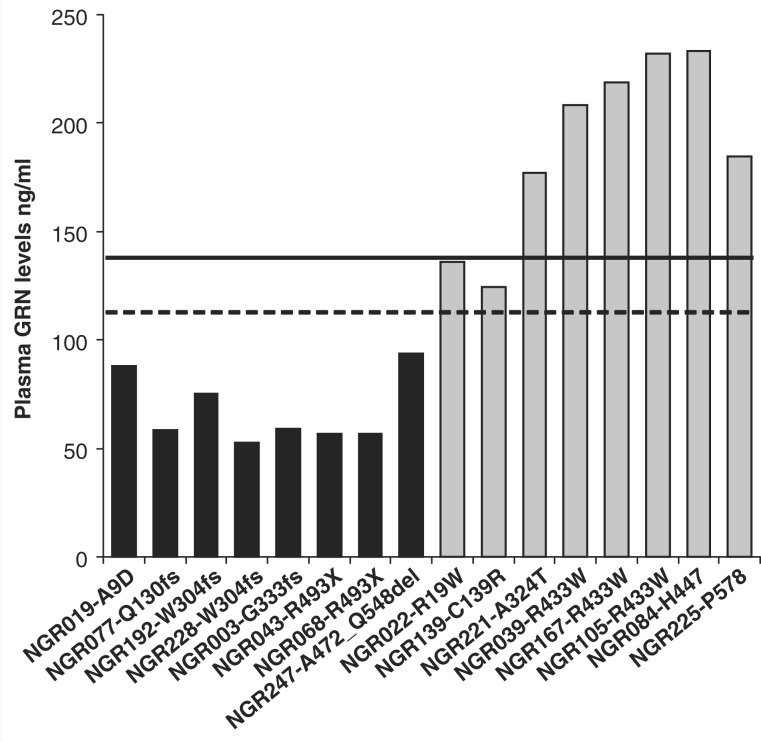
Figure 2.
Specific plasma GRN levels in all types of GRN mutations. GRN expression is plotted for each patient with FTLD from our cohort carrying a GRN mutation. Black bars represent patients with FTLD carrying pathogenic GRN loss-of-function mutations; grey bars represent patients with FTLD carrying GRN mutations with unknown significance. The dashed line is the cut-off value for pathogenic GRN loss-of-function mutations based on the complete FTLD series and the black line indicates the minimum GRN expression identified in our control cohort (Fig. 1). Missense mutations p.R19W and p.C139R show GRN levels below the range detected in control individuals and may induce a partial loss of GRN function.
Table 3.
Estimated sensitivity and specificity of the GRN ELISA assay using different cut-off values
| Plasma GRN level cut-point (ng/ml) | Sensitivity (95% CI)a | Specificity (95% CI)a | ||
|---|---|---|---|---|
| 100 | 97.42 | (86.90–100) | 99.88 | (99.66–99.97) |
| 105 | 98.64 | (90.67–100) | 99.80 | (99.48–99.94) |
| 110 | 99.32 | (93.37–100) | 99.67 | (99.23–99.89) |
| 112 | 99.49 | (94.62–100) | 99.61 | (99.87–99.10) |
| 115 | 99.67 | (95.35–100) | 99.49 | (98.89–99.81) |
| 120 | 99.85 | (96.75–100) | 99.23 | (98.42–99.70) |
| 125 | 99.93 | (97.79–100) | 98.86 | (97.81–99.53) |
| 130 | 99.97 | (98.56–100) | 98.36 | (97.04–99.26) |
| 135 | 99.99 | (99.11–100) | 97.71 | (96.06–98.89) |
aEstimate based on assumption of normally distributed GRN levels after square root transformation. Confidence intervals constructed by parametric bootstrap method.
Within the group of patients carrying GRN mutations with unknown pathogenic nature, NGR139 (p.C139R) and NGR022 (p.R19W) showed relatively low expression of GRN, in the range of the lowest 3% of patients without GRN mutations (p.C139R: 124.2 ng/ml; p.R19W: 135.9 ng/ml) and below the GRN levels observed in control individuals. Although these GRN levels fall outside the range of the typical loss-of-function mutations, these mutations may induce a partial loss-of-function. In contrast, the GRN levels in the other missense mutation carriers and silent mutation carriers were well within the normal range (Fig. 2).
In the group of non-GRN mutation carriers, a significant gender difference in plasma GRN expression levels was observed with higher levels in females (mean value ± SD: 230 ± 48 ng/ml) compared to males (mean value ± SD: 210 ± 44 ng/ml) (P < 0.001; Table 2). In exploratory analysis of subgroups by age and gender, it appeared that GRN levels may decrease with age in males, while remaining constant in females, however these findings did not reach significance (data not shown). Also, the gender difference was not confirmed in our control cohort. No significant differences in GRN plasma levels were observed between patients with a positive and negative family history of dementia or between patients with each of the five clinical subtypes of FTLD (Table 2). Also, no correlation between GRN expression levels and the age of disease onset in GRN mutation carriers or non-GRN mutation carriers was observed.
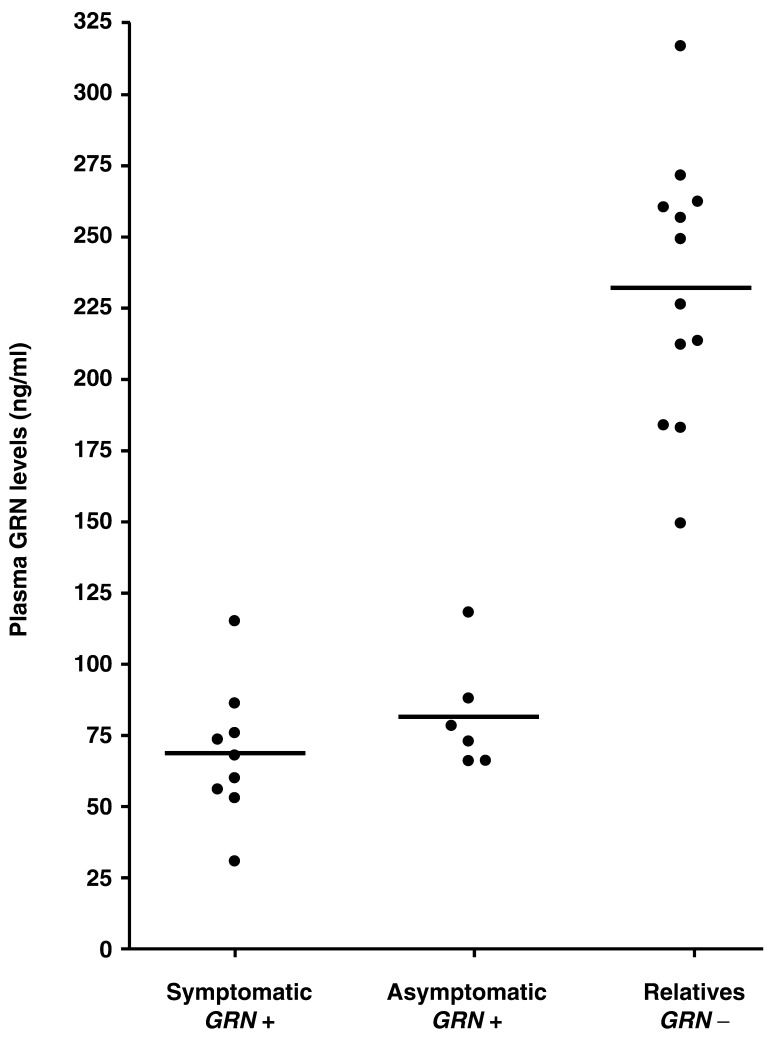
To further characterize the effect of GRN mutations on the levels of GRN in plasma, we performed a GRN ELISA on nine affected (one proband included in the consecutive FTLD series and eight additional patients) and 18 unaffected at-risk individuals from three previously published GRN mutation families with typical loss-of-function mutations (p.T52HfsX2, p.T382SfsX30 and p.R493X) ascertained at Mayo Jacksonville and Mayo Rochester. Six of the 18 asymptomatic at-risk individuals were identified as GRN mutation carriers using DNA sequencing analyses. We observed significantly lower GRN levels in both symptomatic and asymptomatic GRN mutation carriers, than non-GRN mutation carriers (P < 0.001; Fig. 3).
Figure 3.
Plasma GRN levels in symptomatic and asymptomatic GRN mutation carriers in three GRN mutation families. Plasma GRN protein levels (ng/ml) in patients and relatives of three GRN mutation families ascertained in Mayo Jacksonville and Rochester: Symptomatic GRN mutation carriers (n = 9), asymptomatic GRN mutation carriers (n = 6) and relatives without GRN mutations (n = 12). Plasma levels are significantly decreased in symptomatic and asymptomatic GRN mutation carriers compared to relatives without GRN mutations (P < 0.001; Mann–Whitney test). Each data point represents an individual. For each group, the median plasma level is indicated with a wide horizontal line.
All patients in our FTLD cohort with GRN levels <112 ng/ml (n = 8), were explained by pathogenic loss-of-function mutations identified by direct sequencing. Since the presence of a partial or complete GRN gene deletion/duplication is expected to have a similar effect on GRN expression, e.g. the loss of 50% of GRN, these data suggest the absence of GRN copy-number mutations in our series. To further confirm this finding, we performed a GRN ELISA on plasma obtained from the previously published French patient carrying a near complete deletion of GRN including exons 1–11 [patient II.4 from original publication (Rovelet-Lecrux et al., 2008)] and two unrelated French control individuals. These analyses showed a strongly reduced expression of GRN (86.2 ng/ml) in the GRN deletion carrier, compared to control individuals (196.6 and 263.2 ng/ml).
Next, we performed a GRN ELISA in a population of 72 patients with a clinical diagnosis of early-onset Alzheimer's disease. In one patient, strongly reduced GRN expression levels (74 ng/ml) were observed and sequencing analyses showed a novel loss-of-function mutation c.592_593delAG in GRN exon 5, which is expected to result in a shift of the normal reading frame and a premature termination codon (p.R198GfsX19). This patient presented at the age of 63 years with an amnestic syndrome which was confirmed on neuropsychology. MRI showed mild atrophy including frontal atrophy. No family history of dementia was reported. He met NINCDS/ADRDA criteria for probable Alzheimer's disease (McKhann et al., 1984). GRN levels in the other 71 patients with Alzheimer's disease without GRN mutations ranged from 143 to 368 ng/ml (mean value ± SD: 238 ± 53 ng/ml), comparable to GRN levels in control individuals.
Finally, to gain more insight into the processing of GRN and to determine which GRN species are detected by the ELISA, we performed GRN Western blot analyses on plasma samples and brain homogenates of FTLD patients with and without GRN mutations. These studies indicated that the antibodies used in the ELISA could only detect full-length GRN, no intermediate GRN fragments or recombinant GRN cleavage products.
Discussion
The range of mutations identified in GRN over the past 2 years established GRN haploinsuffiency as the uniform disease mechanism underlying FTLD in GRN mutation carriers (Gijselinck et al., 2008a). Microarray data further showed significantly reduced GRN mRNA expression in peripheral blood in GRN loss-of-function mutation carriers compared to non-mutation carriers (Coppola et al., 2008). These findings raised the important question as to whether expression of GRN in plasma could predict GRN mutation status and could be used as a biological marker to identify GRN mutation carriers.
To answer this question, we first studied a large consecutive series of clinical FTLD patients ascertained at Mayo Jacksonville, for which DNA and plasma samples were available. Direct sequencing analyses of GRN identified six different GRN loss-of-function mutations in nine FTLD patients explaining the disease in 4.1% (9/219) of the population and 5.7% (5/87) of FTLD patients with a positive family history. This GRN mutation frequency is very similar to our previously reported mutation frequency of 4.8% in an unbiased subpopulation of US FTLD patients (Gass et al., 2006). GRN mutations included typical nonsense and frameshift mutations as well as the previously reported p.A9D mutation in the signal peptide sequence of GRN, and a partial deletion of GRN exon 11, which extends from 15-bp upstream of exon 11 to 177-bp in exon 11 and is predicted to result in an in-frame deletion of exon 11 (p.A472_Q548del) (Gass et al., 2006; Mukherjee et al., 2006, 2008; Pickering-Brown et al., 2006; Kelley et al., 2007; Spina et al., 2007). GRN expression studies in the complete FTLD cohort using a GRN ELISA demonstrated that plasma GRN levels were strongly reduced in all loss-of-function GRN mutation carriers compared to non-GRN mutation carriers (P < 0.001). Even though we observed a wide range in GRN expression in FTLD patients and control individuals, all mutation carriers showed significantly reduced GRN levels to about one third of the levels observed in non-GRN carriers and control individuals. In our series, a plasma GRN cut-off value of 112 ng/ml distinguished all GRN mutation carriers from non-GRN carriers and we therefore expect it to have close to 100% sensitivity and specificity in future samples. These results are in line with a recently published study by Ghidoni et al. (2008) who established that in an Italian FTLD population a GRN cut-off level of 110.9 ng/ml was 92.8% specific and 100% sensitive to identify GRN mutations. However, since there were only eight GRN mutation carriers in our FTLD population, further carriers will need to be assessed to more definitively decide on the optimal cut-off; in the meantime it may be prudent to use a higher threshold than 112 ng/ml to be more certain of a high sensitivity, without materially lowering specificity. For example, examining Table 3 we see that a cut-off value of 135 ng/ml has a lower confidence limit for sensitivity of >99%, and this is achieved with an estimated specificity of 97.7%.
Interestingly, we showed strongly reduced GRN expression in patients carrying all types of loss-of-function GRN mutations, including patient NGR019 carrying the p.A9D signal peptide mutation and patient NGR247 carrying the partial GRN deletion, p.A472_Q548del. We also confirmed similar low levels of GRN in a French patient with clinical Parkinson's disease carrying a near complete deletion in GRN.
Next, by analysing unaffected relatives carrying GRN mutations, we showed that the GRN reduction is independent of the disease status, suggesting that the GRN ELISA could also function as a diagnostic tool to identify asymptomatic GRN mutation carriers.
GRN sequencing analyses in our cohort of FTLD patients also identified four different GRN missense mutations with unknown pathogenic nature in six FTLD patients. The plasma GRN levels in these missense mutation carriers generally supported the previously predicted effect on GRN function based on in silico structural protein modeling analyses (Brouwers et al., 2008; Gijselinck et al., 2008a). Mutation p.C139R was predicted to be likely pathogenic because of a destabilizing effect on the granulin-fold by disrupting one of the cysteine disulfide bridges. This mutation had been previously reported in an Italian early-onset familial FTD patient and a Belgian late-onset Alzheimer's disease patient, while absent in >900 control individuals that were analysed by us and others (Bernardi et al., 2008; Brouwers et al., 2008). We observed this mutation in a familial FTLD patient diagnosed with FA at the age of 76 years. His problem began with language difficulty including expression and comprehension. Sentence repetition was preserved. Picture description was normal except for some mild anomia. MRI showed mild non-asymmetrical atrophy. PET analyses showed hypometabolism in the left parietotemporal area and neuropsychometric testing indicated deficits in language, frontal function and preserved memory. In line with the predictions, low amounts of GRN were measured in the plasma sample of this patient (124 ng/ml), supporting the pathogenic potential of this mutation through a partial loss of GRN function. In contrast, all four patients carrying missense mutations p.A324T and p.R433W, which were predicted by in silico analyses to be tolerated, and both carriers of the silent GRN mutations (p.H447 and p.P578) showed GRN plasma levels within the normal range. Although we cannot exclude the possibility that these mutations abolish GRN function without reducing the overall expression of GRN, these findings suggested that plasma GRN levels may be a valuable tool in predicting the pathogenic potential of GRN mutations. We propose that individuals carrying intermediate levels of GRN (∼110–140 ng/ml) undergo additional genetic testing to determine the presence of potential pathogenic GRN missense mutations.
Previous studies have reported clinical Alzheimer's disease diagnoses in patients carrying loss-of-function mutations in GRN (Boeve, 2007; Kelley et al., 2007). In a clinicopathological study, we performed on the most common GRN mutation worldwide, p.R493X, 10% of the mutation carriers received a primary clinical diagnosis of Alzheimer's disease and memory impairment was the second most common initial clinical symptom, affecting 30% of the mutation carriers (Rademakers et al., 2007). In this study, using our GRN ELISA as a screening tool, we analysed 72 patients with clinically diagnosed early-onset probable Alzheimer's disease for mutations in GRN and identified one patient (1.4% of the population) carrying a loss-of-function mutation. If a disease modifying treatment for either Alzheimer's disease or FTLD becomes available in the future, then, based on the finding above and knowing that that FTLD usually starts early, one could make the case that it would be reasonable to screen early-onset probable Alzheimer's disease cases with a plasma measure for GRN.
In our study, plasma GRN levels were ∼75% reduced in mutation carriers with respect to non-GRN patients and controls, which is significantly more than expected based on the haploinsufficiency disease mechanism. A similar reduction in GRN expression was recently reported in an Italian series of GRN mutation carriers (Ghidoni et al., 2008). In contrast, only 35–50% reductions in GRN mRNA levels have been reported in GRN mutation carriers (Baker et al., 2006). These findings, together with the fact that we established that only full-length GRN and no granulins or intermediate GRN fragments can be detected by our ELISA, predict an unbalanced GRN metabolism in GRN mutation carriers, whereby the processing of GRN into granulins is increased. This would suggest that patients with GRN mutations may specifically lack full-length GRN, while maintaining normal levels of granulins. This is an interesting finding which may have implications for future therapeutic strategies, especially since GRN and the proteolytically derived granulin fragments each have distinct biological properties, e.g. GRN functions as a growth factor or anti-inflammatory agent, while granulin peptides boost inflammation (Zhu et al., 2002; He and Bateman, 2003; Ahmed et al., 2007). In future experiments, detailed biochemical analysis of the secretion and proteolytic processing of GRN in mutation carriers and control individuals will be essential.
In conclusion, this large study of GRN expression in patients with FTLD, Alzheimer's disease and control individuals provides strong support for the use of a GRN ELISA as a reliable and inexpensive tool to identify GRN mutation patients and carriers. Moreover, compared to classical mutation screening of GRN by sequencing analyses, a GRN ELISA has the important advantage of identifying individuals carrying all types of mutations including GRN deletions.
Funding
National Institutes of Health (P50 AG16574 to R.C.P., D.W.D, B.B., N.R.G-R and R.R.); the Pacific Alzheimer's Disease Research Foundation (C06-01 to D.W.D and R.R.)
Glossary
Abbreviations:
- bvFTD
behavioural variant frontotemporal dementia
- FTLD
frontotemporal lobar degeneration
- PNFA
progressive non-fluent aphasia
- SD
semantic dementia
- SPECT
single photon emission tomography
References
- Ahmed Z, Mackenzie IR, Hutton ML, Dickson DW. Progranulin in frontotemporal lobar degeneration and neuroinflammation. J Neuroinflammation. 2007;4:7. doi: 10.1186/1742-2094-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bennett HP. Granulins: the structure and function of an emerging family of growth factors. J Endocrinol. 1998;158:145–51. doi: 10.1677/joe.0.1580145. [DOI] [PubMed] [Google Scholar]
- Beck J, Rohrer JD, Campbell T, Isaacs A, Morrison KE, Goodall EF, et al. A distinct clinical, neuropsychological and radiological phenotype is associated with progranulin gene mutations in a large UK series. Brain. 2008;131:706–20. doi: 10.1093/brain/awm320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benussi L, Binetti G, Sina E, Gigola L, Bettecken T, Meitinger T, et al. A novel deletion in progranulin gene is associated with FTDP-17 and CBS. Neurobiol Aging. 2008;29:427–35. doi: 10.1016/j.neurobiolaging.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Bernardi L, Tomaino C, Anfossi M, Gallo M, Geracitano S, Costanzo A, et al. Novel PSEN1 and PGRN mutations in early-onset familial frontotemporal dementia. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.01.005. Feb 29 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Parkinson-related dementias. Neurol Clin. 2007;25:761–81. doi: 10.1016/j.ncl.2007.04.002. vii. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Nuytemans K, van der Zee J, Gijselinck I, Engelborghs S, Theuns J, et al. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol. 2007;64:1436–46. doi: 10.1001/archneur.64.10.1436. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Sleegers K, Engelborghs S, Maurer-Stroh S, Gijselinck I, van der Zee J, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008;71:656–64. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- Coppola G, Karydas A, Rademakers R, Wang Q, Baker M, Hutton M, et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol. 2008;64:92–6. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- Ghidoni R, Benussi L, Glionna M, Franzoni M, Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71:1235–9. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Broeckhoven C, Cruts M. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: an update. Hum Mutat. 2008a;29:1373–86. doi: 10.1002/humu.20785. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, van der Zee J, Engelborghs S, Goossens D, Peeters K, Mattheijssens M, et al. Progranulin locus deletion in frontotemporal dementia. Hum Mutat. 2008b;29:53–8. doi: 10.1002/humu.20651. [DOI] [PubMed] [Google Scholar]
- Gorno-Tempini ML, Brambati SM, Ginex V, Ogar J, Dronkers NF, Marcone A, et al. The logopenic/phonological variant of primary progressive aphasia. Neurology. 2008;71:1227–34. doi: 10.1212/01.wnl.0000320506.79811.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003;81:600–12. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- He Z, Ong CH, Halper J, Bateman A. Progranulin is a mediator of the wound response. Nat Med. 2003;9:225–9. doi: 10.1038/nm816. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Ahmed Z, Katsuse O, Parisi JF, Boeve BF, Knopman DS, et al. Neuropathologic features of frontotemporal lobar degeneration with ubiquitin-positive inclusions with progranulin gene (PGRN) mutations. J Neuropathol Exp Neurol. 2007;66:142–51. doi: 10.1097/nen.0b013e31803020cf. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Holton JL, Rossor MN, Godbolt AK, Ozawa T, Strand K, et al. Frontotemporal lobar degeneration and ubiquitin immunohistochemistry. Neuropathol Appl Neurobiol. 2004;30:369–73. doi: 10.1111/j.1365-2990.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, et al. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.08.022. Oct 17 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ber I, van der Zee J, Hannequin D, Gijselinck I, Campion D, Puel M, et al. Progranulin null mutations in both sporadic and familial frontotemporal dementia. Hum Mutat. 2007;28:846–55. doi: 10.1002/humu.20520. [DOI] [PubMed] [Google Scholar]
- Lipton AM, White CL, III, Bigio EH. Frontotemporal lobar degeneration with motor neuron disease-type inclusions predominates in 76 cases of frontotemporal degeneration. Acta Neuropathol. 2004;108:379–85. doi: 10.1007/s00401-004-0900-9. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Shi J, Shaw CL, Duplessis D, Neary D, Snowden JS, et al. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol. 2006;112:551–9. doi: 10.1007/s00401-006-0123-3. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of department of health and human services task force on Alzheimer's Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the work group on Frontotemporal Dementia and Pick's disease. Arch Neurol. 2001;58:1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Mesulam M, Johnson N, Krefft TA, Gass JM, Cannon AD, Adamson JL, et al. Progranulin mutations in primary progressive aphasia: the PPA1 and PPA3 families. Arch Neurol. 2007;64:43–7. doi: 10.1001/archneur.64.1.43. [DOI] [PubMed] [Google Scholar]
- Mesulam MM. Primary progressive aphasia–a language-based dementia. N Engl J Med. 2003;349:1535–42. doi: 10.1056/NEJMra022435. [DOI] [PubMed] [Google Scholar]
- Mukherjee O, Pastor P, Cairns NJ, Chakraverty S, Kauwe JS, Shears S, et al. HDDD2 is a familial frontotemporal lobar degeneration with ubiquitin-positive, tau-negative inclusions caused by a missense mutation in the signal peptide of progranulin. Ann Neurol. 2006;60:314–22. doi: 10.1002/ana.20963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee O, Wang J, Gitcho M, Chakraverty S, Taylor-Reinwald L, Shears S, et al. Molecular characterization of novel progranulin (GRN) mutations in frontotemporal dementia. Hum Mutat. 2008;29:512–21. doi: 10.1002/humu.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Pickering-Brown SM, Baker M, Gass J, Boeve BF, Loy CT, Brooks WS, et al. Mutations in progranulin explain atypical phenotypes with variants in MAPT. Brain. 2006;129:3124–6. doi: 10.1093/brain/awl289. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Baker M, Gass J, Adamson J, Huey ED, Momeni P, et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C–>T (Arg493X) mutation: an international initiative. Lancet Neurol. 2007;6:857–68. doi: 10.1016/S1474-4422(07)70221-1. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Hutton M. The genetics of frontotemporal lobar degeneration. Curr Neurol Neurosci Rep. 2007;7:434–42. doi: 10.1007/s11910-007-0067-6. [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Deramecourt V, Legallic S, Maurage CA, Le Ber I, Brice A, et al. Deletion of the progranulin gene in patients with frontotemporal lobar degeneration or Parkinson disease. Neurobiol Dis. 2008;31:41–5. doi: 10.1016/j.nbd.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Spina S, Murrell JR, Huey ED, Wassermann EM, Pietrini P, Grafman J, et al. Corticobasal syndrome associated with the A9D Progranulin mutation. J Neuropathol Exp Neurol. 2007;66:892–900. doi: 10.1097/nen.0b013e3181567873. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Van Hoecke A, Lambrechts D, Vanacker P, Bogaert E, van Swieten J, et al. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol. 2008;181:37–41. doi: 10.1083/jcb.200712039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swieten JC, Heutink P. Mutations in progranulin (GRN) within the spectrum of clinical and pathological phenotypes of frontotemporal dementia. Lancet Neurol. 2008;7:965–74. doi: 10.1016/S1474-4422(08)70194-7. [DOI] [PubMed] [Google Scholar]
- Zhu J, Nathan C, Jin W, Sim D, Ashcroft GS, Wahl SM, et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–78. doi: 10.1016/s0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]