Abstract
The transcriptional regulation of cardiovascular development requires precise spatiotemporal control of gene expression, and heterozygous mutations of transcription factors has frequently been implicated in human cardiovascular malformations. A novel mechanism involving posttranscriptional regulation by small, noncoding microRNAs (miRNAs) has emerged as a central regulator of many cardiogenic processes. We are beginning to understand the functions that miRNAs play during essential biological processes, such as cell proliferation, differentiation, apoptosis, stress response, and tumorigenesis. The identification of miRNAs expressed in specific cardiac and vascular cell types has led to the discovery of important regulatory roles for these small RNAs during cardiomyocyte differentiation, cell cycle, conduction, vessel formation, and during stages of cardiac hypertrophy in the adult. Here, we overview the recent findings on miRNA regulation in cardiovascular development and report the latest advances in understanding their function by unveiling their mRNA targets. Further analysis of miRNA function during cardiovascular development will allow us to determine the potential for novel miRNA-based therapeutic strategies.
Keywords: microRNA, cardiogenesis, angiogenesis, fetal cardiac gene reactivation, cardiac patterning
Over the last decade, animal studies and advances in human genetics have highlighted the need or precise regulation of key molecular pathways during embryonic development. This is particularly true for the cardiovascular system, where haploinsufficiency of essential genes often causes human cardiac malformations 1. Such congenital heart defects (CHDs) are the most common birth defects in humans, occurring in nearly 1 in 100 live births 2, and are a result of defects in cell lineage decisions or subsequent morphogenesis. The same pathways that regulate early steps of cardiomyocyte determination and differentiation are involved in adaptive processes in adult heart disease and may be harnessed to encourage progenitor cells to adopt a cardiomyocyte cell fate in postnatal life.
The dosage of cardiogenic pathways can be controlled at numerous levels, some of which have been well-studied. In particular, the transcriptional regulation of cardiomyocyte differentiation and cardiac morphogenesis is highly conserved across species, and heterozygous mutations of transcription factors have frequently been implicated in human cardiac malformations 3. Recently, post-transcriptional regulation by small noncoding RNAs, such as microRNAs (miRNAs), has emerged as a central regulator of many cardiogenic processes.
miRNAs are a large class of evolutionarily conserved, small, noncoding RNAs, typically 20–26 nucleotides (nt) in length, that primarily function post-transcriptionally by interacting with the 3' untranslated region (UTR) of specific target mRNAs in a sequence-specific manner (reviewed in Zhao & Srivastava, 20074). The first animal miRNA was described in 1993 as a regulator of developmental timing in Caenorhabditis elegans5, 6 it was not until 2001 that miRNAs were recognized to be widespread in all eukaryotes, including vertebrates 7–9. Over 650 miRNAs are encoded in the human genome, and each is thought to target more than 100 mRNAs, resulting in mRNA degradation or translational inhibition. Interactions between miRNAs and mRNAs are thought to require sequence homology in the 5' end of the miRNA, but significant variance in the degree of complementation in the remaining sequence allows a single miRNA to target a wide range of mRNAs, often regulating multiple genes within a common pathway. As a result, over one-third of mRNAs in the mammalian genome are thought to be regulated by one or more miRNAs 10.
Despite advances in miRNA discovery, the role of miRNAs in physiologic and pathophysiologic processes is just emerging. It has become clear that miRNAs play diverse roles in fundamental biological processes, such as cell proliferation, differentiation, apoptosis, stress response, and tumorigenesis. In many cancers, miRNAs are dysregulated and may act as tumor suppressors; for example, the tumor suppressor gene p53 regulates the miR-34 family 11, and let-7 represses a prevalent oncogene found in a variety of tumors 12. Identification of miRNAs expressed in specific cardiac cell types has led to the discovery of important regulatory roles for these small RNAs during cardiomyocyte differentiation, cell cycle, conduction, and during stages of cardiac hypertrophy in the adult, indicating that miRNAs may be as important as transcription factors in controlling cardiac gene expression.
Here we review the basic mechanisms by which miRNAs function, with a focus on the role of miRNAs during development of the heart and vessels. It appears that a network of miRNAs can be superimposed on well-described signaling and transcriptional networks with considerable intersection between the two. Ultimately, knowledge of the function and regulation of specific miRNAs and their mRNA targets in the heart will lead to a deeper understanding of cardiac cell fate decisions and morphogenesis and ultimately could result in novel therapeutic or preventive approaches for heart disease.
miRNA Organization, Biogenesis, and Target Recognition
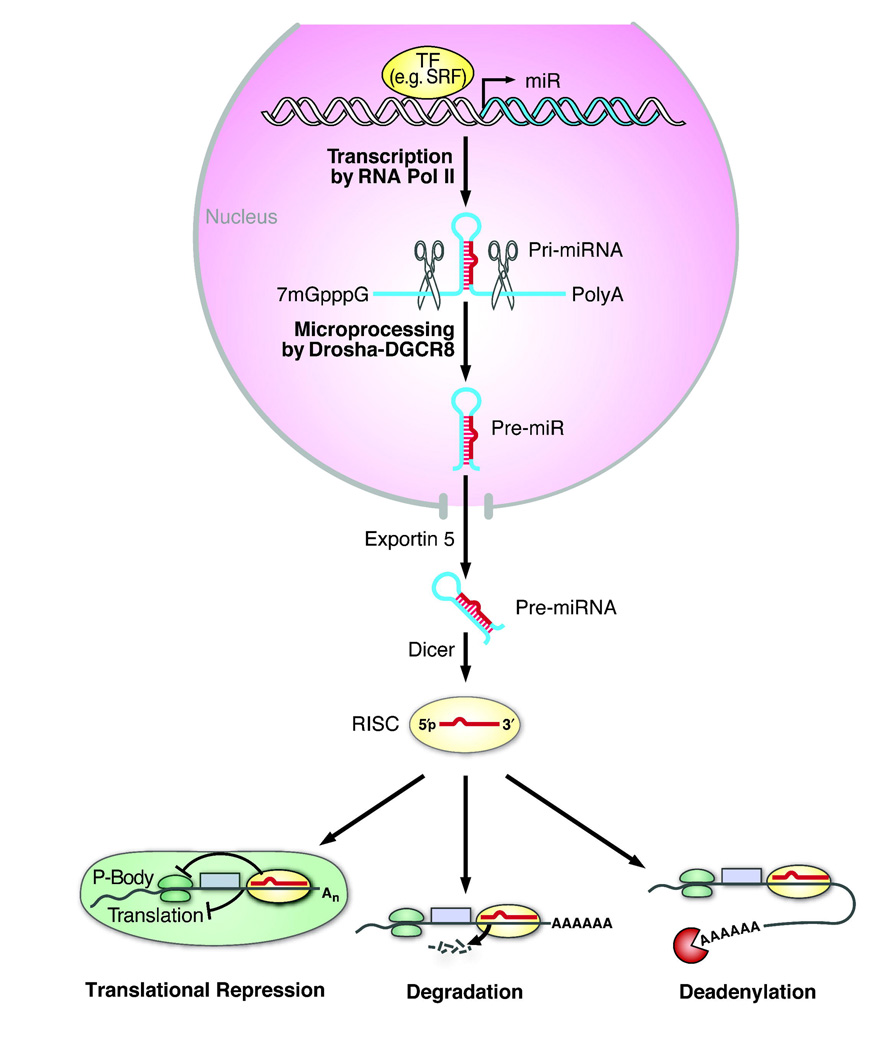
miRNAs regulate gene expression at the post-transcriptional level by mRNA degradation, translation repression, or miRNA-mediated mRNA decay (Fig. 1). Mature miRNAs are formed in a multi-step biological process involving critical endonucleases. miRNAs are initially transcribed from the genome into long (several kilobases) 5' capped polyadenylated (poly(A)) primary transcripts (pri-miRNAs) by RNA polymerase II 13. Some miRNAs interspersed among repetitive DNA elements, such as Alu repeats (5' AG/CT 3'), can also be transcribed by RNA polymerase III 14. The miRNA-encoding portion of the pri-miRNA forms a hairpin structure that is recognized and cleaved in the nucleus by a microprocessing complex. This complex consists of the double-stranded RNA-specific nuclease DROSHA and its cofactor, DiGeorge syndrome critical region 8 (DGCR8) 15. The resulting ~70 nt hairpin precursor miRNA (pre-miRNA) is exported to the cytoplasm by the RAN-GTP-dependent nuclear transport receptor, exportin-5, which acts by recognizing a 2–3 basepair (bp) overhang of the pre-miRNA stem-loop structure 16, 17. The pre-miRNA is further processed by a complex of the RNAse III-like ribonuclease Dicer and the trans-activator RNA-binding protein, which cleaves the pre-miRNA to release the mature miRNA duplex18, 19.
Figure 1.
Schematic representation of miRNA biogenesis and function. Transcription of miRNA genes is typically mediated by RNA polymerase II (pol II). The initial miRNA-containing transcript, termed primary miRNAs (pri-miRNAs), can range from a few hundred nucleotides (nt) to several kilobases long. Inside the nucleus, the pri-miRNA has a characteristic stem-loop structure that can be recognized and cleaved by the ribonuclease III (RNase III) endonuclease Drosha along with its partner DGCR8 (DiGeorge syndrome critical region 8 gene; also known as Pasha). The cleavage product, a ~70-nt stem-loop pre-miRNA, is exported from the nucleus by Exportin 5. In the cytoplasm, another RNase III enzyme, Dicer, further cleaves the pre-miRNA into a double-stranded mature miRNA (~21-nt), which is incorporated into the RNA-induced silencing complex (RISC) allowing preferential strand-separation of the mature miRNA to repress mRNA translation or destabilize mRNA transcripts through cleavage or deadenylation. Abbreviations: SRF, serum response factor; TF, transcription factor (adapted from 4).
An asymmetry in the relative thermodynamic stability of the 5' ends of the miRNA duplex results in preferential loading of the less stable ~22-nt strand into the RNA-induced silencing complex (RISC); the other strand is degraded, although in some cases both strands are incorporated into the RISC 20–22. The RISC helps mediate miRNA:mRNA interactions and subsequent mRNA repression or destabilization 23. miRNAs typically bind to the 3' UTRs of their mRNA targets with imprecise complementarity. Typically, the degree of Watson-Crick base-pairing between bases 2 and 7 (the “seed region”) at the 5' end of the miRNA is critical for binding mRNA targets 24, 25 and mediating repression. RISC-bound miRNAs may also be sequestered away from translational machinery in processing bodies (P-bodies) that act by recruiting poly(A) nucleases to help modulate deadenylatation of mRNA and thereby prevent translation 26–28.
miRNAs can be found in exons or introns of noncoding transcripts with independent enhancer regulation and in the introns and 3' UTRs of protein coding transcripts. They can also overlap with either an exon or an intron, depending on the alternative splicing pattern. In flies and worms, some miRNAs in intronic regions bypass Drosha processing and enter the miRNA biogenesis pathway as pre-miRNAs 29. In many cases, miRNAs are clustered near other miRNAs, and are transcriptionally coregulated and share cooperative regulatory roles.
Among the hundreds of miRNAs identified thus far, only a limited number have been assigned target mRNAs. Several algorithmic databases have been designed for miRNA target prediction that rely, for the most part, on the following criteria: (1) conservation across species, (2) complementarity of the 5' miRNA ‘seed match’ to the 3' UTR (~ 7 nt) 1, 25, 30, (3) G:U wobbles in the seed 31, (4) the thermodynamic context of target mRNA binding sites (i.e., mRNA targets located in regions of high free energy and unstable secondary structure are favored) 1, 32, and (5) multiple miRNA binding sites in 3'UTR 33. These computational programs are continuously updated to integrate new knowledge from validated miRNA:mRNA interactions (reviewed in 34).
miRNA Function During Cardiogenesis
The heart is derived from multiple cell lineages and must differentiate into unique regions, each possessing different physiologic, electrical, and anatomic properties. A group of mesodermal progenitors known as the first heart field (FHF), coalesces along the midline to form a straight heart tube that begins to be patterned in an anterior posterior (AP), dorsal-ventral (DV), and left-right (LR) fashion. A second population of mesodermal cardiac progenitors, known as the second heart field (SHF) migrates from behind the heart tube into the anterior and posterior poles of the heart tube as it begins to undergo rightward looping. Cells from the SHF contribute to most of the outflow tract and right ventricle (RV), along with part of the atria; FHF cells contribute to most of the left ventricle (LV) and part of the atria3, 35. Distinct patterns of gene expression define each region of the heart including individual chambers and valves. Proper looping and ballooning of the outer curvature of the heart tube is necessary for correct alignment of the chambers with one another and with the inflow and outflow tract valves. In higher organisms, septation leads to a four-chambered heart with two atrioventricular valves and two outflow vessels. Cells derived from the neural crest migrate into the outflow tract and are required for outflow septation36, while cells from the proepicardium contribute to the coronary vessels, fibroblasts, and even some muscle cells in the heart37, 38.
One approach to study the comprehensive requirements of miRNAs during vertebrate development has been to create mutations in Dicer, the enzyme required to process miRNAs into their active mature forms. Dicer is encoded by a single locus in vertebrates. Zebrafish lacking maternal and endogenous Dicer die from defects in gastrulation, brain morphogenesis, somitogenesis, and heart development 39, 40. In mice, targeted deletion of Dicer causes lethality at embryonic day 7.5 (E7.5), before body axis formation 41. Cardiac-specific deletion of Dicer using Cre recombinase expressed under the control of the endogenous Nkx2.5 regulatory elements resulted in embryonic lethality at E12.5 1. The Nkx2.5-Cre is active from E8.5 during heart patterning and differentiation, but only after initial commitment of cardiac progenitors 42. It will be important to determine whether Dicer is required for earlier stages of cardiogenesis (before E8.5), such as cardiac lineage specification, since Dicer is required for embryonic stem cell differentiation 43, 44. Dicer activity is also required for normal function of the mature heart as adult mice lacking Dicer in the myocardium have a high incidence of sudden death, cardiac hypertrophy, and reactivation of the fetal cardiac gene program45. Deletions of Dicer in specific heart populations will reveal the importance of miRNA function in distinct aspects of heart development (e.g., cardiomyocyte commitment, chamber morphogenesis, outflow tract remodeling).
Organization and regulation of miR-1 and miR-133
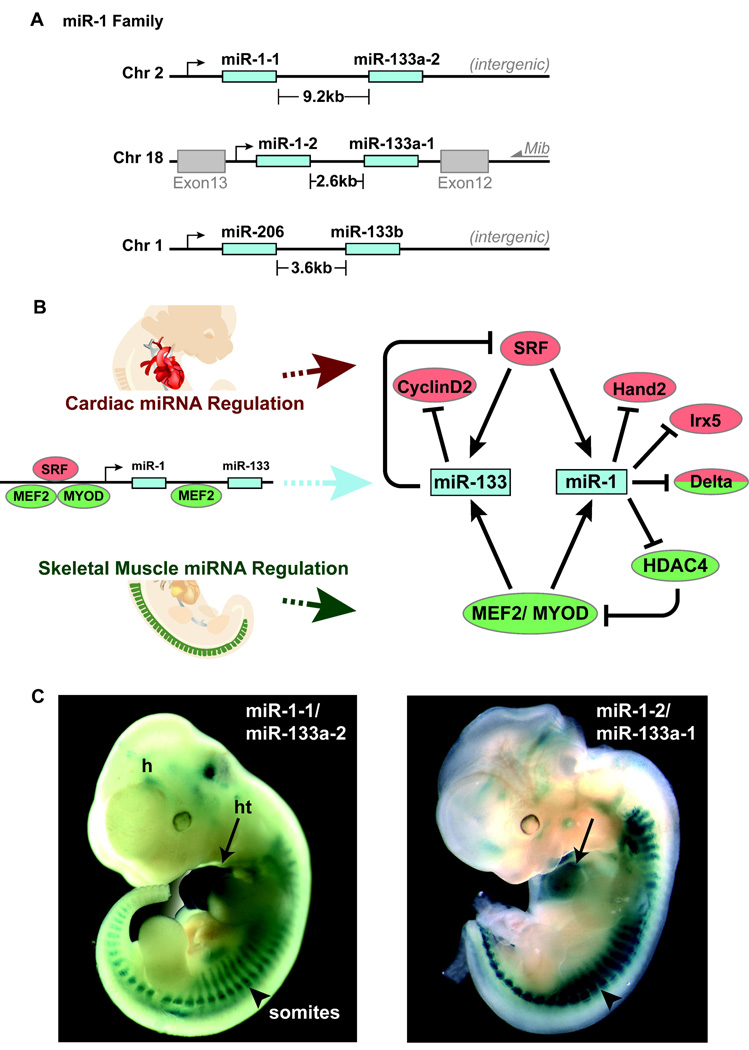
Two widely conserved miRNAs that display cardiac- and skeletal muscle–specific expression during development and in the adult are miR-1 and miR-133, which are derived from a common precursor transcript (bicistronic) 32, 46. Multiple highly conserved loci encode the mature miR-1 (miR-1-1 and miR-1-2) and miR-133 (miR-133a-1, miR-133a-2, and miR-133b) transcripts, which appear to be genetically redundant47. The mature forms of miR-1 derived from the distinct loci are identical, as are the miR-133a forms. The miR-1-1/miR-133a-2 cluster is located in an intergenic region, whereas the miR-1-2/miR-133a-1 is transcribed in an antisense orientation between exons 12 and 13 of the Mindbomb 1 (Mib1) gene (Fig. 2A), involved in Delta-mediated Notch signaling 48. The related miR-1 family member, miR-206, shares extensive sequence homology to miR-1, but is expressed exclusively in skeletal muscle with the co-transcribed miR-133b49 (Fig. 2A).
Figure 2.
Summary of miR-1 and miR-133 genomic organization, regulation, and expression during mouse cardiogenesis. A, Chromosome locations of miR-1 and miR-133a orthologs and miR-206/133b. The miR-1-2/miR-133a-1 cluster is intragenic, and the miR-1-1/miR-133a-2 and miR-206/133b clusters are intergenic. The miR-1/ 133a and miR-206/133b clusters are transcribed as a bicistronic transcript. B, Cardiac (red) and muscle (green) -specific expression of miR-1 and miR-133 clusters is regulated by SRF and myogenic transcription factors, Mef2 and Myod. Targets of miR-1 and miR-133 that regulate cardiac or skeletal muscle are shown. C, LacZ directed by an upstream enhancer of the miR-1-2/ miR-133a-2 cluster and the miR-1-1/miR-133a-1 cluster, respectively, shows expression in the heart (ht) and somites (arrowhead) at mouse embryonic day 11.5.
Transcription of the miR-1/miR-133 bicistronic precursors are directly regulated by the major myogenic differentiation factors, MyoD, myocyte enhancer factor-2 (Mef2), and serum 49response factor (SRF) 32. MyoD functions exclusively in skeletal muscle, while Mef2 and SRF regulate gene expression in cardiac, skeletal and smooth muscle development (Fig. 2B) 50–52. SRF binds to CArG motifs in promoters and enhancers of muscle-specific genes that regulate differentiation, cell-cycle progression, and tissue-specific gene expression 53. In the heart, SRF binds and activates the enhancer regions of miR-1/miR-133 in vitro and in vivo through a serum response element conserved from fly to human 32. Similarly, SRF regulates the cardiac expression of miR-1 in flies, and the bHLH transcription factor Twist and Mef2 regulate somatic muscle expression 47, 54. Mef2 can also activate transcription of the bicistronic miR-1/miR-133 transcript via an intragenic muscle-specific enhancer, which provides cooperative temporospatial regulation of miRNA expression 52. In contrast to the upstream miR-1/133 enhancer, which directs expression within the ventricular chambers 32, the intragenic miR-1/133 enhancer is active in the atrial and ventricular chambers 52. This suggests there may be differential regulation of miR-1 and miR-133 in order to modulate their regulation on target mRNAs in the muscle differentiation pathway downstream of Mef2. Concordant with their common cis- and trans-regulation, both miR-1 and miR-133 are coexpressed in cardiac and skeletal muscle throughout mouse development and are robustly expressed in the adult (Fig. 2B–C) 32, 46, 52.
Function of miR-1 and miR-133 during cardiogenesis and cardiac cell fate decisions
miR-1 expression directed by the enhancers described above commences at approximately E8.5 in mouse and increases throughout development. However, in Drosophila, miR-1 transcripts are detectable during early mesoderm formation as early as the onset of mef2 expression47. This may also be the case in mouse through as yet undescribed enhancers. Overexpression of miR-1 under the control of the β-MHC promoter diminishes the pool of proliferating ventricular myocytes by inducing a premature exit from the cell cycle. This negatively regulates cardiac growth, in part by inhibiting translation of the heart and neural crest derivative-2 protein, Hand2 32, a basic helix-loop-helix protein involved in ventricular myocyte expansion. In mice, Hand2 is initially expressed throughout the linear heart tube, and then becomes restricted to the developing atrial and ventricular myocardium with highest expression in the right ventricle. Mice that lack Hand2 die at E10.5 from right ventricular hypoplasia and decreased trabeculation in the left ventricle 55–57. In mice overexpressing miR-1, trabeculation is also decreased, consistent with the Hand2 mutant phenotype, corroborating Hand2 as a direct target of miR-1 32.
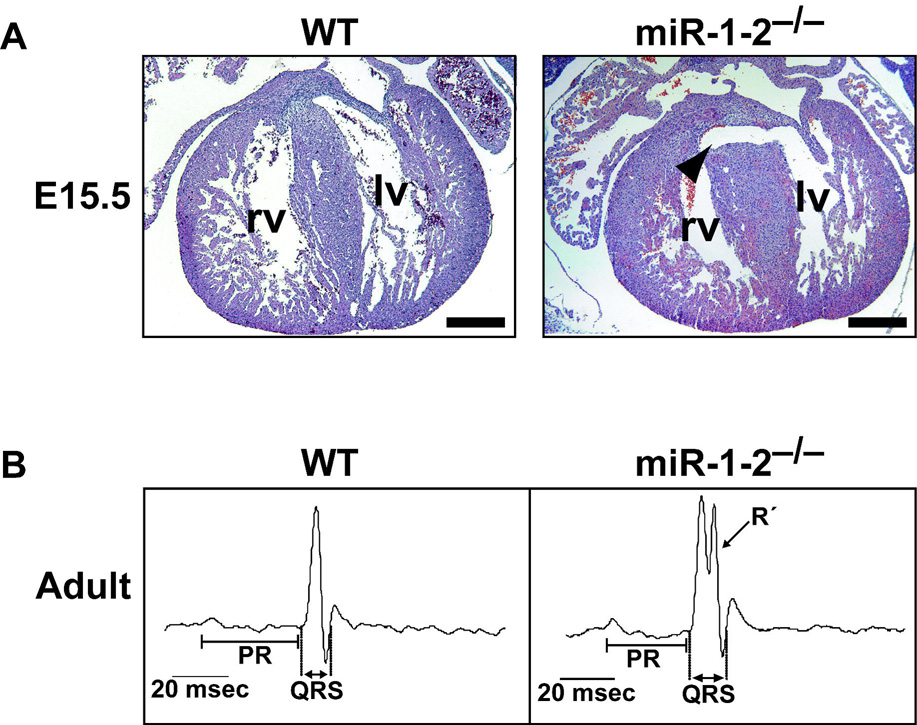
Surprisingly, loss of miR-1-2 in mice, resulting in only a 50% decrease in total miR-1, results in partial embryonic death between E15.5 and birth due to apparent ventricular septal defects and cardiac dysfunction (Fig. 3A–B). These defects can occur from dysregulation of a multitude of events during cardiogenesis, and it is likely that miR-1-2 regulates numerous genesduring this process. Precise dosage of Hand2 is crucial for normal cardiomyocyte proliferation and development, and elevated levels of Hand2 may contribute to the ventricular septal defects and cardiac death 1.
Figure 3.
Cardiac defects in the miR-1-2 mutants. A, Transverse sections of wild-type (wt) or miR-1-2−/− hearts at E15.5 showing ventricular septal defect (arrowhead). B, Representative diagrams of electrocardiograms indicate the location of PR and QRS intervals. The second peak in the QRS complex (R’) was observed in the majority of mutant mice representing delay of electrical conduction. RV, right ventricle; LV, left ventricle; bpm, beats per minute; msec, milliseconds (adapted from 1)
Postnatal mouse cardiomyocytes terminally exit the cell cycle after the first 10 days of life. However, miR-1-2-null adult mice have an increase in mitotic cardiac myocytes along with cardiac hyperplasia. In addition, genome-wide profiling of miR-1-2 mutant adult hearts suggests a broad upregulation of positive regulators of the cell cycle and downregulation of tumor suppressors, indicating a shift in the “threshold” of cells to re-enter the cell cycle 1. Whether this change promotes cardiac repair after injury remains to be determined. The consequence of complete loss of miR-1 in cardiac morphogenesis and adult cardiomyocytes awaits compound loss of miR-1-1 and miR-1-2.
In Drosophila, miR-1 functions to pattern the dorsal vessel (i.e., aorta/heart tube). Moreover, the deletion of the single miR-1 gene (dmiR-1), results in a muscle differentiation defect 47, 54. In a subset of dmiR-1-null flies, muscle progenitors are arrested in a proliferative state and accumulate ectopically. Drosophila hand does not seem to be a target of miR-1, since the fly hand ortholog lacks miR-1 binding sites in its 3’UTR, suggesting that miRNA:mRNA interactions may differ somewhat between species. Instead, dmiR-1 targets transcripts encoding the Notch ligand, Delta, which regulates the expansion of cardiac and muscle progenitor cells 47, suggesting that miR-1 promotes muscle differentiation through down-regulation of the Notch signaling pathway. This is consistent with the known function of the Notch/ Delta signaling pathway during developmental cell fate decisions, including those involving cardiac specification 58.
In cultured myoblasts, miR-1 promotes myoblast differentiation, whereas miR-133 stimulates myoblast proliferation 46. miR-1 targets the histone deacetylase 4 (HDAC4) mRNA, a transcriptional repressor of Mef2-dependent activation of muscle-specific gene expression, suggesting that translational repression of HDAC4 by miR-1 enhances gene activation of Mef2-dependent promoters. Furthermore, miR-133 targets SRF, which is important in muscle proliferation, differentiation and activation of the miR-1/miR-133 transcript, and thus creates a negative feedback loop of regulation46 (Fig. 2B). When rat ventricular cells are subjected to oxidative stress, miR-1 and miR-133 have opposing effects on apoptosis. miR-1 targets the anti-apoptotic heat shock proteins HSP60 and HSP70 and is apoptotic, whereas miR-133 represses caspase-9, a regulator of mitochondria-mediated apoptosis 59, and is anti-apoptotic. Concordantly, compound loss of miR-133a-1 and miR-133a-2 in mice results in enhanced apoptosis, although the in vivo data does not show an upregulation of caspase-9 or other proapoptotic genes 60.
During early cell fate decisions of mouse and human embryonic stem (ES) cells, miR-1 and miR-133 are expressed just as mesoderm emerges and function in concert to promote mesoderm induction, while suppressing differentiation into the ectodermal or endodermal lineages 61. However, miR-1 and miR-133 have antagonistic effects on further adoption of muscle lineages: miR-1 promotes differentiation of mouse and human ES cells toward a cardiac fate, while miR-133 inhibits differentiation into cardiac muscle. miR-1 appears to exert this effect, in part, by translationally repressing the mammalian ortholog of delta, Delta-like-1 (Dll-1), similar to the repression seen in the fly 61. Thus, the bicistronic miR-1/miR-133 transcript encodes distinct mature miRNAs that likely share common targets yet complement each other by balancing the differentiation and proliferation of cardiac and skeletal muscle lineages.
In contrast to in vitro data showing that miR-133 promotes proliferation in cultured myoblasts and cardiac progenitors46, 62, mice lacking miR-133a-1 and miR-133a-2 had excessive cardiac proliferation60. In addition, compound mutants had partial embryonic lethality due to large ventricular septal defects, similar to the miR-1-2 knockout mice1. Dysregulation of cell-cycle control genes and aberrant activation of the smooth muscle gene program were observed in double-mutant mice, which may be due to the upregulation of the miR-133a mRNA targets, cyclinD2 and SRF, respectively.
miR-1 and miR-133 function in the cardiac conduction system
Deletion of miR-1-2 in mice also revealed an essential function of miR-1 in the cardiac conduction system. Mutant mice that survived until birth often suffered sudden death and electrophysiologic testing revealed a spectrum of cardiac arrhythmias in mutant mice1. The heart’s electrical activity begins in the sinoatrial node and propagates impulses to the ventricles, resulting in depolarization, ventricular contraction, and subsequent repolarization of the heart to initiate cardiac relaxation. The miR-1-2 mutants have a shortened P-R interval (the time from the beginning of atrial excitation to the beginning of ventricular excitation) and a prolonged QRS complex (the duration of ventricular depolarization) (Fig. 3B). Ventricular depolarization occurs by rapid conduction through the atrioventricular bundle, bundle branches, and Purkinje fibers. A prolonged QRS often corresponds to a bundle-branch block and can increase the risk of sudden death in humans 63, 64. The cardiac arrhythmias in the miR-1-2 mutants may be caused, in part, by elevated levels of the transcription factor Iroquois homeobox 5 (Irx5), a direct target repressed by miR-11. Irx5 regulates the cardiac ventricular repolarization gradient by negatively regulating the expression of potassium channel genes, such as Kcnd2 65.
Additional studies showed that cardiac electrophysiology is sensitive to miR-1 and miR-133 dosage. In humans with coronary heart disease, miR-1 expression is elevated, and in normal rats or rats subjected to myocardial infarction, overexpression of miR-1 increased the occurrence of arrhythmias 59. Arrhythmias are common after a heart attack, and delivery of an antisense oligonucleotide to decrease miR-1 in the rat infarct model reverses the predisposition to arrhythmias 59. Furthermore, Yang and colleagues showed that miR-1 targets the 3' UTRs of two ion channels prevalent in the adult heart, GJA1 and KCNJ2, which encode the cardiac gap junction connexin 43 (Cx43) and the potassium channel subunit Kir2.1, respectively. GJA1 is responsible for intercellular conductance in the ventricles, while KCNJ2 is responsible for setting and maintaining the cardiac resting membrane potential.
Interestingly, miR-133 also participates in regulating cardiac electrophysiology, as it translationally represses the ion channels KCNQ1 and KCNH2, which encode potassium channel subunits and are responsible for producing the cardiac repolarization current 59, 66. Luo and colleagues showed that miR-133 gain-of-function animals displayed a prolonged QT interval, with increased ventricular depolarization and repolarization times. Prolonged QT interval is characteristic of the congenital long QT syndrome, which is associated with an increased risk of fatal ventricular tachyarrhythmias. Thus, miR-1 and miR-133 appear to regulate numerous genes that control proper cardiac conduction, providing an example of a miRNA’s role as a “master” regulator by virtue of the multiple mRNAs it can target. As such, manipulation of these miRNAs may have therapeutic value in prevention of cardiac arrhythmias, particularly during the high-risk period immediately following myocardial infarction.
miR-138 Regulation of Cardiac Patterning
Intricate transcriptional networks establish chamber-specific gene expression, and these patterning events are highly conserved across species from zebrafish to human 3. Zebrafish are useful models to study cardiac patterning events because of their simple two-chambered heart consisting of a single atrium and ventricle separated by the atrioventricular canal (AVC). The atrial and ventricular chambers express unique myosin genes, whereas the AVC expresses distinct genes such as cspg2, encoding versican, notch1b, and tbx2 67, 68. miR-138 is a highly conserved miRNA found in many parts of the embryo, but within the zebrafish heart is specifically expressed in the ventricular chamber 61. Disruption of miR-138 function led to expansion of AVC gene expression into the ventricle and failure of ventricular cardiomyocytes to fully mature. miR-138 normally restricts AVC gene expression by directly repressing cspg2 in the ventricle. This event is reinforced by ventricular repression of retinoic acid dehydrogenase, resulting in decreased retinoic acid, which is a positive regulator of cspg2 61. It is likely that other region-specific miRNAs will reinforce known signaling and transcriptional networks that establish patterns of gene expression throughout the developing heart tube.
miR-208 Regulation of Fetal-Adult Myosin Isoform Switching
Reactivation of the fetal cardiac gene program, including switching of the major myosin heavy chain (MHC) isoform from α-MHC to β-MHC in mice (the opposite is true in humans), is a hallmark of the failing and hypertrophied post-natal heart. Similarly, fetal cardiac miRNAs are also re-activated in the stressed adult heart 69. Interestingly, miR-208 is found within the intron of the cardiac-specific gene encoding the highly abundant α-MHC, the predominant MHC isoform in the adult mouse heart. miR-208 is involved in regulating MHC isoform switching, as mice lacking miR-208 fail to upregulate β-MHC upon stress and do not develop hypertrophy 70.Furthermore, disruption of miR-208 resulted in ectopic activation of the fast skeletal muscle gene program within the heart. These gene expression effects were in part due to direct inhibition of the thyroid receptor–associated protein, THRAP1, a transcriptional coregulator of the thyroid receptor 70. Thus, nature appears to have embedded an elegant control mechanism within myosins that can regulate abundance of specific isoforms during fetal and adult stages.
miRNA Regulation of Angiogenesis
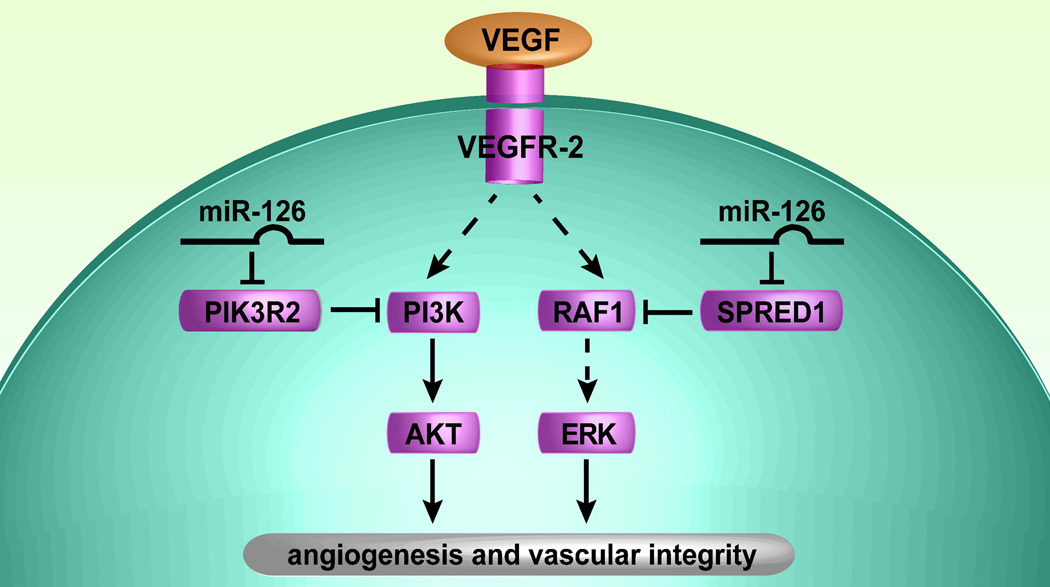
In addition to miRNA regulation of cardiomyocytes, recent reports illustrate what will likely be a broader function of tissue-specific miRNAs during vascular development (reviewed in 71) 61, 72. In particular, miR-126, which is located in the intron of an endothelial-specific gene, Egfl7, is the most highly enriched miRNA in endothelial cells derived from embryonic stem cells or developing embryos63. miR-126 is a key positive regulator of angiogenic signaling in endothelial cells and of vascular integrity in vivo61, 72. Knock-down of miR-126 during zebrafish embryogenesis or deletion of miR-126 in mice resulted in defects in vascular development. For example, collapsed blood vessels and cranial hemorrhages occurred in zebrafish with reduced levels of miR-126 63, and delayed angiogenic sprouting, widespread hemorrhaging and partially embryonic lethality were observed in mice deficient in miR-12672, 73. MiR-126 mutant mice that successfully completed embryogenesis displayed diminished angiogenesis and increased mortality after coronary ligation, a model for myocardial infarction 64. Molecular analysis revealed that miR-126-deficient endothelial cells failed to respond to angiogenic factors, including VEGF, epidermal growth factor (EGF) and bFGF 61, 72. Two direct targets of miR-126 were the Sprouty-related protein 1 (Spred1) 61, 72 and the regulatory subunit of PI3K, p85β (also known as PIK3R2) 63. Because Spred1 and PIK3R2 are negative regulators of cellular signaling cascades, affecting the MAPK and PI3K signaling pathways, respectively, miR-126 promotes VEGF and other growth factor signaling (Fig. 4). By targeting multiple signaling pathways, miR-126 may fine-tune angiogenic responses. Because of miR-126’s central role in vascular development, miR-126-mediated regulation of angiogenesis may be a valuable therapeutic target to promote new blood vessel formation in ischemic conditions as well to inhibit angiogenesis during tumor growth.
Figure 4.
Putative model of miR-126 function in endothelial cells. miR-126 represses SPRED1 and PIK3R2, which negatively regulate VEGF signaling (and possibly other growth factor signaling pathways) via the MAP kinase and PI3 kinase pathways, respectively. Thus, miR-126 promotes VEGF signaling, angiogenesis and vascular integrity by inhibiting protein production of endogenous VEGF repressors within endothelial cells (adapted from61).
Summary and Future Directions
The function of miRNAs in cardiovascular development reviewed here likely foreshadows a much broader role of dozens of miRNAs in regulating most aspects of cardiovascular development. Through their ability to post-transcriptionally regulate mRNA levels, and thus manage protein dosage, miRNAs provide finer regulation within the complex molecular networks that regulate cardiogenesis. The importance of this fine regulation is highlighted by the recognition that most known genetic causes of heart malformations in humans result from haploinsufficiency or heterozygous point mutations. Thus, modulation of miRNA levels may be useful in titrating the dose of critical pathways and may allow restoration of protein levels to a point that crosses the disease threshold. With further characterization, elucidating the function of cardiac-enriched miRNAs may provide us with new diagnostic, prognostic, and therapeutic targets for many forms of cardiovascular disease.
Despite the exponentially increasing knowledge regarding miRNA function, many questions and challenges remain. One of the most critical challenges is the ability to accurately and efficiently determine mRNA targets of specific miRNAs. Current bioinformatics approaches have been very useful, but rely on some pre-conceived knowledge of the miRNA function to sort through the hundreds of predicted targets. A better understanding of the integration of miRNA control with known regulatory networks will be necessary place the function of specific miRNAs in context of current knowledge. The recent, yet controversial, recognition the miRNAs may function both as downregulators and upregulators of translation may add additional complexity to the miRNA:mRNA relationship and miRNA function in specific cellular contexts. It is likely that the cellular context also dictates when a miRNA may engage its mRNA targets and regulates the efficiency with which a miRNA is processed from its precursor forms. These types of events will represent multiple layers of regulation that will themselves titrate the activity of specific miRNAs. It is clear that discovery of the small RNA world has fundamentally altered the way in which biologists view cellular functions, but the field is only in its infancy. Undoubtedly, there will be a logarithmic growth in understanding miRNA function in coming years and it will become clear that virtually every cellular event is titrated by one or more miRNAs. As such, the potential to harness miRNA biology for the betterment of human disease may represent the next frontier in human therapeutics.
Acknowledgements
We thank B. Taylor for assistance with manuscript preparation, members of the Srivastava laboratory for discussions, insights and reviewing of the manuscript, and S. Ordway and G. Howard for editorial assistance. D.S. is supported by grants from the NHLBI/NIH and the March of Dimes Birth Defects Foundation; D.S. is an Established Investigator of the American Heart Association.
Footnotes
This is an un-copyedited author manuscript accepted for publication in Circulation Research, copyright The American Heart Association. This may not be duplicated or reproduced, other than for personal use or within the “Fair Use of Copyrighted Materials” (section 107, title 17, U.S. Code) without prior permission of the copyright owner, The American Heart Association. The final copyedited article, which is the version of record, can be found at http://circres.ahajournals.org/. The American Heart Association disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties.
Disclosures
None
References
- 1.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhuri K, Chatterjee R. MicroRNA detection and target prediction: Integration of computational and experimental approaches. DNA Cell Biol. 2007;26:321–337. doi: 10.1089/dna.2006.0549. [DOI] [PubMed] [Google Scholar]
- 11.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. Rna. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 15.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 19.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 21.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 22.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38 Suppl:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 33.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 35.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nature reviews. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 36.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circulation research. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 37.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 40.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–888. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 42.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 43.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 46.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 49.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 51.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 52.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catala F, Wanner R, Barton P, Cohen A, Wright W, Buckingham M. A skeletal muscle-specific enhancer regulated by factors binding to E and CArG boxes is present in the promoter of the mouse myosin light-chain 1A gene. Mol. Cell. Biol. 1995;15:4585–4596. doi: 10.1128/mcb.15.8.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev. Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- 56.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 57.Yamagishi H, Yamagishi C, Nakagawa O, Harvey RP, Olson EN, Srivastava D. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev. Biol. 2001;239:190–203. doi: 10.1006/dbio.2001.0417. [DOI] [PubMed] [Google Scholar]
- 58.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 59.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 60.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes & development. 2008 doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turrini P, Corrado D, Basso C, Nava A, Bauce B, Thiene G. Dispersion of ventricular depolarization-repolarization: a noninvasive marker for risk stratification in arrhythmogenic right ventricular cardiomyopathy. Circulation. 2001;103:3075–3080. doi: 10.1161/01.cir.103.25.3075. [DOI] [PubMed] [Google Scholar]
- 64.Desai AD, Yaw TS, Yamazaki T, Kaykha A, Chun S, Froelicher VF. Prognostic significance of quantitative QRS duration. Am. J. Med. 2006;119:600–606. doi: 10.1016/j.amjmed.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 65.Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, Guerchicoff A, Pollevick GD, Chan TY, Kabir MG, Cheng SH, Husain M, Antzelevitch C, Srivastava D, Gross GJ, Hui CC, Backx PH, Bruneau BG. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo X, Xiao J, Lin H, Li B, Lu Y, Yang B, Wang Z. Transcriptional activation by stimulating protein 1 and post-transcriptional repression by muscle-specific microRNAs of IKs-encoding genes and potential implications in regional heterogeneity of their expressions. J. Cell. Physiol. 2007;212:358–367. doi: 10.1002/jcp.21030. [DOI] [PubMed] [Google Scholar]
- 67.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development (Cambridge, England) 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chi NC, Shaw RM, De Val S, Kang G, Jan LY, Black BL, Stainier DY. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes & development. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 70.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 71.Fish JE. D. S. microRNAs: Opening a new vein in angiogenesis research. Science Signaling. 2008 doi: 10.1126/scisignal.252pe1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development (Cambridge, England) 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]