Abstract
BTG3/ANA/APRO4 has been reported to be a tumor suppressor gene in some malignancies. It constitutes important negative regulatory mechanism for Src-mediated signaling, a negative regulator of the cell cycle and inhibits transcription factor E2F1. We report that BTG3 is downregulated in renal cancer and that the mechanism of inactivation is through promoter hypermethylation. Quantitative real-time polymerase chain reaction (PCR) showed that BTG3 was downregulated in cancer tissues and cells. Genistein and 5-aza-2′-deoxycytidine (5Aza-C) induced BTG3 messenger RNA (mRNA) expression in A498, ACHN and HEK-293 renal cell carcinoma (RCC) cell lines. Bisulfite-modified PCR and DNA sequencing results showed complete methylation of BTG3 promoter in tumor samples and cancer cell lines. Genistein and 5Aza-C treatment significantly decreased promoter methylation, reactivating BTG3 expression. Chromatin immunoprecipitation assay revealed that genistein and 5Aza-C increased levels of acetylated histones 3, 4, 2H3K4, 3H3K4 and RNA polymerase II at the BTG3 promoter indicative of active histone modifications. Enzymatic assays showed genistein and 5Aza-C decreased DNA Methyltransferase, methyl-CpG-binding domain 2 activity and increased HAT activity. Cell cycle and 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide cell proliferation assays showed that genistein has antiproliferative effect on cancer cell growth through induction of cell cycle arrest. This is the first report to show that BTG3 is epigenetically silenced in RCC and can be reactivated by genistein-induced promoter demethylation and active histone modification. Genistein had similar effects to that of 5Aza-C, which is a potent demethylating agent with high toxicity and instability. Genistein being a natural, non-toxic, dietary isoflavone is effective in retarding the growth of RCC cells, making it a promising candidate for epigenetic therapy in renal carcinoma.
Translational Relevance: Genistein being a natural, non-toxic, dietary isoflavone, is effective in regulating RCC growth through epigenetic pathways, making it a promising candidate for renal cancer therapy.
Introduction
The incidence of renal cell carcinoma (RCC) has increased in recent years with ∼54 390 new cases in 2008 (1). RCCs are heterogenous in histology, genetics and clinical behavior. Clear cell (75–80%) and papillary (10–15%) carcinoma are the two most common subtypes of RCC (2). Tumorigenesis is a multistep process that results from the accumulation and interplay of genetic and epigenetic changes. Increased DNA methylation of CpG islands in the promoter region of genes is well established as a common epigenetic mechanism for the silencing of tumor suppressor genes (TSGs) in cancer cells (3). Several TSGs have been identified as being hypermethylated with associated loss of expression in renal cancer. The VHL and p16INK4a TSGs are inactivated by promoter hypermethylation in upto 20% of clear cell (4) and 10% of all RCC (5). The RASSF1A gene is hypermethylated in 27–56% (6) and the Timp-3 gene is hypermethylated in 58–78%, of primary RCCs (6). There are probably many additional tumor suppressor and cancer genes important in renal tumorigenesis remaining to be identified. A systematic approach to the identification of epigenetically-silenced genes in renal tumor cells could provide methylation signatures for early detection and prognosis. This could also lead to identification of novel targets for therapy and lead to a further understanding of the biology of this disease.
Epigenetic silencing of a gene can be reversed by drugs such as 5-aza-2′-deoxycytidine (5Aza-C) that forms a covalent complex with the active sites of methyltransferase resulting in generalized demethylation (3). Unfortunately, the applicability of this commonly used drug is hampered by its high toxicity and instability in physiological solutions (7).
One of the agents that has recently attracted attention in the treatment and prophylaxis of cancer is genistein. Genistein (4′,5,7-trihydroxyisoflavone) is an abundant isoflavone found in soybeans. Accumulating studies have shown that genistein inhibits a number of cancers in multiple ways. At the cellular level these include inhibition of cell proliferation, induction of apoptosis (8), induction of differentiation (9) and modulation of cell cycle progression (8). At the enzymatic level, genistein has been reported to inhibit the activity of protein tyrosine kinase (10), topoisomerase II (11), aromatase (12) and 17β-hydroxysteroid oxidoreductase (13). Therefore, genistein may be a promising agent for the treatment and prevention of RCC.
BTG3/ANA/APRO4 is a member of the antiproliferative BTG (B-cell translocation gene)/Transducer of ErbB2 gene family, which also includes BTG1, BTG2/TIS21/PC3, Transducer of ErbB2, Tob2 and PC3b (14). These proteins are characterized by a BTG1/APRO homology domain in their N-terminal regions, within that reside two highly conserved motifs, box A and box B. BTG3 is thought to be a negative regulator of the cell cycle and it has been shown that its antiproliferative action is through inhibition of transcription factor E2F1 (15). More recently, BTG3 has been found to associate with and inhibit Src tyrosine kinase (16). Other studies have shown that BTG3 gene is a candidate TSG in human non-small cell lung cancer cell and oral squamous cell carcinomas (17). In oral squamous cell carcinomas, the repression of the BTG3 gene expression frequently accompanied tumor development and this repression was reversed by treatment with demethylating agent (5Aza-C) suggesting that the promoter of the BTG3 gene was hypermethylated (17).
In this study, we examined the promoter methylation status of TSG BTG3 in renal cancer tissues and cancer cell lines and compared it with normal tissues and a non-malignant-immortalized cell line. We further evaluated the effects of genistein on promoter hypermethylation, histone modifications, enzyme activities [DNA Methyltransferase (DNMTase), histone acetylase (HAT), histone deacetylase (HDAC) and methyl-CpG-binding domain (MBD)-2] and compared the results with that of 5Aza-C. In addition, we investigated the effect of genistein on cell cycle, apoptosis and cell proliferation in RCC cell lines and non-malignant-immortalized renal cells.
Materials and methods
Tissue samples and cell culture
Tissue samples from radical nephrectomy were obtained from the Veterans Affairs Medical Center, San Francisco, CA. Informed consent was obtained from all patients. A board-certified pathologist processed the specimens according to protocol and samples were stored at −80°C.
Human RCC cell lines A498, ACHN and HEK-293 and the non-malignant-immortalized renal cell line HK-2 were obtained from the American Type Culture Collection (Manassas, VA). The cancer cell lines were cultured as monolayers in minimum essential medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 50 μg/ml penicillin, 50 μg/ml streptomycin (Invitrogen, Carlsbad, CA) and maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C. The HK-2 cells were cultured in keratinocyte growth medium supplemented with 5 ng/ml human recombinant epidermal growth factor, 0.05 mg/ml bovine pituitary extract (Gibco/Invitrogen, Carlsbad, CA) and maintained in an incubator under the conditions described above. Subconfluent cells (60–70% confluent) were treated with varying concentrations of genistein (0, 10, 25 and 50 μmol/l) (Sigma, St Louis, MO) dissolved in dimethyl sulfoxide or 5Aza-C (5 μmol/l). Cells treated only with vehicle served as control. Fresh genistein or 5Aza-C was administered everyday along with a change of media and the cells were grown for 3 (genistein) or 5 days (5Aza-C).
DNA cell cycle and 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide cell viability assay
The cells were harvested, washed with cold phosphate-buffered saline and processed for cell cycle analysis. Briefly, 1 × 106 cells were resuspended in 1 ml of cold saline GM (4°C) to that cold ethanol (3 ml) was added, and the cells were incubated overnight at 4°C. After centrifugation, the pellet was washed with 2 ml cold phosphate-buffered saline + 5 mM ethylenediaminetetraacetic acid, resuspended in 1 ml phosphate-buffered saline containing 30 μg/ml propidium iodide, 0.3 mg/ml RNase A and incubated at 25°C for 1 h in the dark. The cell cycle distribution of the cells of each sample was then determined using a fluorescence-activated cell sorting caliber instrument (Becton Dickinson FACScan, Biosciences, San Jose, CA) equipped with CellQuest 3.3 software in the fluorescence-activated cell sorting core facility of the Veterans Affairs Medical Center and University of California, San Francisco, CA. CellQuest cell cycle analysis software was used to determine the percentage of cells in the different cell cycle phases.
For growth assay, renal cancer cells were plated in 96-well plates at a density of 5 × 103 cells per well in 100 μl medium. Before treatment, cells were allowed to adhere to the plate for 24 h. Afterward, the medium was replaced with the same volume of medium containing genistein or 5Aza-C including the untreated control. After 24, 72 and 120 h, cell viability was determined by using the CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI) according to the manufacturer's protocol.
RNA/DNA extraction from clinical samples and cell lines
Total RNA was extracted using a combination of TRIzol reagent (Invitrogen) and RNeasy columns (Qiagen, Valencia, CA). Fresh renal tissues were homogenized in 1 ml TRIzol reagent. After the addition of 0.2 ml chloroform, samples were centrifuged for 15 min at 14 000 r.p.m. The aqueous phase was moved to a new centrifuge tube and resuspended with one half volume of 100% ethanol. Samples were then applied to an RNeasy mini-column. For DNA digestion, an Ambion DNA-Free kit was used according to the manufacturer's protocol. RNA quality was assessed using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmingon, DE) spectrophotometer. Extracted RNA was stored at −80°C. Genomic DNA was extracted from paraffin-embedded non-cancerous and cancerous microdissected renal tissues that were obtained from the Veterans Affairs Medical Center. A DNA mini kit (Qiagen) was used to extract DNA from tissue according to the manufacturer's protocols. Genomic DNA and RNA were extracted from 80% confluent plates of cultured cells using AllPrep DNA/RNA Mini Kit (Qiagen) according to the manufacturer's directions.
Quantitative real-time polymerase chain reaction
First strand complementary DNA was prepared from total RNA (1 μg) using the reverse transcription system (Promega). In the real-time polymerase chain reaction (PCR) step, complementary DNA was amplified with Inventoried Gene Assay Products containing two gene-specific primers and one TaqMan MGB probe (6-FAM dye-labeled) using the TaqMan Universal Fast PCR Master Mix in a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Thermal cycling conditions included 95°C for 20 s, 40 cycles of 95°C for 3 s and 60°C for 30 s according to the TaqMan Fast Universal PCR protocol. GAPDH was used as an endogenous control and vehicle control was used as a calibrator. Each sample was run in four wells. The comparative Ct method was used to calculate the relative changes in gene expression in the 7500 Fast Real-Time PCR System. The relative changes of gene expression were calculated using the following formula: fold change in gene expression, 2−ΔΔCt = 2−[ΔCt (genistein-/5Aza-C-treated samples) − ΔCt (untreated control)], where ΔCt = Ct (detected genes) − Ct (GAPDH) and Ct represents threshold cycle number.
Sodium bisulfite modification and sequencing
Bisulfite modification of DNA was performed using the Epi-Tect Bisulfite kit (Qiagen) following the manufacturer's directions. The basic principle of bisulfite modification of DNA is that in the bisulfite reaction, all unmethylated cytosines are deaminated and sulfonated, converting them to thymines, whereas methylated cytonies (5-methylcytosines) remain unaltered. Thus, the sequence of the treated DNA will differ depending on whether the DNA is originally methylated or unmethylated. Primers for bisulfite genomic sequencing PCR were designed by using the online program MethPrimer (18). The primer sequences are shown in Figure 3C. All reactions for tissue samples were subjected to two rounds of amplifications using a nested primer approach. Bisulfite-modified DNA (1 μl) was amplified using a primer pair in a total volume of 20 μl. Aliquots (2 μl) of the first PCR reactions were subjected to second round amplifications using a pair of nested primer pairs in a total volume of 30 μl. The amplification products were confirmed by electrophoresis on a 2% agarose gel and sequenced directly by an outside vendor (McLab, South San Francisco, CA).
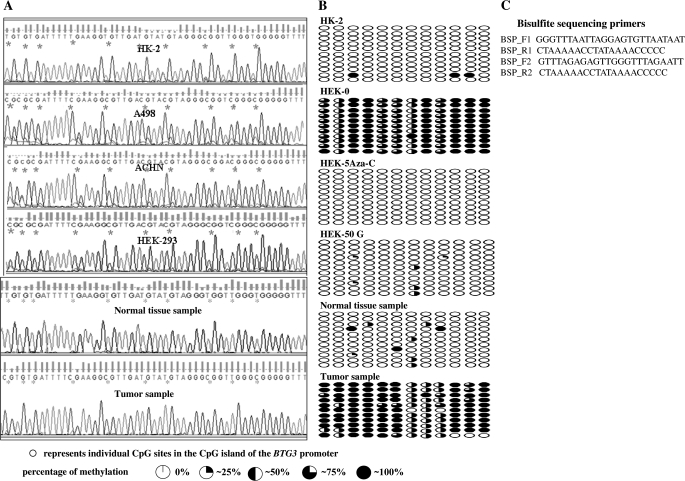
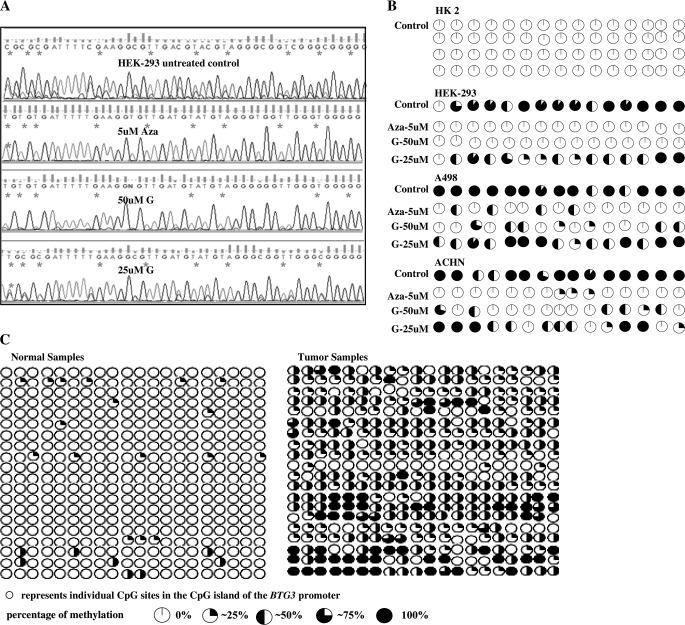
Fig. 3.
BTG3 promoter methylation status. (A) DNA sequencing results showing the BTG3 promoter methylation status in untreated HK2, A498, ACHN, HEK-293 cell lines and representative figures showing promoter methylation status in normal and cancer tissues. The symbol “*” indicates individual CpG sites. Bisulfite modification of the DNA was performed that changes the unmethylated cytosines to thymine thus represented by ‘TG’ in the sequence, whereas the methylated cytosines remain unchanged and thus represented by ‘CG’ in the sequences. For cell lines, a single primer set (BSP_F2–R2) was used for bisulfite modified polymerase chain reaction (BS-PCR), whereas a nested PCR was performed for tissue samples using two primer sets BSP_F1–R1 and BSP_F2–R2. (B) Sequencing results showing methylation status in 10 clones of HK-2, treated and untreated cancer cell line (HEK-293), normal tissue sample and a tumor sample. First row represents the parent PCR product and the remaining rows represent 10 clones of this product. Primer set F1R1 and F3R3 were used for all the samples that were cloned. (C) Sequences of primers used for BS-PCR.
Cloning for methylation confirmation
Bisulfite modification of DNA from selected samples was performed as described above. The modified DNA was amplified using nested PCR with primer sets F1R1 and F3R3 and the products were confirmed by electrophoresis. Amplified products were cloned into the pCR2.1-Topo vector using TOPO TA Cloning Kit (Invitrogen). Ten to fifteen colonies were randomly chosen for culture and DNA was purified using PureLink Quick Plasmid Miniprep Kit (Invitrogen) and sequenced by an outside vendor (Quintara Biosciences, Albany, CA).
Chromatin immunoprecipitation analysis
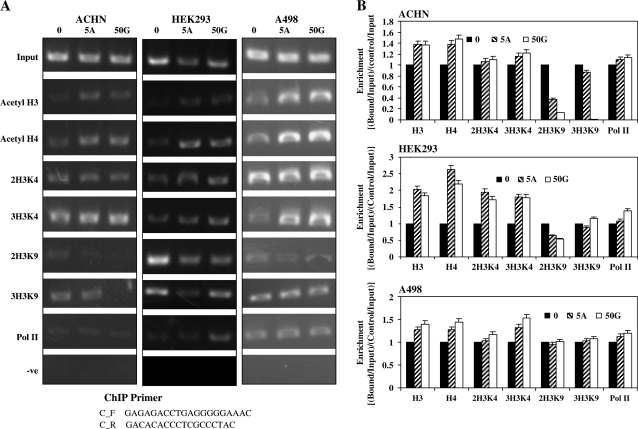
Chromatin immunoprecipitation analysis was performed using the EZ-ChIP kit (Upstate Biotechnology, Charlottesville, VA) according to the manufacturer's directions. Antibodies used in the immunoprecipitations were purchased from Upstate Biotechnology and Ambion (Austin, TX) and were specific for acetyl histone H3 (06–599), acetyl histone H4 (06–866), dimethyl-histone H3 lysine 4 (07–030), trimethyl-histone H3 lysine 4(07–473), dimethyl-histone H3 lysine 9 (07–441), trimethyl-histone H3 lysine 9 (ab8898) and anti-polymerase II (05–623). The immunoprecipitated DNA was eluted in a total volume of 50 μl and 2 μl were used for PCR that was performed with an annealing temperature of 60°C for a total of 28 cycles. The amplified DNA was electrophoresed in a 2% agarose gel and visualized by staining with ethidium bromide. The ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/) was used for optical densitometry. For enrichment calculation, each sample was normalized with their respective input samples and then the treated samples were compared with their untreated controls for each used antibody. In short, enrichment was calculated as the ratio between the net intensity of each bound sample normalized to its input sample and the vehicle control sample normalized to vehicle control input samples (Bound sample/Bound sample Input)/(Vehicle control sample/Vehicle control Input). The sequence of primers is given in Figure 6.
Fig. 6.
Histone modifications. (A) Effect of genistein and 5Aza-C on the histone modifications of the BTG3 promoter. ChIP assay was performed on cells after treatment with 50 μM/l genistein and 5 μM/l 5Aza-C. Untreated control (0), 5Aza-C (5A), 50 μM Genistein (50G). (B) Histone modification enrichment data calculated from the corresponding DNA fragments amplified by PCR at annealing temperature of 60°C for a total of 28 cycles; bars, error ± standard deviation. The amplified DNA was electrophoresed in a 2% agarose gel and visualized by staining with ethidium bromide. The ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/) was used for optical densitometry. Enrichment was calculated as the ratio between the net intensity of each bound sample normalized to its input sample and the vehicle control sample normalized to vehicle control input samples (Bound sample/Bound sample Input)/(Vehicle control sample/Vehicle control Input).
DNMTase analysis
Total DNMTase activity was measured using EpiQuik DNA Methyltransferase Activity/Inhibition Assay kit (Epigentek, Brooklyn, NY). Nuclear extracts were isolated using the EpiQuik Nuclear Extraction Kit (Epigentek) and 3 μl of nuclear extract was added to each reaction well, according to the manufacturer's protocol. The final volume of nuclear extract yield was used to normalize the assay results for differences in cell number. Nuclear extracts were incubated with methylation substrate for 1 h at 37°C and then exposed to the capture antibody for 60 min and the detection antibody for 30 min at room temperature. Absorbance was determined using a microplate spectrophotometer at 450 nm, and DNMT activity (OD/h/ml) was calculated according to the formula: (Sample OD − blank OD)/(sample volume) × 1000, according to the manufacturer's instructions.
MBD2 binding activity assay
Total MBD2 activity was measured using the EpiQuik MBD2 Binding Activity Assay Kit (Epigentek). Nuclear extracts were isolated using the EpiQuik Nuclear Extraction Kit (Epigentek) and 3 μl of nuclear extract was added to each reaction well, according to the manufacturer's protocol. Nuclear extracts were incubated with methylated DNA for 1 h at 37°C and then exposed to the affinity antibody for 60 min and the detection antibody for 30 min at room temperature. Absorbance was determined using a microplate spectrophotometer at 450 nm, and MBD2 binding percentage was calculated according to the following formula: (OD (treated sample − blank)/(O.D (untreated control − blank) × 100%, according to the manufacturer's instructions.
DNMT1, DNMT3a and DNMT3b assays
Nuclear extracts were assayed for individual DNMT proteins of interest (DNMT1, DNMT3a or DNMT3b) using the Epiquik DNMT1, -3a and -3b assay kits, respectively (Epigentek). Protein standards of known concentration (30, 20, 10 and 2 ng) were included to generate a standard curve. The amount of DNMT protein was calculated as follows: DNMT protein (ng/ml) = (Sample OD − blank OD/standard slope) × sample dilution, according to the manufacturer's instructions.
HAT and HDAC analysis
Total HAT and HDAC activities were measured using the EpiQuik HAT and EpiQuik HDAC Activity/Inhibition Assay Kits, respectively (Epigentek). In brief, the nuclear extracts were incubated with specific substrate for 1 h at 37°C, followed by capture antibody for 60 min and then detection antibody for 30 min at room temperature. Absorbance was determined using a microplate spectrophotometer at 450 nm. HAT activity (ng/h/mg) [(Sample OD − blank OD)/(slope × h × protein amount (μg) added into the assay) × 1000] and HDAC activity (ng/h/ml) {[O.D (control − blank) – O.D (sample − blank)/Slope × h] × sample dilution} were measured according to the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using StatView version 5.0 for Windows as needed. Data were analyzed using StatView and a statistically significant difference was considered to be P < 0.05 and is represented by “*” on the bars in the figures. For all the results where applicable, the expression levels were quantified by optical densitometry using ImageJ Software version 1.36b (http://rsb.info.nih.gov/ij/).
Results
Induction of cell cycle arrest
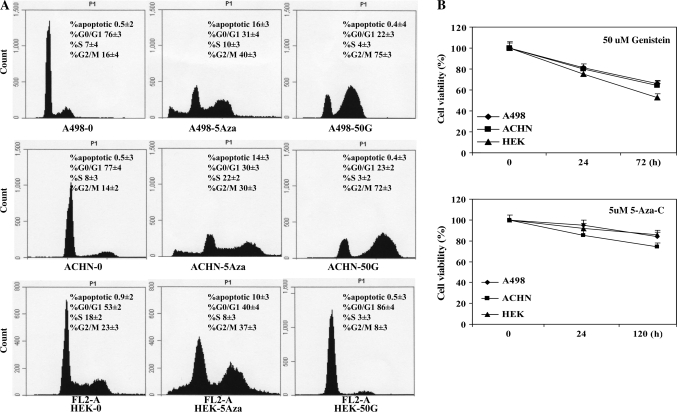
Fluorescence-activated cell sorting analysis was done to test the effect of genistein or 5Aza-C on cell cycle distribution. As summarized in Figure 1A, both agents resulted in a significantly higher number of cells in the G2–M phase (30–72%) compared with vehicle treated control (14–23%) suggesting that genistein and 5Aza-C caused a G2–M phase arrest in all renal cancer cell lines except HEK-293 where 50 μM genistein caused a G0–G1 arrest (86%). 5Aza-C induced apoptosis (10–16%) in all three renal cancer cells whereas this effect was not observed in the genistein-treated cells.
Fig. 1.
Effect of genistein on cell cycle progression. DNA content and cell cycle progression were analyzed by flow cytometry. (A) The two peaks in the fluorescence-activated cell sorting diagrams indicate G0–G1 and G2–M cells with S phase cells between peaks. Sub G1 fractions represent cells with fragmented DNA or apoptotic cells. The main panel shows a single representative result, whereas the numerical values are the mean ± SD of three experiments. (B) The 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide cell proliferation assay results. The data expressed in the graph is the mean ± SE of three independent experiments. Declines in cell viability were statistically significant (*P < 0.05).
In order to examine the effect of genistein and 5Aza-C on renal cancer cell proliferation, we performed the 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide cell viability assay. As shown in Figure 1B, genistein (50 μM for 72 h) and 5Aza-C (5 μM for 5 days) significantly decreased cell viability in a time-dependent manner confirming that both agents have antiproliferative effects on cancer cells. Genistein's antiproliferative effect on RCC cells may be due to induction of cell cycle arrest rather than apoptosis.
BTG3 expression profile
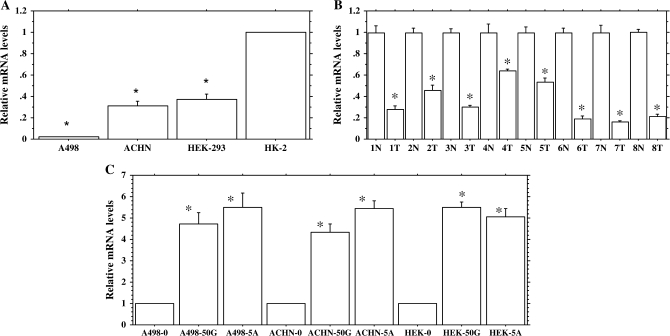
In order to determine relative expression levels of BTG3 in RCC, we performed TaqMan quantitative real-time PCR analysis for A498, ACHN and HEK-293 cell lines and compared them with the non-malignant-immortalized HK-2 cell line. We also compared the mRNA expression levels of tumor samples and normal tissue samples (Figure 2A and B). The results showed that relative mRNA expression was significantly lower in tumor samples and RCC cell lines when compared with normal tissue samples and the HK-2 cell line. These results show that the BTG3 gene is transcriptionally downregulated in RCC.
Fig. 2.
BTG3 expression profile. (A) Expression profile of BTG3 in renal cancer (RCC) and non-malignant-immortalized kidney cells. (B) Expression profile of BTG3 in renal carcinoma (T) and normal (N) clinical samples. (C) Relative expression profile of BTG3 gene following treatment with 0 and 50 μM genistein (50G) and 5 μM 5-Aza-2′-deoxycytidine (5A). Relative quantification was performed by quantitative real-time PCR using the following formula: Fold change in gene expression, 2−ΔΔCt = 2−[▵Ct (genistein-/5Aza-C-treated samples) − ΔCt (untreated control)], where ΔCt = Ct (detected genes) − Ct (GAPDH) and Ct represents threshold cycle number. For untreated controls data were normalized to 1. Data are in triplicate from three independent experiments. All data are expressed as the mean ± SE (bars). *Statistically significant at P < 0.05.
Effect of genistein and 5Aza-C treatment on the expression of BTG3
Genistein significantly upregulated the relative expression level of BTG3 in a dose-dependent manner over vehicle control (Figure 2C, Data not shown for lower doses). Maximum increase was observed with 50 μM genistein (5- to 6-fold) followed by 25 μM (3- to 4-fold, Data not shown). 5Aza-C treatment also increased the BTG3 mRNA expression level by 4- to 6-fold in all RCC cell lines compared with vehicle control. The results indicate that the BTG3 gene can be significantly induced by genistein and 5Aza-C treatment.
Methylation status of BTG3 promoter
To check whether transcriptional silencing of the BTG3 gene is due to promoter hypermethylation, we analyzed the methylation status of the BTG3 promoter in 20 pairs of tumor and normal tissue samples and cell lines by bisulfite-modified PCR followed by direct sequencing of the modified DNA samples. We used MethPrimer software (18) to select primers in the CpG rich region of the BTG3 promoter around the transcription start site. Primers were designed with no CpG sites in either the forward or reverse primer and thus, amplification proceeds in a manner unbiased by promoter methylation status. Selected amplicons were subsequently subcloned, and the recombinants were identified and subjected to automated DNA sequencing. Resulting sequences were compared with the parent promoter sequence from that the clones were made and the methylation status of the CpG dinucleotides within this amplicon was determined by characteristic chemical changes associated with cytosines existing in either a methylated or an unmethylated state (Figure 3B). DNA sequencing results revealed the promoter of the BTG3 gene in tumor samples was hypermethylated in comparison with normal tissue samples (Figures 3A and 4C). The BTG3 promoter in RCC cell lines was completely methylated whereas there was absence of CpG island methylation in HK-2 (Figure 3A). Genistein treatment (50 μM) significantly demethylated the hypermethylated BTG3 promoter followed by 25 μM genistein (Figure 4A and B). The 5Aza-C also significantly demethylated the BTG3 promoter. These results indicate that transcriptional silencing of the BTG3 gene is due to promoter hypermethylation that can be reversed by genistein and 5Aza-C treatment.
Fig. 4.
Effect of genistein and 5Aza-C on BTG3 methylation. (A). Representative sequencing results showing the effect of 5Aza-C (AZ) and genistein (G) on promoter methylation of the BTG3 gene. (B) Summarized results showing demethylation of CpG sites by 5Aza-C (Aza) and genistein (G) treatments in three RCC cell lines and a HK-2. (C) Summarized results showing promoter methylation status in 20 pairs of normal and tumor samples. Each pair was obtained from the same patient and microdissected by a certified pathologist into normal and tumor. The symbols open circle and asterisk indicate individual CpG sites in the CpG island. Bisulfite modification of the DNA was performed that changes the unmethylated cytosines to thymine thus represented by ‘TG’ in the sequence whereas the methylated cytosines remain unchanged and thus represented by ‘CG’ in the sequences. For cell lines, a single primer set (BSP_F2–R2) was used for BS-PCR whereas a nested PCR was performed for tissue samples using two primer sets BSP_F1–R1 and BSP_F2–R2.
Enzymatic activity assays
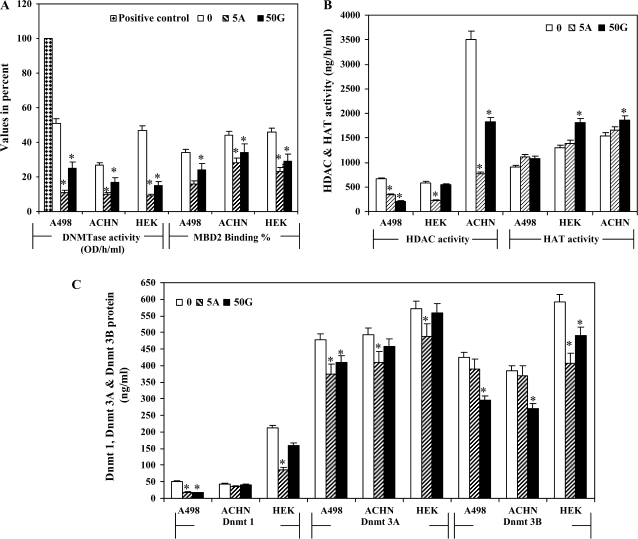
We performed different enzyme activity assays related to methylation and histone modifications. The DNMTase activity was downregulated by treatment with 50 μM genistein and 5Aza-C (Figure 5A). There was a significant decrease in DNMT1 protein levels by genistein and 5Aza-C treatments except in ACHN cells (Figure 5C). A decrease in DNMT 3a protein was observed with 5Aza treatment, whereas DNMT 3b levels were significantly decreased by both genistein and 5Aza-C in all three RCC cell lines (Figure 5C). MBD2 binding activity was decreased by genistein and 5Aza-C 20–40% compared with untreated controls (Figure 5A). We also checked HAT and HDAC activity and observed that both genistein and 5Aza-C treatment increased HAT but decreased HDAC activity compared with untreated controls (Figure 5B).
Fig. 5.
Enzymatic activity assays. (A) Percent methyltransferase activity and MBD2-binding activity expressed as binding percent. Positive control was provided with the assay kit. (B) HAT and HDAC activity (ng/h/ml). (C) DNMT proteins expressed in ng/ml were calculated by formula [OD (sample−blank)/slope) × sample dilution]. Untreated control (0), 50 μM Genistein (50G), 5 μM 5Aza-2′-deoxycytidine (5A).
Changes in chromatin modifications
To determine whether there were covalent chromatin changes after genistein and 5Aza-C treatment at the BTG3 locus, we did chromatin immunoprecipitation analysis with various antibodies as described in Materials and Methods. Genistein and 5Aza-C treatment resulted in enrichment of acetylated histones H3, H4 and H3 di- and tri-methylated lysine 4 close to the BTG3 transcription start site in all RCC cell lines (Figure 6A and B). There was also decrease in repressive modifications (2H3K9 and 3H3K9) in ACHN and HEK cell lines (Figure 6A and B). These chromatin changes are indicative of gene activation. Therefore, the promoter demethylation and increased BTG3 expression caused by genistein and 5Aza-C correlated with active hisone modifications at the transcription start site in RCC cell lines.
Discussion
Our study clearly demonstrates that TSG BTG3 is transcriptionally downregulated in RCC and that this is due to promoter CpG island methylation. We showed that the methylation-silenced BTG3 gene can be reactivated by genistein that causes CpG demethylation, inhibition of DNMT and MBD2 activity and induction of active histone modifications in renal cancer cells. In addition, we have shown that genistein's antiproliferative effect is through cell cycle arrest. To our knowledge, this is the first report showing that the BTG3 gene is epigenetically silenced in RCC and that it can be reactivated by genistein-induced promoter demethylation and histone modification.
Control of cell cycle progression in cancer cells is considered to be a potentially effective strategy for the control of tumor growth since molecular analysis of human cancers has revealed that cell cycle regulators are frequently mutated in many common malignancies (19). Our in vitro data indicated that treatment of renal cancer cells with genistein resulted in a significant G2–M phase arrest of cell cycle progression. Genistein has been found to induce G2–M cell cycle arrest in breast, gastric, human melanoma (20), PC3 prostate cancer (21) and lung cancer cells (22). Thus, it is generally accepted that genistein can cause G2–M cell cycle arrest. The 3-(4,5-dimethylthiazole-2-yl)-2,5-biphenyl tetrazolium bromide assays showed that genistein has antiproliferative effects on RCC cells and this may be due to induction of cell cycle arrest since we did not observe induction of apoptosis by genistein. Our results also showed that in the HEK-293 cell line, genistein caused G0–G1 arrest. Genistein has been previously reported to arrest mouse fibroblasts, melanoma cells (23) and prostate cancer (LNCaP) cells (24) at the G0–G1 phase of the cell cycle.
Tumorigenesis is a multistep process and one mechanism in the multistep model is the promoter methylation of specific TSGs. If methylation occurs within the promoter region of a suppressor gene, epigenetic silencing of this gene may lead to functional inactivation, a mechanism reported for various tumor entities (25). Our results show that the BTG3 gene is hypermethylated in all the three RCC cell lines and tumor samples compared with normal tissues and HK-2. Promoter hypermethylation subsequently leads to the transcriptional silencing of the BTG3 gene (Figure 2A and B). Yamamoto et al. (17) have also reported that the BTG3 gene is silenced by hypermethylation in primary oral squamous cell carcinomas.
If the methylation of a TSG is relevant for gene silencing, then reversal of methylation by demethylating agents may lead to reactivation of the gene. Unfortunately, the applicability of the commonly used 5Aza-C is hampered by its high toxicity and instability in physiological solutions (7). In this study, we used genistein, a natural, non-toxic dietary isoflavone and compared its effect with that of 5Aza-C. Our results revealed that genistein significantly induced BTG3 mRNA levels and its effects were similar to that of 5Aza-C in RCC cell lines (Figure 2C). This is consistent with other reports that have shown that genistein upregulated mRNA expression of the BRAC1 gene, p16INK4a, RARβ, MGMT genes and p16INK4a, p21CIP1/WIF1 (24). Further, we tried to determine if transcriptional repression of BTG3 is a consequence of promoter methylation. We found that the CpG island in the promoter of the BTG3 gene was hypermethylated in cancer cell lines and tumor samples (Figure 3A) compared with normal tissues and HK-2. Treatment with 50 μM genistein caused demethylation of these sites though to a lesser extent than that of 5Aza-C (Figure 4A and B). Direct inhibition of transcription may be through blocking the binding of transcription factors to promoters-containing methylated CpG sites (26), whereas indirect repression may involve proteins such as MePC2 that specifically bind to methylated DNA via a MBD (27). The MBD-containing proteins are thought to mediate transcriptional repression by recruiting HDAC activity to methylated DNA, resulting in a deacetylated repressive chromatin structure (27). It is probably that inhibition of DNMTase and enhanced histone acetylation can also prevent the hypermethylation and silencing of these key genes and thereby contributing to the prevention of carcinogenesis. The prevention of intestinal tumorigenesis by DNMT1 deficiency and 5Aza-C has been demonstrated in Min mice, which carry a mutated Apc gene (28). Our enzymatic activity assays revealed that genistein and 5Aza-C treatment inhibited DNMTase, decreased MBD2 binding and also increased HAT activity. However, there was little change in HDAC activity with genistein treatment (Figure 5A–C). The 5Aza-C is a potent inhibitor of DNMTase activity through irreversible binding of DNMTs to 5Aza-C-substituted DNA (29).
It has recently been suggested that DNMT1 is capable of de novo methylating activity as well as having a maintenance function (30). Therefore, it is feasible that, in cancers, DNMT1 participates in the de novo methylation of CpG islands. Some studies have reported that DNMT1 overexpression resulted in CpG island methylator phenotype of cancers derived from various organs (31–33). Once overexpressed DNMT1 induces de novo DNA hypermethylation on multiple CpG islands at an early stage of renal tumorigenesis, the aberrant DNA methylation status may be maintained even if DNMT1 expression is diminished during malignant progression. The level of DNMT1 protein expression was higher in sarcomatoid carcinomas than in grade 2 or 3 RCCs, suggesting that increased DNMT1 protein expression may participate in the progression to the most malignant form of RCC. Alternatively, only cancer cells that maintain DNMT1 overexpression during the grade 2 and 3 stages are able to progress to sarcomatoid carcinomas (32,34). Although DNMT1 is a major DNMT in humans, two other enzymes, DNMT3a and DNMT3b, have also been shown to possess DNMTase activity. Genomic methylation patterns may be established through cooperation among these three DNMTs, even in cancer cells. Our current results showed a significant decrease in DNMT1 protein levels by genistein and 5Aza-C treatment except in ACHN cells (Figure 5C). In general, a decrease in DNMT 3a and 3b was observed with 5Aza-C and genistein in all three RCC cell lines used in this study but varied in each individual cell line (Figure 5C).
It is generally accepted that aberrant DNA methylation and histone modifications work together to silence many TSGs in human cancers (34,35). There are reports that show hyper-acetylation of histone lysine residues facilitates transcriptional activation (35,36) and induction of gene expression (36). Our results revealed that both genistein and 5Aza-C caused an increase in acetyl H3, acetyl H4, 2H3K4 and 3H3K4 histones compared with untreated control in all three RCC cell lines (Figure 6A and B). Enrichment of these active chromatin modifications near the transcription start site of the BTG3 gene are associated with active gene expression. Furthermore, we found a decrease in repressive modifications (2H3K9 and 3H3K9) that are indicative of gene silencing.
In conclusion, our study is the first report to show that the BTG3 TSG is epigenetically silenced in RCC and can be reactivated by genistein-induced promoter demethylation and active histone modification. In addition, genistein showed antiproliferative effects on cancer cells through cell cycle arrest. Genistein showed similar effects to that of 5Aza-C, which is a potent demethylating agent but has high toxicity and instability in physiological solutions. Since genistein is a natural, non-toxic, dietary isoflavone and is effective in RCC growth retardation, it may be a promising candidate for epigenetic therapy in RCC.
Funding
Veterans Affairs Merit Review, Veterans Affairs Research Enhancement Award Program; National Institutes of Health (RO1CA111470, T32DK007790 to R.D.).
Acknowledgments
We thank Dr Roger Erickson for his support and assistance with the preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- 5Aza-C
5-aza-2′-deoxycytidine
- DNMTase
DNA Methyltransferase
- HAT
histone acetylase
- HDAC
histone deacetylase
- MBD
methyl-CpG-binding domain
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- RCC
renal cell carcinoma
- TSG
tumor suppressor gene
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Zambrano NR, et al. Histopathology and molecular genetics of renal tumors toward unification of a classification system. J. Urol. 1999;162:1246–1258. [PubMed] [Google Scholar]
- 3.Jones PA, et al. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 4.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl Acad. Sci. USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herman JG, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 6.Dulaimi E, et al. Promoter hypermethylation profile of kidney cancer. Clin. Cancer Res. 2004;10:3972–3979. doi: 10.1158/1078-0432.CCR-04-0175. [DOI] [PubMed] [Google Scholar]
- 7.Bender CM, et al. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 8.Shao ZM, et al. Genistein inhibits proliferation similarly in estrogen receptor-positive and negative human breast carcinoma cell lines characterized by P21WAF1/CIP1 induction, G2/M arrest, and apoptosis. J. Cell. Biochem. 1998;69:44–54. [PubMed] [Google Scholar]
- 9.Constantinou A, et al. Induction of differentiation and DNA strand breakage in human HL-60 and K-562 leukemia cells by genistein. Cancer Res. 1990;50:2618–2624. [PubMed] [Google Scholar]
- 10.Akiyama T, et al. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 11.Okura A, et al. Effect of genistein on topoisomerase activity and on the growth of [Val 12]Ha-ras-transformed NIH 3T3 cells. Biochem. Biophys. Res. Commun. 1988;157:183–189. doi: 10.1016/s0006-291x(88)80030-5. [DOI] [PubMed] [Google Scholar]
- 12.Adlercreutz H, et al. Inhibition of human aromatase by mammalian lignans and isoflavonoid phytoestrogens. J. Steroid Biochem. Mol. Biol. 1993;44:147–153. doi: 10.1016/0960-0760(93)90022-o. [DOI] [PubMed] [Google Scholar]
- 13.Makela S, et al. Inhibition of 17beta-hydroxysteroid oxidoreductase by flavonoids in breast and prostate cancer cells. Proc. Soc. Exp. Biol. Med. 1998;217:310–316. doi: 10.3181/00379727-217-44237. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda S, et al. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 15.Ou YH, et al. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 2007;26:3968–3980. doi: 10.1038/sj.emboj.7601825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahmani Z. APRO4 negatively regulates Src tyrosine kinase activity in PC12 cells. J. Cell Sci. 2006;119:646–658. doi: 10.1242/jcs.02778. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto N, et al. Analysis of the ANA gene as a candidate for the chromosome 21q oral cancer susceptibility locus. Br. J. Cancer. 2001;84:754–759. doi: 10.1054/bjoc.2000.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li LC, et al. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 19.Molinari M. Cell cycle checkpoints and their inactivation in human cancer. Cell Prolif. 2000;33:261–274. doi: 10.1046/j.1365-2184.2000.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casagrande F, et al. p21CIP1 is dispensable for the G2 arrest caused by genistein in human melanoma cells. Exp. Cell Res. 2000;258:101–108. doi: 10.1006/excr.2000.4914. [DOI] [PubMed] [Google Scholar]
- 21.Davis JN, et al. Genistein-induced upregulation of p21WAF1, downregulation of cyclin B, and induction of apoptosis in prostate cancer cells. Nutr. Cancer. 1998;32:123–131. doi: 10.1080/01635589809514730. [DOI] [PubMed] [Google Scholar]
- 22.Lian F, et al. Genistein-induced G2-M arrest, p21WAF1 upregulation, and apoptosis in a non-small-cell lung cancer cell line. Nutr. Cancer. 1998;31:184–191. doi: 10.1080/01635589809514701. [DOI] [PubMed] [Google Scholar]
- 23.Kuzumaki T, et al. Genistein induces p21(Cip1/WAF1) expression and blocks the G1 to S phase transition in mouse fibroblast and melanoma cells. Biochem. Biophys. Res. Commun. 1998;251:291–295. doi: 10.1006/bbrc.1998.9462. [DOI] [PubMed] [Google Scholar]
- 24.Majid S, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68:2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 25.Jones PA, et al. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 26.Iguchi-Ariga SM, et al. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 27.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 28.Eads CA, et al. Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic Mice. Cancer Res. 2002;62:1296–1299. [PubMed] [Google Scholar]
- 29.Creusot F, et al. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J. Biol. Chem. 1982;257:2041–2048. [PubMed] [Google Scholar]
- 30.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 31.Kanai Y, et al. DNA methyltransferase expression and DNA methylation of CPG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int. J. Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Arai E, et al. Regional DNA hypermethylation and DNA methyltransferase (DNMT) 1 protein overexpression in both renal tumors and corresponding nontumorous renal tissues. Int. J. Cancer. 2006;119:288–296. doi: 10.1002/ijc.21807. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa T, et al. DNA hypermethylation on multiple CpG islands associated with increased DNA methyltransferase DNMT1 protein expression during multistage urothelial carcinogenesis. J. Urol. 2005;173:1767–1771. doi: 10.1097/01.ju.0000154632.11824.4d. [DOI] [PubMed] [Google Scholar]
- 34.Kondo Y, et al. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell. Biol. 2003;23:206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristeleit R, et al. Histone modification enzymes: novel targets for cancer drugs. Expert Opin. Emerg. Drugs. 2004;9:135–154. doi: 10.1517/eoed.9.1.135.32947. [DOI] [PubMed] [Google Scholar]
- 36.Archer SY, et al. Histone acetylation and cancer. Curr. Opin. Genet. Dev. 1999;9:171–174. doi: 10.1016/s0959-437x(99)80026-4. [DOI] [PubMed] [Google Scholar]