Abstract
OBJECTIVE: To conduct clinical and molecular genetic analyses of the members of an extended family in Central Indiana with a high prevalence of restless legs syndrome (RLS).
PARTICIPANTS AND METHODS: From February 1, 2006, through August 31, 2008, we collected data from members of this family, which is of English descent. Genealogical methods were used to expand the family tree, and family members were screened with an RLS questionnaire. Telephone interviews and personal examinations were performed at Mayo Clinic and during a field trip to Central Indiana. Blood samples were collected for molecular genetic analysis. A follow-up telephone interview was conducted 1 year later.
RESULTS: The family tree spans 7 generations with 88 living members, 30 of whom meet the criteria for diagnosis of RLS established by the International Restless Legs Syndrome Study Group. Three affected family members also have Parkinson disease or essential tremor. The mode of RLS inheritance is compatible with an autosomal dominant pattern. The affected family members do not exhibit linkage to the 5 known RLS loci or mutations in the RLS susceptibility genes MEIS1 and BTBD9.
CONCLUSION: Of 88 members of this single extended family in Central Indiana, 30 were diagnosed as having RLS. Because our analysis shows that the disease is not linked to any of the known RLS loci or risk-associated genes, we postulate that members of this family may carry a gene mutation in a novel genetic locus.
Of 88 members of a single extended family, 30 were diagnosed as having restless legs syndrome; because this analysis shows that the disease is not linked to any of the known restless legs syndrome loci or risk-associated genes, members of this family may carry a gene mutation in a novel genetic locus.
BTBD9 = BTB (POZ) domain containing 9; LOD = logarithm of the odds; MEIS1 = Meis homeobox 1; PD = Parkinson disease; PLM = periodic leg movement; RLS = restless legs syndrome
Restless legs syndrome (RLS) is a neurologic disorder of combined sensory and motor dysfunction first described by Ekbom1 in 1945. The prevalence of idiopathic RLS is estimated to be 1.2% to 15.0%.2,3 All studies report an increase in prevalence with age, and some suggest that RLS may occur more frequently in women.4 Common treatments include levodopa, dopamine agonists, opiates, benzodiazepines, anticonvulsants, and iron supplements.5 Symptoms can appear intermittently and later become persistent. Secondary forms of RLS have been described and are associated with other conditions, including pregnancy, iron deficiency, peripheral neuropathies, diabetes, or chronic renal failure.3
The clinical diagnosis of RLS is made on the basis of 4 essential criteria: (1) an urge to move the legs that is usually accompanied by uncomfortable and unpleasant sensations in the legs, (2) a worsening of the urge to move or the unpleasant sensations during periods of rest or inactivity such as lying down or sitting, (3) partial or total relief of the symptoms by movements such as walking or stretching, and (4) occurrence or worsening of the unpleasant sensations in the evening or at night. A family history of RLS and response to dopaminergic agents are additional features that support a diagnosis of RLS. Most patients also present with periodic leg movements (PLMs) during sleep and quiet wakefulness, resulting in difficulty falling or staying asleep.6
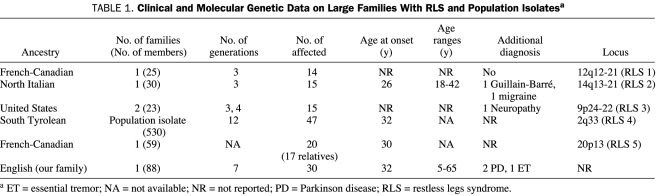
Familial aggregation of RLS has been recognized since Ekbom1 formally described the condition in 1945. Studies in twins have consistently shown that RLS has a genetic etiology7,8 and suggest that the condition might best be explained by an oligogenic, multifactorial model, with 1 or more genes of major effect.3 To date, 5 loci for RLS have been reported (Table 1). These include 1 autosomal recessive locus in a French-Canadian pedigree that was mapped to chromosome 12q12-21 (RLS1)9 and 4 autosomal dominant loci: 1 in a North Italian pedigree (RLS2: 14q13-21),10 1 in 2 families from the United States (RLS3: 9p24-22),11 1 in a population isolate from South Tyrol (RLS4: 2q33),12 and 1 in a large French-Canadian pedigree (RLS5: 20p13).13
TABLE 1.
Clinical and Molecular Genetic Data on Large Families With RLS and Population Isolatesa
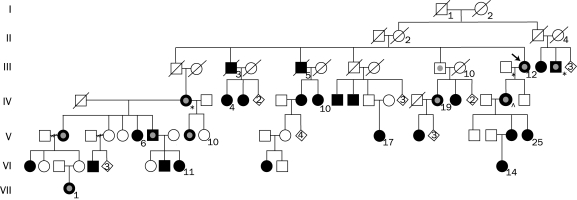
We describe a large kindred (Figure) with autosomal dominant RLS and report the results of the genetic analysis of the 5 previously known RLS loci and other previously identified susceptibility genes.
FIGURE.
Abbreviated pedigree of the Central Indiana restless legs syndrome family. Square = male. Circle = female. Diamonds with numbers inside indicate a number of descendents regardless of sex. A diagonal line through a square or circle indicates a deceased person. An arrow indicates proband. Phenotypes of affected individuals: * = Parkinson disease; ⋂ = essential tremor. Fully darkened symbols indicate restless legs syndrome; darkened symbols outlined with inner light gray circle indicate restless legs syndrome and periodic leg movement while awake; and symbol with inner gray only indicates periodic movement while awake.
PARTICIPANTS AND METHODS
Our RLS proband was identified in 2005. Because this person reported an extensive family history of RLS, we performed a genealogical investigation to expand the family tree. After receiving approval for the study from the Mayo Clinic Institutional Review Board, we performed genealogical, clinical, and molecular investigations. The study was conducted from February 1, 2006, through August 31, 2008.
Genealogical Studies
Historical material and family and medical records were collected, and interviews with family members were conducted.
Clinical Studies
The study was designed in 4 phases. In the first phase, family members were mailed an approved, customized RLS questionnaire and the International Restless Legs Syndrome Study Group rating scale. The RLS questionnaire included information regarding patient demographics, sleep onset and duration, RLS diagnostic criteria, PLM, age at RLS onset and disease duration, past RLS diagnosis, RLS treatment and its efficacy, other sleep disorders, past sleep studies, secondary RLS conditions, current medications, and family history of RLS or neurodegenerative disease. In the second phase, family members who completed an RLS questionnaire were interviewed by telephone (by S.-C.L.) to substantiate the RLS diagnosis. In the third phase, the investigators traveled to Central Indiana to examine an available subset of family members. During the fourth phase, all affected persons were contacted by telephone 1 year later for follow-up questioning. During this interview, the affected patients were asked questions outlined in the RLS questionnaire to reconfirm the diagnosis of RLS. Blood samples were collected from 39 family members during phases 1, 2, and 3.
Molecular Genetic Investigations
Genomic DNA was extracted from peripheral blood lymphocytes using standard protocols. To interrogate the segregation of the chromosomal regions linked to RLS, simple tandem repeat, microsatellite markers providing coverage at approximately 2.2-cM resolution were genotyped in a subset of samples collected in phases 1 and 2 and large enough to discriminate linkage to known RLS loci (estimated maximum logarithm of the odds [LOD] score = 2.26). This subset consisted of 9 affected persons (III-12, IV-2, IV-5, IV-9-12, IV-19, V-17), 2 unaffected persons (III-9, IV-13) and 1 person who had married into the family (IV-14). Fluorescent-labeled forward primers flanked informative dinucleotide, trinucleotide, or tetranucleotide repeats with heterozygosity greater than 0.75. Polymerase chain reactions included 20 pmol of each primer, 15 ng of DNA template, 5 mM of nucleoside triphosphates (deoxyadenosine triphosphate, deoxycytidine triphosphate, deoxyguanosine triphosphate and deoxythymidine triphosphate), 1x Qiagen buffer (Qiagen, Valencia, CA), 1x Q solution, and 0.02 units Taq polymerase (Qiagen) per reaction. Thermocycling was performed on a Hybaid MultiBlock Satellite Thermal Cycler System (Thermo Fisher Scientific, Waltham, MA) with a 1-cycle denaturation step at 93°C for 2 minutes, followed by 35 cycles at 93°C for 20 seconds, touchdown from 57°C to 52°C from cycles 3 through 15 (30 seconds each) and 72°C (45 seconds), with a final elongation step at 72°C for 10 minutes. Single tandem repeat microsatellites were then paneled into groups of products having compatible allele sizes and fluorescent dyes and were analyzed on an Applied Biosystems 3730 capillary array system using GeneMapper Software 4.0 (Applied Biosystems, Foster City, CA). The DNA genotyping standards CEPH 1331-01 and 1331-02 (for more information on genotyping standards, see http://www.cephb.fr/) were genotyped as size controls and included on every plate to ensure consistency on allele calls. Data from the GeneMapper files were exported for analysis.
Parametric 2-point LOD scores were calculated using MLINK from the FASTLINK package, and haplotypes were generated using SIMWALK.14-16 The disease was assumed to have an autosomal dominant inheritance, a gene frequency of 0.0001, and penetrance of 100%. The genetic distances of the markers were obtained from the Marshfield sex-averaged map, and allele frequencies were estimated from within the family.
Polymerase chain reaction was used to sequence BTB (POZ) domain containing 9 (BTBD9) and Meis homeobox 1 (MEIS1) exons and exon-intron boundaries in 2 affected persons (III-12 and IV-10) (primer pair sequences are available on request). These 2 persons were selected because of their affected status and genealogical distance. Products of polymerase chain reaction were purified from unincorporated nucleotides using Solid Phase Reversible Immobilization technology (Agencourt Bioscience, Beverly, MA) with a Biomek FX Laboratory Automation Workstation (Beckman Coulter, Fullerton, CA). Sequence analysis was performed as previously described.3
RESULTS
Genealogical Investigations
The family is most likely of English ancestry and traceable to the early 19th century. The pedigree contains 88 persons spanning 7 generations with 30 members affected with RLS (28 alive, 2 deceased). Historical records of the 2 deceased affected family members confirmed their RLS diagnosis.
Clinical Studies
The mean ± SD age of patients at RLS onset was 32±16.32 years (range, 5-65 years). The mean ± SD disease duration was 18.63±16.78 years (range, 1-54 years).
The disease was present in both sexes, with a male-to-female ratio of 8:22. Male-to-male transmission was present 1 time; female-to-female, 9 times; and male-to-female, 5 times. Of the 28 living affected patients, 10 were personally examined by study investigators and the remaining 18 were contacted by telephone to confirm their self-reported RLS diagnosis. All 28 family members with RLS were contacted a second time for a 1-year follow-up interview that confirmed their diagnosis of RLS. Each affected patient who was either personally examined or interviewed over the telephone fulfilled the criteria for diagnosis of RLS of the International Restless Legs Syndrome Study Group. Two family members with RLS (III-14, IV-2) had Parkinson disease (PD), 1 (IV-24) had RLS and essential tremor, and 2 (V-7, VI-1) had sleep apnea. Of the affected patients, 10 reported symptoms of PLM in addition to RLS, and 1 reported PLM without RLS (III-9); 7 reported RLS symptoms affecting their upper extremities and torso (III-12, III-14, IV-2, IV-9, IV-24, V-2, and VII-1).
Six of the family members were diagnosed as having RLS alone by their primary care physician before the study was initiated and were taking RLS medication with beneficial response. Of those, 3 were treated with pramipexole, 1 with ropinirole, 1 with clonazepam, and 1 with zolpidem. Of the 2 family members previously diagnosed as having RLS and PD, 1 (IV-2) was treated with carbidopa-levodopa and pramipexole with benefit, and 1 (III-14) was treated for RLS but not for PD. The family member previously diagnosed as having RLS and essential tremor (IV-24) was treated with klonopin with benefit. Additional family members were diagnosed as having RLS after enrolling in the study.
Of the family members, 7 reported having additional illnesses at the onset of their RLS: 2 reported fibromyalgia (IV-2, V-2); 1, neuropathy (III-14); 1, iron deficiency (VI-12); and 1, unspecified anemia (IV-2). Symptoms of RLS during pregnancy were reported by 2 family members (V-4, V-2). Although V-4 had no RLS symptoms after childbirth, V-2 reported that the RLS became chronic after childbirth. None of the family members reported kidney failure, low back injury, chronic heart failure, or poor leg circulation at the onset of their RLS symptoms. No autopsies were performed in deceased family members with RLS.
Molecular Genetic Investigations
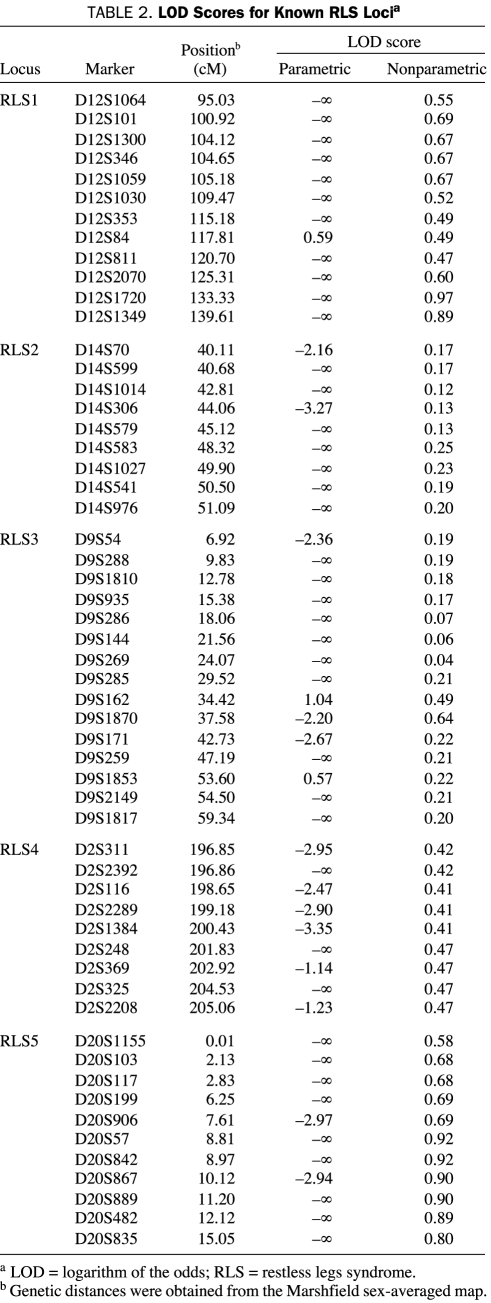
Parametric and nonparametric analyses did not reveal any significant or suggestive LOD score for known RLS loci (Table 2). Haplotype segregation did not identify a common haplotype segregating with RLS disease. Direct sequencing of MEIS1 and BTBD9 did not identify any unknown variant in coding regions or within 100 bases from the exon-intron boundaries.
TABLE 2.
LOD Scores for Known RLS Locia
DISCUSSION
The Central Indiana kindred reported here comprises 88 family members spanning 7 generations. The family is white and most likely of English descent, but we were unable to confirm their geneology. Of these 88 family members, 30 have been diagnosed as having RLS; of these, 7 have severe RLS involving the upper extremities and torso and 6 have been treated with RLS medication with positive response. The pattern of RLS disease inheritance is compatible with an autosomal dominant model. All affected family members fulfilled diagnostic criteria for RLS. Because there are no confirmed pathognomonic biomarkers for RLS, our 4-phase study design was used to substantiate the diagnosis of RLS. The family was initially screened using an RLS questionnaire. Family members were then interviewed over the telephone or personally examined and were later contacted again for a 1-year follow-up telephone interview.
Although our study design attempted to distinguish family members with primary RLS from those with secondary RLS, our study had its limitations. An inherent limitation in using the questionnaire is the potential unreliability of self-report. The diagnosis of RLS is challenging because of the lack of objective measures (ie, tests, pathology, imaging) to prove a person has RLS, the subjective nature of the RLS diagnostic criteria, the fluctuating nature of RLS onset, the subjective interpretation and reporting of RLS criteria, and the lack of pathognomonic biomarkers. To further substantiate diagnosis, polysomnography could have been included to assess sleep and motor symptoms.
Genetic analysis in this pedigree excluded all known RLS loci through linkage analysis and haplotype segregation. Therefore, a novel locus harboring a gene with mutations leading to the RLS phenotype may be present in members of this family. Data from 39 family members have been collected (22 affected with RLS). Power analysis has shown that a maximum 2-point LOD score of 5.02 may be obtained assuming a disease penetrance of 80%, a phenocopy rate of 5%, and a disease allele frequency of 0.01 (more realistic parameters than those used in most studies). Data derived from this RLS kindred are sufficiently powerful to map a novel gene for RLS.
Pathogenic and susceptibility variants for a disease have been found to be present in the same gene. For example, the leucine-rich repeat kinase 2 gene (LRRK2) was found to harbor pathogenic mutations resulting in PD17,18 and was later identified as a risk factor for that disease.19,20 Variants in 3 loci (MEIS1, BTBD9, and mitogen-activated protein kinase kinase 5 [MAP2K5]/LBXCOR1 homolog (mouse) [LBXCOR1]) have been identified recently as risk factors for the development of RLS21,22; however, only the results for MEIS1 and BTBD9 have been replicated.23 To assess the presence of pathogenic mutations in MEIS1 and BTBD9, all exon and exon-intron boundaries were sequenced, but novel variants were not identified.
CONCLUSION
In a large family with 30 members diagnosed as having RLS, linkage to all known RLS loci was excluded, and mutations in the susceptibility genes MEIS1 and BTBD9 were not detected. This family highlights the genetic heterogeneity of RLS and may carry a novel RLS locus.
Acknowledgments
We thank the family members, whose participation made this research project possible.
Footnotes
Ms Young and Drs Lin and Wszolek received support from The Mayo Foundation Research Committee (CR program). Drs Wszolek and Farrer received support from the Morris K. Udall National Institutes of Health/National Institute of Neurological Disorders and Stroke Parkinson Disease Center of Excellence grant awarded to Mayo Clinic's site in Jacksonville, FL (P05 NS40256) and the Pacific Alzheimer Research Foundation Centre grant (C06-01).
REFERENCES
- 1.Ekbom KA. Restless legs: a clinical study. Acta Med Scand. 1945;158(suppl):1-123 [Google Scholar]
- 2.Allen RP, Walters AS, Montplaisir J, et al. Restless legs syndrome prevalence and impact: REST General Population Study. Arch Intern Med. 2005;165(11):1286-1292 [DOI] [PubMed] [Google Scholar]
- 3.Mata IF, Bodkin CL, Adler CH, et al. Genetics of restless legs syndrome. Parkinsonism Relat Disord. 2006;12(1):1-7 [DOI] [PubMed] [Google Scholar]
- 4.Winkelmann J. Genetics of restless legs syndrome. Curr Neurol Neurosci Rep. 2008;8(3):211-216 [DOI] [PubMed] [Google Scholar]
- 5.Thorpy MJ. New paradigms in the treatment of restless legs syndrome. Neurology 2005;64(12, suppl 3):S28-S33 [DOI] [PubMed] [Google Scholar]
- 6.Allen RP, Picchietti D, Hening WA, Trenkwalder C, Walters AS, Montplaisi J. Restless legs syndrome: diagnostic criteria, special considerations, and epidemiology: a report from the restless legs syndrome diagnosis and epidemiology workshop at the National Institutes of Health. Sleep Med. 2003;4(2):101-119 [DOI] [PubMed] [Google Scholar]
- 7.Desai AV, Cherkas LF, Spector TD, Williams AJ. Genetic influences in self-reported symptoms of obstructive sleep apnoea and restless legs: a twin study. Twin Res. 2004;7(6):589-595 [DOI] [PubMed] [Google Scholar]
- 8.Ondo WG, Vuong KD, Wang Q. Restless legs syndrome in monozygotic twins: clinical correlates. Neurology 2000;55(9):1404-1406 [DOI] [PubMed] [Google Scholar]
- 9.Desautels A, Turecki G, Montplaisir J, Sequeira A, Verner A, Rouleau GA. Identification of a major susceptibility locus for restless legs syndrome on chromosome 12q. Am J Hum Genet. 2001December;69(6):1266-1270 Epub 2001 Nov 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonati MT, Ferini-Strambi L, Aridon P, Oldani A, Zucconi M, Casari G. Autosomal dominant restless legs syndrome maps on chromosome 14q. Brain 2003;126(part 6):1485-1492 [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Ondo WG, Rao S, Li L, Chen Q, Wang Q. Genomewide linkage scan identifies a novel susceptibility locus for restless legs syndrome on chromosome 9p. Am J Hum Genet. 2004May;74(5):876-885 Epub 2004 Apr 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichler I, Marroni F, Volpato CB, et al. Linkage analysis identifies a novel locus for restless legs syndrome on chromosome 2q in a South Tyrolean population isolate. Am J Hum Genet. 2006October;79(4):716-723 Epub 2006 Aug 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levchenko A, Provost S, Montplaisir JY, et al. A novel autosomal dominant restless legs syndrome locus maps to chromosome 20p13. Neurology 2006;67(5):900-901 [DOI] [PubMed] [Google Scholar]
- 14.Cottingham RW, Jr, Idury RM, Schäffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53(1):252-263 [PMC free article] [PubMed] [Google Scholar]
- 15.Schäffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44(4):225-237 [DOI] [PubMed] [Google Scholar]
- 16.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58(6):1323-1337 [PMC free article] [PubMed] [Google Scholar]
- 17.Paisán-Ruíz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 2004;44(4):595-600 [DOI] [PubMed] [Google Scholar]
- 18.Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004;44(4):601-607 [DOI] [PubMed] [Google Scholar]
- 19.Di Fonzo A, Wu-Chou YH, Lu CS, et al. A common missense variant in the LRRK2 gene, Gly2385Arg, associated with Parkinson's disease risk in Taiwan. Neurogenetics 2006July;7(3):133-138 Epub 2006 Apr 22 [DOI] [PubMed] [Google Scholar]
- 20.Ross OA, Wu YR, Lee MC, et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson's disease. Ann Neurol. 2008;64(1):88-92 [DOI] [PubMed] [Google Scholar]
- 21.Stefansson H, Rye DB, Hicks A, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007August16;357(7):639-647 Epub 2007 Jul 18 [DOI] [PubMed] [Google Scholar]
- 22.Winkelmann J, Schormair B, Lichtner P, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007August;39(8):1000-1006 Epub 2007 Jul 18 [DOI] [PubMed] [Google Scholar]
- 23.Vilariño-Güell C, Farrer MJ, Lin SC. A genetic risk factor for periodic limb movements in sleep [letter]. N Engl J Med. 2008;358(4):425-427 [DOI] [PubMed] [Google Scholar]