Abstract
Although physiological and biochemical data since long suggested that Na+/H+ and K+/H+ antiporters are involved in intracellular ion and pH regulation in plants, it has taken a long time to identify genes encoding antiporters that could fulfil these roles. Genome sequencing projects have now shown that plants contain a very large number of putative Cation/Proton antiporters, the function of which is only beginning to be studied. The intracellular NHX transporters constitute the first Cation/Proton exchanger family studied in plants. The founding member, AtNHX1, was identified as an important salt tolerance determinant and suggested to catalyze Na+ accumulation in vacuoles. It is, however, becoming increasingly clear, that this gene and other members of the family also play crucial roles in pH regulation and K+ homeostasis, regulating processes from vesicle trafficking and cell expansion to plant development.
Key words: NHX-type ion transporters, pH regulation, plant membrane vesicles, potassium homeostasis, salt tolerance
Introduction
Potassium and Sodium, constituting the seventh and sixth most abundant elements on earth play essential roles for all living organisms. Inside living cells, potassium plays a key role in the maintenance of electrostatic balance and is essential for the activity of many enzymes.1 In plants, physiological studies and thermodynamic considerations have indicated the presence of K+/H+ antiporter systems at the plasma membrane, tonoplast, mitochondrial and chloroplast membranes and intracellular membranes of the secretory pathway.2–4 K+/H+ antiporters are suggested to be responsible for the active accumulation of K+ inside vacuoles, essential to maintain turgor and drive cell expansion.1,2 At the same time, although high cytoplasmic Na+ concentrations are toxic, plants activate high affinity Na+ uptake mechanisms in conditions of K+ deficiency, indicating that the more ubiquitous Na+ can to some extend functionally replace K+,5,6 at least as osmoticum inside the vacuole. Clearly in conditions of high salinity this becomes evident, as an important mechanism to survive salt stress relies on the accumulation of excess cytoplasmic Na+ in vacuoles, reducing the amount in the cytoplasm and providing osmotic pressure.7,8
Measurements of Na+/H+ and K+/H+ antiport activity in tonoplast vesicles represented one of the first demonstrations of secondary active transport in plants, and have been reported for many plant species.8–12 In spite of this early discovery, the genes encoding these intracellular Na+/H+ and K+/H+ antiport systems could not be identified by heterologous complementation or other approaches that were successful for many other transporters, and only the plasma membrane Na+/H+ antiporter SOS1 was identified by a mutant screen for salt sensitivity in Arabidopsis.13 A gene, encoding a protein with homology to animal plasma membrane Na+/H+ antiporters of the NHE family and the yeast ScNHX1 gene was first identified in the, at that time, partially sequenced Arabidopsis genome and named AtNHX1.14 These proteins, together with the human NHE6 and NHE7 proteins were shown to constitute a new NHE subfamily of intracellular Na+/H+antiporters.15 Heterologous expression of AtNHX1 in yeast complemented the salt sensitivity caused by disruption of the corresponding yeast homolog ScNHX1.14,16 Overexpression of AtNHX1 was shown to confer salt tolerance to Arabidopsis plants17 and various other plant species.18,19 Subsequently, many more members of the intracellular NHE antiporters, now called NHX, were identified in plants, fungi and animals (see below).
Phylogenetic Analysis
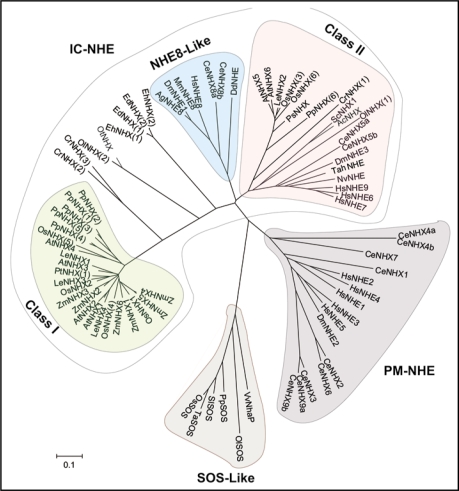
According to the classification made by Saier et al.20 (http://www.tcdb.org/index.php), Cation/Proton antiporters can be grouped into the CPA1 and CPA2 families. The CPA1 family has evolved from ancestral NhaP genes in prokaryotes15 (Fig. 1, Table 1). The Arabidopsis plasma membrane Na+/H+ antiporter AtSOS1 gene is related to the NhaP genes, and representative SOS or NhaP like sequences can be found in all phylae of the plant kingdom (SOS-Like). The most extensively studied family of the CPA1 proteins are the plasma membrane NHE antiporters present only in vertebrates (PM-NHE). Related sequences can also be found in other lower animals like C. elegans, but not in plants or fungi. The more recently discovered intracellular NHE/NHX sequences that can be found in plants, animals and fungi, have evolved separately from the plasma membrane NHE sequences, and constitute a very diverse group (IC-NHE/NHX). This family was subdivided into Class-I and Class-II sequences,21 that share only about 20–25% identity, as well as the NHE8-like family found in animals only.15 Class-I sequences are very divergent from other IC-NHE/NHX sequences and have so far been identified in monocotyledonous and dicotyledonous angiosperms, gymnosperms as well as the moss Physcomitrella patens. Distantly related single sequences can be found in the green algae Chlamydomonas reinhardtii, Osteococcus lucimarinus and Osteococcus tauri, which might point to specialized function of the Class-I anti-porters in land plants. The grouping of the green algal proteins might also have been affected by sequence errors, notably insertions or deletions or erroneously predicted splice-sites, as they are predicted from translated genomic sequences that have not been experimentally verified, although resulting misaligned portions, characterized by gaps in the alignement, were removed from the analysis. The Studied Class-I NHX isoforms were shown to have a vacuolar membrane localization, which appears to be a unique feature of this Class. The most closely related non-plant sequences are found in the primitive parasitic eukaryotes Entamoeba histolytica and Entamoeba dispar. The related protein DdNHE1 of the NHE8-like family in the social amoeba Dictyostelium discoideum has been shown to be involved in cell polarity and chemotaxis through cytoplasmic pH regulation, and was predicted to be a recycling plasma membrane protein like the other members of the NHE-8 family.22 Plant Class-II sequences that constitute a separate subclade within the Class-II sequences, were identified in angiosperms and the gymnosperm Picea sitchensis with slightly more distant members in the moss Physcomitrella. Founding members of the IC-NHE/NHX family ScNHX1 and the human NHE6 and NHE7 group together with the plant Class-II Localization and Gene Expression sequences, but are still rather divergent in sequence, as are the sequences found in the green algae Ostreococcus lucimarinus and Chlamydomonas reinhardtii. Studied antiporters of this class were shown to be expressed in various endosomal compartments.
Figure 1.
Phylogenetic tree of 79 proteins of the monovalent cation proton antiporter CPA1 family. Phylogeneitic relationships were inferred using the Neighbor-Joining method [Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–25]. The bootstrap consensus tree inferred from 500 replicates [Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985; 39:783–91], is taken to represent the evolutionary history of the proteins analyzed [Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985; 39:783–91]. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V and Vogel HJ, eds. Evolving Genes and Proteins. New York: Academic Press 1965; 97–166] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 93 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–9]. A list of included sequences is provided in Table 1.
Table 1.
UniProtKB/TrEMBL accession numbers (www.uniprot.org/uniprot/) and length of the proteins shown in Figure 1
| Length (AA) | Length (AA) | ||||
| Bacteria | Viridiplantae | ||||
| VvNhaP | Q8D8X5 | 831 | AtNHX1 | Q68KI4 | 538 |
| Eucaryota | AtNHX2 | Q56XP4 | 546 | ||
| Amoebozoa | AtNHX3 | Q84WG1 | 503 | ||
| DdNHE1 | Q86IA5 | 775 | AtNHX4 | Q8S397 | 529 |
| EhNHX(1) | Q50XA0 | 604 | AtNHX5 | Q8S396 | 517 |
| Eh(NHX(2) | B0EJB5 | 561 | AtNHX6 | Q8RWU6 | 535 |
| EdNHX(1) | B0E8R9 | 328 | AtSOS1 | Q9LKW9 | 1146 |
| EdNHX(2) | Q50XG9 | 561 | LeNHX1 | Q93YH2 | 534 |
| Fungi | LeNHX2 | Q93YH1 | 531 | ||
| ScNHX1 | Q04121 | LeNHX3 | Q1JRA3 | 537 | |
| AcNHX1 | A1C9W3 | 701 | LeNHX4 | Q1JRA2 | 536 |
| Viridiplantae | SlSOS1 | Q4W3B5 | 1151 | ||
| CrNHX(1) | A8J0T9 | 497 | Animalia | ||
| CrNHX(2) | A8J1K5 | 297 | NvNHE | A7S6K9 | 554 |
| CrNHX(3) | A8J5G2 | 589 | TahNHX | B3S5H9 | 493* |
| OlNHX(1) | A4RQC8 | 357* | CeNHX1 | Q8T5S2 | 497* |
| OlNHX(2) | A4S1Z7 | 427* | CeNHX2 | Q8T5S1 | 644 |
| OtNHX1 | Q012P4 | 292 | CeNHX3 | O16452 | 670 |
| OlSOS1 | A4RRY8 | 1247 | CeNHX4a | Q8T5R9 | 749 |
| PpNHX(1) | A9SSI2 | 563 | CeNHX4b | Q19444 | 684 |
| PpNHX(2) | A9T5K8 | 561 | CeNHX5a | Q20944 | 630 |
| PpNHX(3) | A9THT5 | 546 | CeNHX5b | Q8T5R7 | 611 |
| PpNHX(4) | A9RVH1 | 545 | CeNHX6 | Q8T5R6 | 533* |
| PpNHX(5) | A9TD24 | 534 | CeNHX7 | Q21386 | 783 |
| PpNHX(6) | A9SH77 | 479 | CeNHX8a | Q8T5R4 | 681 |
| PpSOS1 | A9RIV6 | 1161 | CeNHX8b | Q8T5R3 | 655 |
| PsNHX | A9NW71 | 594 | CeNHX9a | P35449 | 667 |
| PtNHX | DR058123** | 280* | CeNHX9b | P35449-2 | 667 |
| ZmNHX1 | Q84MI0 | 540 | DmNHE1 | Q8SZX8 | 649 |
| ZmNHX2 | Q84MH9 | 540 | DmNHE2 | Q9VIF9 | 1291 |
| ZmNHX3 | Q7XYX3 | 539 | DmNHE3 | Q8IPJ4 | 751 |
| ZmNHX4 | Q7XYX2 | 538 | DmNHE8 | A2A465 | 576 |
| ZmNHX5 | Q7XYX1 | 545 | AgNHE8 | Q7QKG3 | 650 |
| ZmNHX6 | Q7XYX0 | 541 | MmNHE8 | A2A465 | 576 |
| OsNHX1 | Q9SXJ8 | 535 | HsNHE1 | B1ALD6 | 815 |
| OsNHX2 | Q6UUW2 | 544 | HsNHE2 | Q9UBY0 | 812 |
| OsNHX(3) | Q0J2X1 | 535 | HsNHE3 | P48764 | 834 |
| OsNHX(4) | Q2R0E9 | 545 | HsNHE4 | Q6AI14 | 798 |
| OsNHX(5) | Q5ZA11 | 528 | HsNHE5 | A5PKY7 | 896 |
| OsNHX(6) | A2Z2G5 | 383 | HsNHE6 | Q92581 | 669 |
| OsSOS1 | Q5ICN3 | 1148 | HsNHE7 | Q96T83 | 725 |
| TaSOS1 | Q4L224 | 1142 | HsNHE8 | Q9Y2E8 | 577 |
| HsNHE9 | Q8IVB4 | 645 |
Annotated as partial sequences.
genebank accession number for EST sequence. The used abreviations correspond to the following species: Vv, Vibrio vulnificus; Dd, Dictyostelium discoideum; Eh, Entamoeba Histolytica; Ed, Entamoeba dispar SAW760; Sc, Saccharomyces cerevisiae; Ac, Aspergillus clavatus; Cr, Chlamydomonas reinhardtii; Ol, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Pp, Physcomitrella patens; Ps, Picea sitchensis; Pt, Pinus taeda; Zm, Zea mays; Os, Oryza sativa; Ta, Triticum aestivum; At, Arabidopsis thaliana; Sl and Le, Solanum lycopersicon; Nv, Nematostella vectensis; Tah, Trichoplax adhaerens; Ce, Caenohabditis elegans; Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Mm, Mus musculus; Hs, Homo sapiens. The numbering of the sequences is according to published reports. Numbers in between brackets are used only to ditinguish the presented proteins.
Localization and Gene Expression
The tonoplast localization of the plant Class-I NHX antiporters is well documented. Subcellular localization studies have been performed by immunoblotting using polyclonal antibodies against native proteins,17,23–27 by transitory expression of fluorescent fusion proteins in onion epidermal cells,27–29 by stable expression of fluorescent fusion protein in BY2 cells30 and by immunogold labelling.27 The only exception was found by Vera-Estrella et al.31 who detected a 50 kDa protein that cross-reacted with a polyclonal antibody raised against the AtNHX1 protein in Plasma membranes but not in tonoplast from Thellungiella roots. For the Class-II antiporters, subcellular localization studies have only been reported for the tomato LeNHX2 and the Arabidopsis AtNHX5 proteins.21,32 For both proteins, transitory expression in onion epidermal cells of fluorescent fusion proteins shows localization in small vesicles, indicative of a prevacuolar or endosomal localization clearly distinct from the central vacuole or ER/Golgi membranes, although the exact localization was not yet determined. The localization is reminiscent of the prevacuolar localization of the yeast ScNHX1 protein which suggests similar functions for the yeast and plant proteins as opposed to the human isoforms found in recycling endosomes (NHE6, NHE9,15,33), and trans or mid-trans Golgi (NHE7, NHE8,33,34).
In all plants, several isoforms of NHX proteins are found. Most of the isoforms are expressed in the absence of stress throughout the plant,16,23,26,28,35,36 and induced by salt stress in leafs,16,37,38 both roots and leafs,23,39–41 stems42 or roots.43,44 Some isoforms are also reported to be induced by ABA,28,38 KCl,14,26,35,40,44 dehydration stress30 or hyper-osmotic stress.26,28,40,44 The AtNHX1 isoform was reported not to be induced by cold or drought.45 Finally, in Citrus, an isoform was discovered based on its induction by heat.46
In Arabidopsis, the expression of all 6 isoforms was studied in more detail.28 The predominant isoforms are AtNHX1 and AtNHX2, found in roots, shoots and seedlings.28,45,47 Expression levels of AtNHX3, 4 and 6 was much lower in these tissues. AtNHX1 expression was shown to be upregulated in leaves but not roots by NaCl or ABA.16 In seedlings, AtNHX1 and AtNHX2 were shown to be induced by salt stress, hyper-osmotic shock and ABA treatment, whilst AtNHX5 was induced by salt stress only.8 AtNHX1 and AtNHX2 were not induced by NaCl in ABA deficient aba2-1 mutants, showing that NaCl induction of these isoforms is dependent on ABA signalling.28,45 The tissue distribution of AtNHX1 was further studied by promotor-GUS analysis in transgenic Arabidopsis45 and by in situ hybridization,47 showing that the gene is expressed in all tissues except the root tip. Especially high expression levels were observed in guard cells suggesting a role for AtNHX1 in K+ accumulation in these cells.45 High GUS activity was also induced in response to salt stress not only in leaves, but also in root hair cells, suggesting a role in Na+ accumulation in the enlarged vacuoles of these cells in response to salt stress.45 High levels of expression were also observed in floral tissues, and in cells closely associated to the vascular tissue in leaves and inflorescence stems.45,47
Some NHX isoforms have been shown to have a more specific expression pattern in flower or fruit, related to specific function. This is the case for the Ipomea Nil InNHX1 protein, found mainly in flower limbs where it determines flower colour through vacuolar pH changes29 and the grape berry VvNHX1 protein that is highly expressed in mature fruit where it is supposed to be involved in K+ accumulation and vacuolar expansion during ripening.48 Also in tomato, the expression of LeNHX4 is mostly detected in fruits and flowers (Gálvez FJ, Jiang XJ and Venema K, unpublished data).
Information about gene expression can also be obtained by exploring microarray data available from high-throughput projects. User friendly interphases to these data are provided at for instance http://wardlab.cbs.umn.edu/arabidopsis/ or http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi.49 These microarray data confirm the induction of Class-I AtNHX antiporters in leaves (AtNHX1, 2 and 4) or roots (AtNHX3) by salt or osmotic stress. Like other tissue specific isoforms from Ipomea Nil, grape or tomato (see above), AtNHX4 expression is detected mainly in mature pollen and seeds. High levels of AtNHX1 are found in guard cells compared to surrounding mesophyl cells (http://www-biology.ucsd.edu/labs/schroeder/index.html), in accordance with the promotor-GUS fusion experiments.45 Also AtNHX2, 5 and 6 show a high level of expression in guard cells as compared to surrounding mesophyl cells. Furthermore, AtNHX1 and AtNHX5 are induced by ABA in these cells.
It can also be observed that several NHX isoforms are induced or repressed by biotic stresses. The reason for this has not been investigated, but could be related to a role for these proteins in cytoplasmic pH regulation.21,50
Biochemical Properties
Detection of cation/proton antiport in vesicles.
Measurements of acidification of isolated membrane vesicles or intact vacuoles have been fundamental to demonstrate primary active transport of protons by the plasma membrane or tonoplast proton ATPases or PPases.51–53 Inside acid pH gradients can be monitored following the accumulation of membrane permeant weak bases like amonia, immidazole, radioactive amines like [14C]methylamine or fluorescent amines like quinacrine or acridine orange, that are freely permeant in the neutral form, but become trapped iside the vesicles in the protonated form.51 Fluorescence quenching of these dyes upon accumulation inside the vesicles offers a very sensitive measurement of vesicle acidification, even when using membrane vesicles in which only a small proportion consists of tight vesicles containing active ATPase enzymes. However, estimation of real pH gradients is normally not possible.54,55
As plant H+ pumps are electrogenic, the formation of the inside acid pH gradient also depends on the presence of permeant anions at the outside or permeant cations at the inside of the vesicles, to facilitate charge balance. For this reason, assays are normally performed in the presence of relatively permeant Cl− ions, or K+ ions plus the K+ ionophore valinomycin, facilitating electrically coupled H+:Cl− cotransport, or H+:K+ antiport. Indications for the presence of Cation/Proton antiporter systems have been obtained by monitoring the effect of salts on the establishment or dissipation of such pH gradients. Cation/Proton antiporters are expected to diminish the establishment of a pH gradient by proton ATPases,9,10 or to induce dissipation of the gradient when the salt is added once a stable pH gradient is formed.8,56,57 The speed of pH gradient formation or dissipation is however also critically dependent on the relative conductivity of anions and cations. Since isolated membrane vesicles are relatively leaky to protons, it is often difficult to distinguish between electrically coupled or genuine enzymatic Cation/Proton antiport.
First biochemical descriptions of cation/proton antiporter activity in membrane vesicles.
Despite these difficulties, the kinetics of dissipation of a preestablished pH gradient in isolated vacuolar vesicles and intact vacuoles from red beet could be resolved in a saturable and amiloride sensitive electroneutral Na+/H+ antiport component and electrically coupled non-saturable Na+/H+ or K+/H+ exchange.56,57 Similar tonoplast Na+/H+ antiport activity was detected afterwards in various plant species, with Km values for Na+ ranging from 2,4 to 51 mM.56–62 The activity was shown to be induced rapidly by growing plants in saline conditions especially in glycophytes.58 Evidence for K+/H+ antiport activity was also obtained studying the effects of salts on proton gradient formation or dissipation in plasma membranes of tobacco callus,9 tonoplast and plasma membranes isolated from cotton and Atriplex roots12 and zucchini hypocotyls.10 The K+/H+ antiport activity appears to be much less selective for K+, exhibiting considerable activity with other monovalent cations.
Measurement of the activity of the proteins of the NHX antiporter family.
None of the biochemical approaches has lead to the identification of the proteins responsible for antiport activity. It is now clear that the situation is much more complex than originally anticipated, with a total of at least 44 sequences with homology to Na+/H+ or K+/H+ antiporters identified in the Arabidopsis genome, some of which are expressed in tonoplast, plasma membrane or internal membranes of the endosomal pathway.4
Sequence homology of AtNHX1 with amiloride sensitive animal NHE antiporters suggests that this is the protein responsible for the amiloride sensitive and salt stress induced specific Na+/H+ antiport found in tonoplast vesicles.17 The activity of AtNHX1 was assayed in vacuolar membranes obtained from transgenic Arabidopsis overexpressing the protein.17 Na+/H+ antiport activity could be measured in the transgenic plants, showing a Km of 7 mM for Na+ whilst activity in wild type plants was very low. Disruption of AtNHX1 resulted in an even lower Na+/H+ exchange activity.47 Although at first suggested to be specific for Na+, later studies have shown that AtNHX1 expressed in plants also catalyzes K+/H+ antiport, albeit with lower affinity.17,18,47 The fact that null mutants in AtNHX1 or lines overexpressing the gene have substantially altered total antiporter activity, indicate that AtNHX1 is the major contributor to vacuolar Na+/H+ or K+/H+ antiporter activity, in spite of the presence of 5 more NHX isoforms and other proteins of the CPA1 and CPA2 family in Arabidopsis.
To be able to evaluate the structure-function relationships of the plant NHX antiporters in more detail, avoiding interference with other plant antiporters, a more convenient system is provided by heterologous expression in yeast. Darley et al.63 showed that AtNHX1 activity could be measured in vacuolar membrane vesicles obtained from a Saccharomyces cerevisiae yeast strain in which the only endogenous NHX isoform ScNHX1 gene was disrupted. The activity of the plant enzyme appeared to be slightly higher than that of the endogenous yeast protein, with Km values of 11 and 16 mM for Na+ respectively, whilst no antiport activity could be detected in vesicles obtained from the strain not expressing yeast or plant antiporter. The antiport appeared also electroneutral as judged from experiments with the membrane potential probe Oxonol V, and was shown to be sensitive to amiloride. Using a similar yeast strain, Yamaguchi et al.64 found that K+/H+ antiport activity was about two times higher than Na+/H+ antiport activity in vacuolar vesicles obtained from a yeast strain overexpressing the AtNHX1 protein, whilst Km values for K+ and Na+ where 12 and 24 mM respectively. The discrepancies between specificity measurements in plant or yeast could be due to plant specific regulatory mechanisms not present in the heterologous system. In support of this observation it was found that, removal of the C-terminal domain increases the Na+/H+ antiport activity, whilst binding of the AtCaM15 protein has the opposite effect.64,65 It was proposed that in plants, in normal conditions AtNHX1 functions in K+ accumulation, but that salt stress would activate the Na+/H+ exchange mode, releasing interacting partners like CaM15 from the C-terminal domain. Overexpression of AtNHX1 would also induce mainly Na+/H+ antiporter mode due to lack of interacting partners. In yeast, the enzyme would be present in the unactivated K+/H+ antiporter mode. However, differences in expression levels for the mutant enzymes might also have affected the results.64 More importantly, it was shown that the main contributor to Na+/H+ and K+/H+ antiport activity in the yeast vacuolar vesicles is the Vnx1 protein, a protein with homology to Ca2+/H+ and Ca2+/Na+ antiporters.66 The earlier reports describing AtNHX1 activity in yeast should thus be reconsidered as they have been affected by this major background activity. Finally, ScNHX1 was shown to be involved in protein targeting and prevacuolar/vacuolar biogenesis67,68 which complicates the obtention of comparable vacuolar membrane preparations from wild type and ScNHX1 null mutants.
For these reasons, the use of membrane vesicles or intact vacuoles in which many unidentified ion transporters are still functional is not ideal for structure function studies on the plant NHX antiporters. In this respect, heterologous expression in yeast also facilitates protein purification using suitable affinity tags. To avoid interference with other ion transporters such purified protein can be reconstituted in artificial liposomes. Finally, the encapsulation of impermeant pH indicator dyes inside the proteoliposomes during reconstitution, permits real quantitative measurement of pH within the range of responsiveness of the dye. Using this approach it was shown that the AtNHX1 protein catalyzes both Na+/H+ and K+/H+ antiport with similar affinity of about 40 mM.69 This antiport could be inhibited by the amiloride analogs EIPA and benzamil.
The only member of the plant Class-II NHX family that has been studied and for which activity measurements are available is de tomato protein LeNHX2. This protein was also purified and reconstituted into liposomes, showing that the protein catalyzes relatively specific K+/H+ antiport,38 coinciding with ion accumulation studies in yeast.28,38 Unlike AtNHX1 activity, LeNHX2 K+/H+ antiport can actually be blocked by low concentrations of Na+ (Venema K, et al. unpublished results). The activity appeared insensitive to amiloride or its derivatives, in spite of the conservation of amino acids involved in drug sensitivity. Moreover, Arabidopsis plants expressing the tomato protein showed an increased K+/H+ antiport in a subcellular fraction corresponding to internal membranes, coinciding with K+ accumulation and reduced Na+ content in these plants.32
Activity measurements of other NHX isoforms are scarce. Activity of the grape berry Class-I antiporter VvNHX1 was assayed in yeast vacuolar vesicles, indicating similar affinities for K+ and Na+.48 The overexpression of AtNHX3 in salt tolerant sugar beet enhanced above all K+/H+ antiport in tonoplast membrane vesicles and K+ accumulation,70 very similar to the results reported for LeNHX2 overexpression in Arabidopsis32 The purified and reconstituted human NHE8 protein, a mid/trans-Golgi expressed antiporter, somewhat similar to the plant vacuolar clade of antiporters, was also shown to catalyze above all K+/H+ and to a lesser extent Na+/H+ antiport.33 Similarly, the mosquito NHE8 isoform was shown to catalyze Na+/H+ and K+/H+ and Li+/H+ antiport in reconstituted vesicles, whilst 22Na+ uptake was sensitive to amiloride in NHE8 expressing NHE-deficient fibroblast cells.71 Inhibition of Rb+ influx into endosomal compartments by K+, Na+ or Li+, indicates K+/H+ antiport activity and to a lesser extent Na+/H+ and Li+/H+ antiport activity for the human protein NHE7.34 In yeast, a rapid efflux of 22Na+,72 or 86Rb+,68 in plasma membrane permeabilized cells is observed in strains lacking NHX1, whilst a vacuolar pool of these ions remains present in the wild type strain, indicating that the yeast ScNHX1 protein also catalyzes Na+/H+ and K+/H+ exchange. Alltogether these data show that the intracellular NHX family of proteins catalyze relatively non-specific K+/H+ and Na+/H+ antiport, and are much less specific for Na+ than the plasma membrane NHE protein.
Structural Organization and Regulatory Properties
Topological analysis and structure-function studies have so far only been performed with the AtNHX1 protein. Hydropathy analysis of NHX indicates a domain organization similar to NHE isoforms, suggesting that structural features are conserved across the families. Typically, 12 hydrophobic regions that potentially constitute transmembrane helices are predicted in the conserved hydrophobic N-terminal domain, with a divergent hydrophilic C-terminal domain that would be involved in regulatory interactions. To date, two different topological models are proposed.64,73 Detailed in vitro translation experiments indicate that AtNHX1 topology closely resembles the model proposed for human NHE1,73,74 with 11 transmembrane helices and an intramembraneous loop corresponding to hydrophobic region 9. The plant NHX isoforms, contrary to most intracellular NHX isoforms of other organisms lack the first H1 hydrophobic stretch that in NHE1 or NHE6 were shown to represent an N-terminal signal peptide required for endoplasmic reticulum insertion.73 Even so, the first transmembrane helix of AtNHX1, corresponding to transmembrane helix 2 in NHE1, is inserted in the same orientation into the membrane, whilst the C-terminus is exposed to the cytoplasm.73 Hydrophobic region 9 has a very similar topology as compared to the characteristic H10 loop in the human NHE1 isoform, and wouldn't cross the membrane completely.73,74 Mutagenesis of the corresponding region in the yeast ScNHX1 protein has revealed that several amino acids that are uniquely conserved amongst the intracellular NHX are essential for function.75 Still care has to be taken with these data, as a 3D homology model of NHE1 based on the crystal structure of NhaA gives slightly different results, notably concerning NHE1 TM helix 9 and the intramembraneous loop H10.76
A different membrane topology for the Arabidopsis AtNHX1 protein was found based on insertion mutagenesis with a 3xHA epitope.64 In this model the C-terminal domain would be exposed to the vacuolar lumen, whilst the N-terminus would be cytoplasmic. In accordance with this topology it was found that in the yeast ScNHX1 protein some amino acids in the C-terminal domain are N-glycosylated, which indicates that at least part of the C-terminal domain of ScNHX1 is exposed to the endosomal lumen at some stage.77 In this new topology model of AtNHX1, which predicts only 9 transmembrane helices, hydrophobic domain 3, containing the putative amiloride binding domain, and the hydrophobic domains 5 and 6, containing residues that are likely involved in Na+ or H+ binding and transport, would not cross the membrane. This would result in several transmembrane helices being inserted in the opposite direction in the membrane, which was related to the fact that plant NHX enzymes have an opposite transport direction as compared to the plamsa membrane located NHE proteins.64 Whilst animal plasma membrane NHE proteins are activated by cytoplasmic acidification, and normally catalyze entry of Na+ coupled to the extrusion of protons,76 the plant enzymes are suggested to be involved in extrusion (to the vacuole) of Na+ or K+, causing cytoplasmic acidification. The related bacterial (2H+/Na+) NhaA protein is activated by internal alkalinization and catalyzes the entry of protons coupled to the extrusion of Na+.78 Electroneutral 1:1 Cation/Proton exchangers could also be fully reversible, as was shown for instance for amiloride sensitive Na+/H+ exchange in mamalian cells79 and the Schizosaccharomyces pombe plasma membrane SOD2 Na+/H+ antiporter.80 Detailed mutagenesis studies for the human NHE1 protein and structural resolution of individual transmembrane helices, have pinpointed residues in transmembrane segments 4, 7 and 9 (corresponding to 3, 6 and 8 in Arabidopsis AtNHX1) that could be directly involved in ion transport.81 These transmembrane regions are strongly conserved also in the intracellular NHX family. A mechanism for ion translocation in NHE1 was proposed, based on these mutagenesis data and an NHE1 homology model build according to the structure of the bacterial (2H+/Na+) antiporter NhaA.76 This model shows essential roles for P167, P168, E262, D267 and S351, which correspond to amino acids P88, P89, E179, D185 and S271 in the Arabidopsis AtNHX1 sequence, and that are conserved throughout the intracellular NHX family. Also most other residues in NHE that are important for drug binding or activity, are conserved in the NHX sequences. These data strengthen the idea that structure and functioning of the NHX and NHE families is very similar, and that the transport direction is imposed by regulatory domains.
The activity of animal NHE proteins can be regulated by a variety of regulatory mechanisms involving the long C-terminal tail. Preliminary experiments have indicated that removal of the last 82 amino acids in the Arabidopsis AtNHX1 protein modifies the transport specificity of the protein, increasing especially Na+/H+ antiport activity but not K+/H+ antiport activity, indicating a regulatory role of this domain.64 Later it was shown, using a two-hybrid screen and immuno precipitation assays, that the C-terminal domain interacts with a CaM-Like protein AtCaM15.65 AtCaM15 was also found inside the vacuole of transiently transformed Arabidopsis protoplasts and in yeast cells expressing the protein. This localization would permit interaction with the C-terminal domain of AtNHX1 within the vacuoles. Activity measurements using yeast vacuoles obtained from cells expressing AtNHX1 and AtCaM15 indicated that the CaM15 binding inhibits Na+/H+ antiport by AtNHX1, without a significant effect on the Km of the transport reaction. Inhibition of K+/H+ antiport activity was less pronounced, resulting in an increased specificity for K+.
AtNHX1 activity is possibly also regulated through interaction with the protein kinase SOS2.82 SOS2 is the pivotal kinase of the SOS (Salt Overly Sensitive) pathway involved in regulation of ion transport under salt stress and in regulation of several other stress responses.83 It was reported that amiloride sensitive specific vacuolar Na+/H+ antiporter activity in Arabidopsis membrane vesicles was lower in vesicles obtained from sos2 knockout mutants, and that this activity could be stimulated in vitro by the addition of activated SOS2 protein.82 The activity was further inhibited by AtNHX1 antibodies. It was later shown by tandem affinity purification and yeast two-hybrid assays that SOS2 also interacts with several vacuolar V-ATPase subunits and that vesicles isolated from sos2 knockout mutants show considerably lower V-ATPase dependent acidification.83 Comparison of antiport activity in vesicles obtained from wild-type and sos2 mutant plants is thus difficult, as the V-ATPase mediated vesicle acidification and thus driving force for the antiport is not the same in the two cases.
Structural or regulatory mechanisms have not been studied for other plant NHX isoforms. The C-terminus of the yeast NHX1 protein was shown to interact with the small GTPase activating protein Gyp6.84 A model was proposed in which Gyp6 functions as a negative regulator of NHX. Inhibition of NHX1 would result in a more acidic endsosome/prevacuolar compartment limiting retrograde traffic from the prevacuolar compartment to the trans Golgi network or Golgi compartment. Such inhibition would be relieved upon delivery by anterograde traffic of the small GTPase protein GTP-Ypt6, as it will compete with NHX1 for Gyp6 binding. This would result in endosome/prevacuolar alkalinization and termination of the Ypt6 signal, stimulation of retrograde traffic permitting reactivation of Gyp6 by Ric1/Rgp1 in the trans Golgi network or Golgi.84 The C-terminus of the human trans Golgi network localized NHE7 protein was shown to interact with several SCAMP (secretory carrier membrane protein) proteins, which would affect shutteling of NHE7 between recycling vesicles and the trans Golgi network.85 The C-terminus of the NHE7 isoform was also shown to interact with caveolins, facilitating association of NHE7 to caveole/lipd rafts.86 The C-terminal domains of the human isoforms NHE6, 7 and 9, but not NHE8 were found to interact with RACK1 (Receptor for activated C Kinase 1). This interaction was suggested to be important for luminal pH of endocytic recycling compartments and distribution of NHE6 between endosomes and the plasma membrane.87 Rat NHE6 was further shown to interact with the G protein coupled receptor AT-2.88
Function of NHX Antiporters
Salt tolerance.
When grown in saline environment all plants will accumulate Na+ ions to some extent, due to the strong driving force for its entry. Except for some halophytic species that are able to effectively maintain very low Na+ net influx,41,89,90 the accumulation of Na+ inside vacuoles is a strategy used by many plants to survive salt stress.7,8,90 At the cellular level, Na+ accumulation in vacuoles will lower the amount of toxic Na+ ions in the cytoplasm, and lower osmotic potential in the vacuole to maintain turgor pressure and cell expansion in saline conditions. In this way, the translocation and storage of Na+ inside vacuoles in the shoot are suggested to be key factors for sustained growth during salt stress in some plant species.42,90,91 Other plant species tend to limit Na+ accumulation in shoots by reduced transport from root to shoot, recirculation of Na+ out of the shoots and storage in root or stem cell vacuoles.43,90,91 Involvement of NHX antiporters in these processes is indicated by the induction of Na+/H+ antiport activity or NHX gene expression in aerial parts or roots of many plant species when grown in saline environments (see above).
Information on the role of NHX antiporters in ion accumulation and salt tolerance can be obtained by overexpression or silencing of the genes, or by comparison of NHX gene expression and ion accumulation in closely related species differing in salt tolerance. In this context, comparing Melilotus indicus, a halophyte growing up to 400 mM NaCl, with a glycophytic relative Medicago intertexta, it was found that the halophytic species accumulated much less Na+ and maintained higher levels of K+. Na+ accumulation and induction of very similar NHX transcripts in response to salt stress could be found only in the glycophytic species, but not in the halophyte, indicating that NHX gene induction is related to the includer phenotype.41 Also comparing different maize varieties differing in salt tolerance, it was observed that NHX transcripts were only induced in roots of a variety known to exclude Na+ from the shoot.43 Similarly, it was observed that HvNHX1 was mostly induced in roots of the relatively salt tolerant monocot barley, whilst in rice, OsNHX1 induction is above all observed in shoots, suggesting that the high salt tolerance in barley is related to accumulation of Na+ in root cell vacuoles in order to limit transport to the shoot.26,44 Expression pattern of other isoforms was however not studied and the result could thus also have been due to the fact that an isoform with a root specific induction pattern was studied in barley, whilst a shoot induced isoform was studied in rice.
Preliminary studies have shown that AtNHX1 null mutants, or tomato plants in which the antiporter LeNHX2 is silenced are more sensitive to salt stress, although no data on ion accumulation are available.32,47 Much more research is to be expected in the future studying individual Arabidopsis NHX knock-out mutants. Ectopic overexpression of NHX genes has however received by far the most attention, as it can be used as a biotechnological tool to improve crop salt tolerance. Care has to be taken however interpreting these data, as expression is no longer tissue specific or stress inducible, and regulatory properties and even cellular localization of the enzymes might be altered by the strong overexpression. Indeed, altered ion specificity was suggested for overexpressed AtNHX1 protein.64,65
Only Yang et al.93 claim that overexpression of AtNHX1 does not improve salt tolerance in transgenic Arabidopsis plants. All other published reports on overexpression of AtNHX1 or other plant NHX isoforms in a variety of plant species show substantially increased salt tolerance.17–19,30,32,35,36,70,94–103 There is no clear difference in efficiency of the different isoforms, or whether they were obtained from glycophytes or halophytes,36,99 and both Class-I and Class-II antiporters seem to have a similar effect on salt tolerance.32,97 Differential salt tolerance appears thus to be related to regulation of NHX gene expression, and not to differential properties of the proteins. Increased salt tolerance is not always accompanied by increased vacuolar Na+ accumulation. In plants, all possible scenarios with respect to Na+ or K+ can be found (higher Na+, lower K+;17,100 higher Na+ and K+;94,96,97 only marginal differences;26,99 higher K+ and lower Na+,32,36,71 ). As NHX transporters also transport K+, an effect on internal K+ concentrations is to be expected, especially for the more K+ specific Class-II antiporters. A decrease in Na+ content is more difficult to explain, but could result from secondary mechanisms triggered by the improved K+ homeostasis in transgenic plants.32 Transgenic plants were also reported to have a lowered leaf water potential, allowing higher water uptake rates in saline or drought conditions.92
In yeast, disruption of ScNHX1 or overexpression of plant antiporters also affects intracellular Na+ and K+ concentrations. Especially yeast in which the main Na+ efflux system ENA1 is disrupted will accumulate large amounts of Na+ in response to salt stress, at the cost of internal K+.14 Disruption of ScNHX1 has some effect on internal Na+ content, but above all causes a further diminution of internal K+.16,38 Overexpression of AtNHX1 or AtNHX2 strongly increases intracellular K+ and Na+ in ENA and ScNHX1 disrupted yeast cells grown in the presence of NaCl.28 Overexpression of the Class-II antiporters AtNHX5 and LeNHX2 increases intracellular K+, but reduces intracellular Na+.28,38 In the case of the tomato antiporter, it was shown that K+ accumulates in internal stores.38 Also Gaxiola et al.14 showed that ScNHX1 mainly affects intracellular K+ accumulation, as overexpression of the plant H+ pyrophosphatase, supposedly increasing the vacuolar/prevacuolar pH gradient and thus the driving force for ScNHX1 mediated cation accumulation, results in increased salt tolerance, increased K+ levels, and reduced intracellular Na+ levels in the presence of endogenous ScNHX1 only.14
Based on these observations it can be stated that the sequestration model, that suggests that NHX mediated salt tolerance is a consequence of the accumulation of toxic Na+ inside the vacuole, away from the cytosol is too simple, and that at least part of the tolerance is due to K+ accumulation or altered K+ homeostasis, although the precise mechanism remains unclear. It was shown that in conditions of salt stress, K+ continues to be accumulated actively especially in leaf mesophyl cells, and NHX antiporters could very well be involved in such K+ accumulation.104 Especially Class-II antiporters, that are suggested to be more specific to K+ seem to increase internal K+, but cause reduction of internal Na+.14,28,32,38 These data on ion accumulation indicate that the role for NHX antiporters in salt tolerance is above all related to osmotic adjustment by ion accumulation inside the vacuole or endosomal compartments. This is also apparent from the observation that the genes are equally induced by salt, KCl or osmotic treatments, and that plant and yeast NHX antiporters also confer resistance to high KCl or hyper-osmotic shock in yeast cells.14,26,105
K homeostasis.
Apart from a role in osmotic adjustment by Na+ or K+ accumulation in conditions of salt stress, NHX proteins were suggested to fulfil a role in K+ homeostasis in normal growth conditions, based on their ion specificity and affinity.18,69 Most of the cellular K+ is present in the vacuole were it has a biophysical function to maintain turgor and drive cell expansion. The smaller cytoplasmic pool has both osmotic and biochemical functions. Whilst K+ is actively included in the vacuole in normal growth conditions, active export of K+ from the vacuole to the cytoplasm is necessary in severe K+ depletion to maintain adequate cytosolic K+ concentrations. An acidification of the cytoplasm is observed in these conditions that could serve as a signal to induce high affinity K+ uptake, or K+ efflux from the vacuole.2 A reduced pH gradient across the tonoplast membrane would also attenuate the driving force for vacuolar K+ accumulation by a K+/H+ antiporter mechanism. Thermodynamically, active K+ influx into the vacuole in K+ replete conditions can be mediated by the operation of a K+/H+ antiporter, but active efflux was suggested to require a K+:H+ symport system,2 provided that the vacuole is more acidic than the cytoplasm, a condition that apparently not always applies.27 Rodríguez-Rosales et al.32 observed that Arabidopsis plants that overexpress the tomato Class-II antiporter LeNHX2 are more sensitive to K+ deplete conditions, and it was hypothized that by strong overexpression of the NHX protein, the increased antiporter activity could counteract the vacuolar K+ efflux necessary in such conditions, decreasing cytoplasmic K+ concentrations and causing growth inhibition. Similarly, overexpression of AtNHX1 in tomato was reported to provoke K+ deficiency symptoms in spite of increased K+ uptake and content.21,106 In this case, the decreased cytoplasmic K+ concentrations could trigger a K+ starvation signal leading to higher K+ uptake.21 Arabidopsis nhx1 null mutants are reported to have smaller leaf area and epidermal cell size,47 which is possibly related to a vacuolar K+ deficit necessary for turgor generation and cell expansion. In accordance it was observed that nhx mutants exhibit lower root K+ uptake rates and shoot K+ content.21,106 The high expression level of some NHX proteins in stomatal guard cells also suggest that the proteins are essential for vacuolar K+ accumulation and rapid turgor changes that occur in these cells, although such effects were not reported for the studied nhx mutants.45,47,106 DNA array analysis of the nhx null mutants showed increased expression of the high affinity K+ uptake system KUP7/HAK7, and decreased expression of the putative K+ transport system AtKEA4, also pointing to a role for AtNHX1 in potassium homeostasis.107 The high expression level of some isoforms in known sinks for potassium like fruits or flowers, where growth is dependent on cell expansion, point to a role of these isoforms in vacuolar K+ accumulation. It this respect, it was suggested that the high expression level of VvNHX1 protein in grape berries during ripening was related to vacuolar K+ accumulation to drive water flow towards the developing fruit48 needed for the berry size increase. Also in Ipomea tricolor, in addition to the role in vacuolar pH determination and flower colour, the simultaneous induction of NHX1, V-ATPase, V-PPase and PM-ATPase was suggested to be required for K+ accumulation to reduce water potential and drive cell enlargement during flower opening.27
Cellular pH regulation.
Cellular pH homeostasis is one of the most important factors for cellular function. In plants cells cytoplasmic pH is determined by the action of primary proton pumps and metabolic processes producing H+ or OH−. Cation/Proton antiporters constitute proton leak pathways permitting rapid cytoplasmic pH adjustment.108 Several biotic and abiotic stresses have been reported to affect cytoplasmic or vacuolar pH and cytoplasmic pH variation has been shown to be at the basis of many signalling pathways involved in stress responses, developmental processes, hormonal control of stomatal movements, gravitropic response and elongation growth.109–114
Involvement of plant NHX genes in vacuolar pH regulation was most clearly demonstrated analysing the dependence of flower colour on vacuolar pH. The colour change in flowers of Ipomea tricolor cv heavenly blue from purplish red to blue, is caused by a vacuolar pH increase from 6.6 to 7.7 during flower opening, as pH determines the colour of anthocyanins inside the vacuole.27,115 It was shown that in the related Ipomea Nil, a purple flowering mutant that carries a mutation in an NHX gene, was unable to increase vacuolar pH to create the normal blue petals.115 The high vacuolar pH suggests that vacuoles are alkaline respective to the cytosol. Development of such alkaline pH by an electroneutral K+/H+ antiporter mechanism would require higher cytoplasmic K+ concentration respective to the vacuole, as has been reported to occur in K+ deplete conditions in barley root epidermal and cortical cells.2
Although involvement of plant NHX transporters in cytoplasmic pH regulation was not yet demonstrated, it was shown that elicitor induced cytoplasmic acidification, responsible for induction of oxidative burst and synthesis of secondary metabolites, is dependent on stimulation by lysophosphatodylcholine of amiloride sensitive tonoplast Na/H antiporter activity.114
In yeast, involvement of ScNHX1 in pH regulation was clearly demonstrated. ScNHX1 disrupted cells are sensitive to acid pH and have more acidic vacuolar and cytoplasmic pH.68 Activation of ScNHX1 in yeast by acidic pH was suggested to be at the basis of enhanced Na+ accumulation in mutant cells with lower plasma membrane PMA1 activity.72
The most important function of yeast ScNHX1 appears to be related to its involvement in protein sorting through endosomal pH regulation. The ScNHX1 gene was shown to be identical to the vacuolar protein sorting gene VPS44,67 and disruption causes CPY secretion, endosomal enlargement and accumulation in endosomes of the G protein coupled receptor Ste3 or the dye FM4–64 that are delivered to the vacuole in wild type cells.67,68 Disruption of ScNHX1 also causes strong Hygromycin B sensitivity,14 a phenotype shared by many other Vacuolar Protein Sorting (VPS) mutants, indicating that the phenotype is dependent on defective vacuolar biogenesis, potentially a site for detoxification of the drug.67,75 Addition of weak bases can suppress these phenotypes in nhx1 null mutants showing that endosomal alkalinisation by ScNHX1 is essential for trafficking out of the endosome.75 Such a fundamental role in pH regulation is likely to depend on the more physiologically relevant K+ and not Na+. Therefore, the ability to catalyze K+/H+ exchange appears to be a universal feature of the intracellular NHX enzymes, as has been discussed above.68,75
Both Class-I and Class-II plant NHX isoforms complement NaCl, KCl and hygromycin sensitivity of the yeast ScNHX1 disruption mutant.16,28,38 It is thus tempting to suggest a role for plant NHX proteins in endosomal pH regulation and protein trafficking as well. The effect of expression of plant NHX genes in yeast on protein trafficking was however not studied, and no information from plant studies is available. Interestingly however it was shown that plants that overexpress the intracellular vesicle trafficking protein AtRab7, involved in vacuolar biogenesis, have increased resistance to salt and osmotic stress and accumulate increased amounts of Na+ inside the vacuole.116 Furthermore, suppression of vesicle-SNARE expression was shown to increase salt tolerance, presumably by inhibition of the delivery of ROS-producing endosomes to the vacuole.117 Regulation of vesicle trafficking by plant NHX could thus represent an alternative or additional route for NHX proteins to enhance salt tolerance. The plant Class-II sequences, more closely related to NHX proteins from other organisms with a demonstrated role in vesicle trafficking, catalyze more specific K+/H+ exchange, and show a subcellular expression pattern similar to the yeast ScNHX1 protein, which makes them the most likely candidates for such role in plants. Silencing of the LeNHX2 gene in tomato plants causes a very severe phenotype which would be in accordance with a fundamental cellular role for the encoded protein.32 The Class-I antiporters could also play a role in intracellular vesicles trafficking, as DNA array analysis of an nhx T-DNA insertional mutant showed changes in the expression of a large number of genes encoding proteins associated with intravesicular trafficking, trafficking to the nucleus and Golgi processing.107
Concluding Remarks
Published research has shown that NHX antiporters play roles in salt tolerance, vacuolar pH regulation and K+ homeostasis. Contrary to general belief, NHX mediated salt tolerance is not strictly related to Na+ accumulation, and reduced Na+ content and increased K+ content are equally often observed. This suggests that the genes function in vacuolar osmotic adjustment via K+ or Na+ accumulation, in accordance with their ion specificity and gene induction pattern, or that salt tolerance is induced by other mechanisms possibly by indirect effects on vesicle trafficking via endosomal pH regulation. In this respect it has to be pointed out that plant salt tolerance mechanisms appear to be plant species or variety specific, determined by differential responses at the tissue, cell-type and subcellular level. Also vacuolar or cytoplasmic pH and K+ concentrations are variable between cell types, and differentially regulated in different species. The differential expression of NHX genes in plants or varieties that accumulate or exclude Na+ indicates that NHX proteins could play crucial roles in such differences, but this has not been studied in much detail. Only in one case, the alkaline pH in vacuoles of Ipomea nil, a species and cell-type specific role of NHX was clearly demonstrated.
Little more information can be gathered from general overexpression experiments and crude experiments comparing species based on total shoot or root ion content and antiporter expression levels. It indeed seems odd, that unregulated overexpression of one gene, promoting vacuolar accumulation of ions in vacuoles across the plant, can substantially improve such a complex trait as salt tolerance in such a wide variety of plants. To describe function of the NHX genes in plants, genetic studies using individual or multiple NHX knock-out mutants are required, as well as detailed studies on tissue distribution and vacuolar or cytoplasmic ion content. Function of plant NHX genes in endosomal vesicle trafficking has not been studied so far and the search for regulatory mechanisms and NHX interacting partners is lacking far behind compared to yeast and animal studies. These studies will be crucial to provide more clues to the real function of the various NHX isoforms in plants.
Acknowledgements
Funding was obtained through grants from Spanish Plan Nacional I + D + I (BIO2005-0878 to Kees Venema and BIO2008-01691 to M. Pilar Rodríguez-Rosales) and from Consejería de Innovación Ciencia y Empresa, Junta de Andalucía (AGR2005-436 to M. Pilar Rodríguez-Rosales).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7919
References
- 1.Leigh RA, Wyn Jones RG. A hypothesis relating critical potassium concentrations to the distribution and functions of this ion in the plant cell. New Phytol. 1984;97:1–13. [Google Scholar]
- 2.Walker DJ, Leigh RA, Miller AJ. Potassium homeostasis in vacuolated plant cells. Proc Natl Acad Sci USA. 1996;93:10510–10514. doi: 10.1073/pnas.93.19.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song CP, Guo Y, Qiu Q, Lambert G, Galbraith DW, Jagendorf A, et al. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:10211–10216. doi: 10.1073/pnas.0403709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sze H, Padmanaban S, Cellier F, Honys D, Cheng NH, Bock KW, et al. Expression patterns of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol. 2004;136:2532–2547. doi: 10.1104/pp.104.046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A. Sodium transport and HKT transporters: the rice model. Plant J. 2003;34:788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- 6.Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, et al. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 9.Sze H. Proton-pumping adenosine triphosphatase in membrane vesicles of tobacco callus. Sensitivity to vanadate and K+ Biochim Biophys Acta. 1983;732:586–594. [Google Scholar]
- 10.Scherer GFE, Martiny-Baron G. K+/H+ exchange transport in plant membrane vesicles is evidence for K+ transport. Plant Sci. 1985;41:161–168. [Google Scholar]
- 11.Sarafian V, Poole R. K+/H+ antiport in tonoplast vesicles. Plant Physiol. 1987;83:329. [Google Scholar]
- 12.Hassidim M, Braun Y, Lerner HR, Reinhold L. Na+/H+ and K+/H+ antiport in root membrane vesicles isolated from the halophyte Atriplex and the glycophyte cotton. Plant Physiol. 1990;94:1795–1801. doi: 10.1104/pp.94.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaxiola RA, Rao R, Sherman A, Grifasi P, Alpier SL, Fink GR. The Arabidopsis thaliana proton transporters, AtNHX1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:223–239. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 16.Quintero FJ, Blatt MR, Pardo JM. Functional conservation between yeast and plant endosomal Na+/H+ antiporters. FEBS Lett. 2000;471:224–228. doi: 10.1016/s0014-5793(00)01412-5. [DOI] [PubMed] [Google Scholar]
- 17.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H-X, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19:765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H-X, Hodson JN, Williams JP, Blumwald E. Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci USA. 2001;98:12832–12836. doi: 10.1073/pnas.231476498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saier MH, Eng BH, Fard S, Garg J, Haggerty DA, Hutchinson WJ, et al. Phylogenic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 21.Pardo JM, Cubero B, Leidi EO, Quintero FJ. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot. 2006;57:1181–1199. doi: 10.1093/jxb/erj114. [DOI] [PubMed] [Google Scholar]
- 22.Patel H, Barber DL. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J Cell Biol. 2005;169:321–329. doi: 10.1083/jcb.200412145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T. Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol. 2001;46:35–42. doi: 10.1023/a:1010603222673. [DOI] [PubMed] [Google Scholar]
- 24.Xia T, Apse MP, Aharon GS, Blumwald E. Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol Plant. 2002;116:206–212. doi: 10.1034/j.1399-3054.2002.1160210.x. [DOI] [PubMed] [Google Scholar]
- 25.Vasekina AV, Yershov PV, Reshetova OS, Tikhonova TV, Lunin VG, Trofimova MS, Babakov AV. Vacuolar Na+/H+ antiporter from Barley: identification and response to salt stress. Biochemistry (Mosc) 2005;70:100–107. doi: 10.1007/s10541-005-0057-8. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, et al. Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 2004;45:146–159. doi: 10.1093/pcp/pch014. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida K, Kawachi M, Mori M, Maeshima M, Kondo M, Nishimura M, et al. The involvement of tonoplast proton pumps and Na+(K+)/H+ exchangers in the change of petal colour during flower opening of Morning Glory, Ipomea tricolor cv. Heavenly Blue. Plant Cell Physiol. 2005;46:407–415. doi: 10.1093/pcp/pci057. [DOI] [PubMed] [Google Scholar]
- 28.Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, et al. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313x.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi M, Fukada-Tanaka S, Hoshino A, Takada J, Inagaki Y, Iida S. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in Japanese Morning Glory. Plant Cell Physiol. 2005;46:259–267. doi: 10.1093/pcp/pci028. [DOI] [PubMed] [Google Scholar]
- 30.Li WY, Wong FL, Tsai SN, Phang TH, Shao G, Lam HM. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environm. 2006;29:1122–1137. doi: 10.1111/j.1365-3040.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 31.Vera-Estrella R, Barkla BJ, García-Ramírez L, Pantoja O. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 2005;139:1507–1517. doi: 10.1104/pp.105.067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Rosales MP, Jiang XJ, Gálvez FJ, Aranda MN, Cubero B, Venema K. Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 2008;179:366–377. doi: 10.1111/j.1469-8137.2008.02461.x. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura N, Tanaka S, Teko Y, Mitsui K, Kanazawa H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J Biol Chem. 2005;280:1561–1572. doi: 10.1074/jbc.M410041200. [DOI] [PubMed] [Google Scholar]
- 34.Numata M, Orlowski J. Molecular cloning and characterization of a novel (Na+, K+)/H+ exchanger localized to the trans-Golgi network. J Biol Chem. 2001;276:17387–17394. doi: 10.1074/jbc.M101319200. [DOI] [PubMed] [Google Scholar]
- 35.Wu CA, Yang GD, Meng QW, Zheng CC. The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 2004;45:600–607. doi: 10.1093/pcp/pch071. [DOI] [PubMed] [Google Scholar]
- 36.Zhang GH, Su Q, An LJ, Wu S. Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiol Biochem. 2008;46:117–126. doi: 10.1016/j.plaphy.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Kagami T, Suzuki M. Molecular and functional analysis of a vacuolar Na+/H+ antiporter gene of Rosa hybrida. Genes Genet Syst. 2005;80:121–128. doi: 10.1266/ggs.80.121. [DOI] [PubMed] [Google Scholar]
- 38.Venema K, Belver A, Marín-Manzano MC, Rodríguez-Rosales MP, Donaire JP. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. J Biol Chem. 2003;278:22453–22459. doi: 10.1074/jbc.M210794200. [DOI] [PubMed] [Google Scholar]
- 39.Brini F, Gaxiola RA, Berkowitz GA, Masmoudi K. Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol Biochem. 2005;43:347–354. doi: 10.1016/j.plaphy.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda A, Nakamura A, Tanaka Y. Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta. 1999;1446:149–155. doi: 10.1016/s0167-4781(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 41.Zahran HH, Marín-Manzano MC, Sánchez-Raya AJ, Bedmar EJ, Venema K, Rodríguez-Rosales MP. Effect of salt stress on the expression of NHX-type ion transporters in Medicago intretexta and Melilotus indicus plants. Physiol Plant. 2007;131:122–130. doi: 10.1111/j.1399-3054.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 42.Chauhan S, Forsthoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ. Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J. 2000;24:511–522. doi: 10.1046/j.1365-313x.2000.00903.x. [DOI] [PubMed] [Google Scholar]
- 43.Zörb C, Noll A, Karl S, Leib K, Yan F, Schubert S. Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress. J Plant Physiol. 2004;162:55–66. doi: 10.1016/j.jplph.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Fukuda A, Chiba K, Maeda M, Nakamura A, Maeshima M, Tanaka Y. Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. J Exp Bot. 2004;55:585–594. doi: 10.1093/jxb/erh070. [DOI] [PubMed] [Google Scholar]
- 45.Shi H, Zhu JK. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol. 2002;50:543–550. doi: 10.1023/a:1019859319617. [DOI] [PubMed] [Google Scholar]
- 46.Porat R, Pavoncello D, Ben-Hayyim G, Lurie S. A heat treatment induced the expression of a Na+/H+ antiport gene (cNHX1) in citrus fruit. Plant Sci. 2002;162:957–963. [Google Scholar]
- 47.Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36:229–239. doi: 10.1046/j.1365-313x.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 48.Hanana M, Cagnac O, Yamaguchi T, Hamdi S, Ghorbel A, Blumwald E. A Grape berry (Vitis vinifera L.) Cation/proton antiporter is associated with berry ripening. Plant Cell Physiol. 2007;48:804–811. doi: 10.1093/pcp/pcm048. [DOI] [PubMed] [Google Scholar]
- 49.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PloS ONE. 2007;8:718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viehweger K, Dordschbal B, Roos W. Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signalling via the activation of Na+-dependent proton fluxes. Plant Cell. 2002;14:1509–1525. doi: 10.1105/tpc.002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scarborough GA. Proton translocation catalyzed by the electrogenic ATPase in the plasma membrane of Neurospora. Biochemistry. 1980;19:2925–2931. doi: 10.1021/bi00554a017. [DOI] [PubMed] [Google Scholar]
- 52.Hager A, Frenzel R, Laible D. ATP-dependent proton transport into vesicles of microsomal membranes of Zea mays coleoptiles. Z Naturforsch. 1980;35:783–793. doi: 10.1515/znc-1980-9-1021. [DOI] [PubMed] [Google Scholar]
- 53.Churchill KA, Holaway B, Sze H. Separation of two types of electrogenic H+-pumping ATPases from oat roots. Plant Physiol. 1983;73:921–928. doi: 10.1104/pp.73.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmgren MG. Acridine orange as a probe for measuring pH gradients across membranes: mechanism and limitations. Anal Biochem. 1991;192:316–321. doi: 10.1016/0003-2697(91)90542-2. [DOI] [PubMed] [Google Scholar]
- 55.Clerc S, Barenholz Y. A quantitative model for using acridine orange as a transmembrane pH gradient probe. Anal Biochem. 1998;259:104–111. doi: 10.1006/abio.1998.2639. [DOI] [PubMed] [Google Scholar]
- 56.Blumwald E, Poole R. Na+/H+ antiport activity in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 1985;78:163–167. doi: 10.1104/pp.78.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blumwald E, Poole R. Salt tolerance in suspension culture of sugar beet. Induction of Na+/H+ antiport activity at the tonoplast by growth in salt. Plant Physiol. 1987;83:884–887. doi: 10.1104/pp.83.4.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garbarino J, Dupont FM. Rapid induction of Na+/H+ exchange activity in barley root tonoplast. Plant Physiol. 1989;89:1–4. doi: 10.1104/pp.89.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staal M, Maathuis FJM, Elzenga JTM, Overbeek JHM, Prins HBA. Na+/H+ antiport activity in tonoplast vesicles from roots of the salt-tolerant Plantago maritima and the salt-sensitive Plantago media. Physiol Plant. 1991;82:179–184. [Google Scholar]
- 60.Matoh T, Ishikawa T, Takahashi E. Collapse of ATP-induced pH gradient by sodium ions in microsomal membrane vesicles prepared from Atriplex gmelini leaves. Plant Physiol. 1988;89:180–183. doi: 10.1104/pp.89.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fukada A, Nakamura A, Tanaka Y. Molecular cloning and expression of the Na+/H+ exchanger gene in Oryza sativa. Biochim Biophys Acta. 1999;1446:149–155. doi: 10.1016/s0167-4781(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 62.Ballesteros E, Blumwald E, Donaire JP, Belver A. Na+/H+ antiport activity in tonoplast enriched vesicles isolated from sunflower roots induced by salt stress. Physiol Plant. 1997;99:328–334. [Google Scholar]
- 63.Darley CP, van Wuytswinkel OCM, van der Woude K, Mager WH, de Boer AH. Arabidopsis thaliana and Saccharomyces cerevisiae NHX1 genes encode amiloride sensitive electroneutral Na+/H+ exchangers. Biochem J. 2000;351:241–249. doi: 10.1042/0264-6021:3510241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamaguchi T, Apse MP, Shi H, Blumwald E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA. 2003;100:12510–12515. doi: 10.1073/pnas.2034966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci USA. 2005;102:16107–16112. doi: 10.1073/pnas.0504437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cagnac O, Leterrier M, Yeager M, Blumwald E. Identification and characterization of Vnx1p, a novel type of vacuolar monovalent cation/H+ antiporter of Saccharomyces cerevisiae. J Biol Chem. 2007;282:24284–24293. doi: 10.1074/jbc.M703116200. [DOI] [PubMed] [Google Scholar]
- 67.Bowers K, Boaz PL, Patel FI, Stevens TH. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:4277–4294. doi: 10.1091/mbc.11.12.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+(K+)/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venema K, Quintero FJ, Pardo JM, Donaire JP. The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J Biol Chem. 2002;277:2413–2418. doi: 10.1074/jbc.M105043200. [DOI] [PubMed] [Google Scholar]
- 70.Liu H, Wang Q, Yu M, Zhang Y, Wu Y, Zhang H. Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively overexpressing an Arabidopsis thaliana vacuolar Na+/H+ antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage root. Plant Cell Environm. 2008;31:1325–1334. doi: 10.1111/j.1365-3040.2008.01838.x. [DOI] [PubMed] [Google Scholar]
- 71.Kang'ethe W, Aimanova KG, Pullikuth AK, Gill SS. NHE8 mediates amiloride-sensitive Na+/H+ exchange across mosquito Malpighian tubules and catalyzes Na+ and K+ transport in reconstituted proteoliposomes. Am J Physiol Renal Physiol. 2007;292:1501–1512. doi: 10.1152/ajprenal.00487.2005. [DOI] [PubMed] [Google Scholar]
- 72.Nass R, Cunningham KW, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 73.Sato Y, Sakaguchi M. Topogenic properties of transmembrane segments of Arabidopsis thaliana NHX1 reveal a common topology model of the Na+/H+ exchanger family. J Biochem. 2005;138:425–431. doi: 10.1093/jb/mvi132. [DOI] [PubMed] [Google Scholar]
- 74.Wakabayashi S, Pang T, Su X, Shigekawa M. A novel topological model of the human Na+/H+ exchanger isoform 1. J Biol Chem. 2000;275:7942–7949. doi: 10.1074/jbc.275.11.7942. [DOI] [PubMed] [Google Scholar]
- 75.Mukerjee S, Kallay L, Brett CL, Rao R. Mutational analysis of the intramembranous H10 loop of yeast Nhx1 reveals a critical role in ion homeostasis and vesicle trafficking. Biochem J. 2006;398:97–105. doi: 10.1042/BJ20060388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landau M, Herz K, Padan E, Ben-Tal N. Model structure of the Na+/H+ exchanger 1 (NHE1). Functional and clinical implications. J Biol Chem. 2007;282:37854–37863. doi: 10.1074/jbc.M705460200. [DOI] [PubMed] [Google Scholar]
- 77.Wells KM, Rao R. The yeast Na+/H+ exchanger Nhx1 is an N-linked glycoprotein. Topological implications. J Biol Chem. 2001;276:3401–3407. doi: 10.1074/jbc.M001688200. [DOI] [PubMed] [Google Scholar]
- 78.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 79.Paris S, Pouysségur J. Biochemical characterization of the amiloride-sensitive Na+/H+ antiport in chinese hamster lung fibroblasts. J Biol Chem. 1983;258:3503–3508. [PubMed] [Google Scholar]
- 80.Hahnenberger KM, Jia Z, Young PG. Functional expression of the Schizosaccharomyces pombe Na+/H+ antiporter gene, sod2, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5031–5036. doi: 10.1073/pnas.93.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy T, Ding J, Li X, Sykes BD, Rainey JK, Fliegel L. Structural and functional characterization of TM IX of the NHE1 isoform of the Na+/H+ exchanger. J Biol Chem. 2008;283:22018–22030. doi: 10.1074/jbc.M803447200. [DOI] [PubMed] [Google Scholar]
- 82.Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly sensitive (SOS) pathway. J Biol Chem. 2004;279:207–215. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- 83.Batelli G, Verslues PE, Agius F, Qiu Q, Fujii H, Pan S, et al. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol Cell Biol. 2007;27:7781–7790. doi: 10.1128/MCB.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ali R, Brett CL, Mukherjee S, Rao R. Inhibition of sodium/proton exchange by Rab-GTPase-activating protein regulates endosomal traffic in yeast. J Biol Chem. 2004;279:4498–4506. doi: 10.1074/jbc.M307446200. [DOI] [PubMed] [Google Scholar]
- 85.Lin PJC, Williams WP, Luu Y, Molday RS, Orlowski J, Numata M. Secretory carrier membrane proteins interact and regulate trafficking of the organellar (Na+,K+)/H+ exchanger NHE7. J Cell Sci. 2005;118:1885–1897. doi: 10.1242/jcs.02315. [DOI] [PubMed] [Google Scholar]
- 86.Lin PJC, Williams WP, Kobiljski J, Numata M. Caveolins bind to (Na+, K+)/H+ exchanger NHE7 by a novel binding module. Cell Signal. 2007;19:978–988. doi: 10.1016/j.cellsig.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 87.Ohgaki R, Fukura N, Matsushita M, Mitsui K, Kanazawa H. Cell surface levels of organellar Na+/H+ exchanger isoform 6 are regulated by interaction with RACK1. J Biol Chem. 2008;283:4417–4429. doi: 10.1074/jbc.M705146200. [DOI] [PubMed] [Google Scholar]
- 88.Pulakat L, Cooper S, Knowle D, Mandavia C, Bruhl S, Hetrick M, et al. Ligand-dependent complex formation between the angiotensin II receptor subtype AT2 and Na+/H+ exchanger NHE6 in mammalian cells. Peptides. 2005;26:863–873. doi: 10.1016/j.peptides.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 89.Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, et al. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Phys. 2004;135:1697–1709. doi: 10.1104/pp.104.039909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Munns R, Tester M. Mechanisms of salinity tolerance. Anu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 91.Maathuis FJM, Amtmann K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot. 1999;84:123–133. [Google Scholar]
- 92.Lohaus G, Hussmann M, Pennewiss K, Schneider H, Zhu JJ, Sattelmacher B. Solute balance of a maize (Zea mays L.) source leaf as affected by salt treatment with special emphasis on phloem retranslocation and ion leaching. J Exp Bot. 2000;51:1721–1732. doi: 10.1093/jexbot/51.351.1721. [DOI] [PubMed] [Google Scholar]
- 93.Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, et al. Overexpression of SOS (Salt Overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant. 2009 doi: 10.1093/mp/ssn058. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brini F, Hanin M, Mezghani I, Berkowitz GA, Masmoudi K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J Exp Bot. 2007;58:301–308. doi: 10.1093/jxb/erl251. [DOI] [PubMed] [Google Scholar]
- 95.Rajagopal D, Agarwal P, Tyagi W, Singla-Pareek SL, Reddy MK, Sopory SK. Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol Breeding. 2007;19:137–151. [Google Scholar]
- 96.Zhao J, Zhi D, Xue Z, Liu H, Xia G. Enhanced salt tolerance of transgenic progeny of tall fescue (Festuca arundinacea) expressing a vacuolar Na+/H+ antiporter gene from Arabidopsis. J Plant Physiol. 2007;164:1377–1383. doi: 10.1016/j.jplph.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 97.Shi LY, Li HQ, Pan XP, Wu GJ, Li MR. Improvement of Torenia fournieri salinity tolerance by expression of Arabidopsis AtNHX5. Functional Plant Biol. 2008;35:185–192. doi: 10.1071/FP07269. [DOI] [PubMed] [Google Scholar]
- 98.He C, Yan J, Shen G, Fu L, Holaday AS, Auld D, et al. Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fibre yield in the field. Plant Cell Physiol. 2005;46:1848–1854. doi: 10.1093/pcp/pci201. [DOI] [PubMed] [Google Scholar]
- 99.Li JY, He XW, Xu L, Zhou J, Wu P, Shou HX, et al. Molecular and functional comparisons of the vacuolar Na+/H+ exchangers originated from glycophytic and halophitic species. J Zhejiang Univ Science B. 2007;9:132–140. doi: 10.1631/jzus.B0710445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen LH, Zhang B, Xu ZQ. Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum) Transgenic Res. 2008;17:121–132. doi: 10.1007/s11248-007-9085-z. [DOI] [PubMed] [Google Scholar]
- 101.Verma D, Singla-Pareek SL, Rajagopal D, Reddy MK, Soory SK. Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. J Biosci. 2007;32:621–628. doi: 10.1007/s12038-007-0061-9. [DOI] [PubMed] [Google Scholar]
- 102.Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM. Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+ Plant Sci. 2004;167:849–859. [Google Scholar]
- 103.Ohta M, Hayashi Y, Nakashima A, Hamada A, Tanaka A, Nakamura T, et al. Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice. FEBS Lett. 2002;532:279–282. doi: 10.1016/s0014-5793(02)03679-7. [DOI] [PubMed] [Google Scholar]
- 104.Cuin TA, Miller AJ, Laurie SA, Leigh RA. Potassium activities in cell compartments of salt-grown barley leaves. J Exp Bot. 2002;54:657–661. doi: 10.1093/jxb/erg072. [DOI] [PubMed] [Google Scholar]
- 105.Nass R, Rao R. The yeast endosomal Na+/H+ exchanger, Nhx1, confers osmotolerance following acute hypertonic shock. Microbiology. 1999;145:3221–3228. doi: 10.1099/00221287-145-11-3221. [DOI] [PubMed] [Google Scholar]
- 106.Leidi EO, Barragán V, Cubero B, Quintero FJ, Pardo JM. Function of endosomal NHX antiporters on plant K nutrition. Society of Experimental Biology Annual Main Meeting. Comparative Biochemistry and Physiology—Part A. 2005;141:342. [Google Scholar]
- 107.Sottosanto JB, Gelli A, Blumwald E. DNA array analyses of Arabidopsis thaliana lacking vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression. Plant J. 2004;40:752–771. doi: 10.1111/j.1365-313X.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- 108.Sakano K. Revision of biochemical pH-Stat: Involvement of alternative pathway metabolisms. Plant Cell Physiol. 1998;39:467–473. [Google Scholar]
- 109.Roos W. Ion mapping in plant cells—Methods and applications in signal transduction research. Planta. 2000;210:347–370. doi: 10.1007/PL00008144. [DOI] [PubMed] [Google Scholar]
- 110.MacRobbie EAC. Signal transduction and ion channels in guard cells. Phil Trans R Soc Lond B. 1998;353:1475–1488. doi: 10.1098/rstb.1998.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao J, Davis LC, Verpoorte R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv. 2005;23:283–333. doi: 10.1016/j.biotechadv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 112.Netting AG. pH, absicic acid and the integration of metabolism in plants under stressed and non-stressed conditions II. Modifications in modes of metabolism induced by variation in the tension on the water column and by stress. J Exp Bot. 2002;53:151–173. [PubMed] [Google Scholar]
- 113.Frohnmeyer H, Grabov A, Blatt MR. A role for the vacuole in auxin-mediated control of cytosolic pH by Vivia mesophyll and guard cells. Plant J. 1998;13:109–116. [Google Scholar]
- 114.Viehweger K, Dordschbal B, Roos W. Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+-dependent proton fluxes. Plant Cell. 2002;14:1509–1525. doi: 10.1105/tpc.002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fukada-Tanaka S, Inagaki Y, Yamaguchi T, Saito N, Iida S. Colour-enhancing protein in blue petals. Nature. 2000;407:581. doi: 10.1038/35036683. [DOI] [PubMed] [Google Scholar]
- 116.Mazel A, Leshem Y, Tiwari BS, Levine A. Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e) Plant Physiol. 2004;134:118–128. doi: 10.1104/pp.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leshem Y, Melamed-Book N, Cagnac O, Ronen G, Nishri Y, Solomon M, et al. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2 containing vesicles with tonoplast and increased salt tolerance. Proc Natl Acad Sci USA. 2006;103:18008–18013. doi: 10.1073/pnas.0604421103. [DOI] [PMC free article] [PubMed] [Google Scholar]